Abstract
Our view of the structure and function of the interphase nucleus has drastically changed in the last years. It is now widely accepted that the nucleus is a well organized and highly compartmentalized organelle and that this organization is intimately related to nuclear function. In this context, chromatin -initially considered a randomly entangled polymer- has also been shown to be structurally organized in interphase and its organization was found to be very important to gene regulation. Relevant and not completely answered questions are how chromatin organization is achieved and what mechanisms are responsible for changes in the positions of chromatin loci in the nucleus. A significant advance in the field resulted from tagging chromosome sites with bacterial operator sequences, and visualizing these tags using green fluorescent protein fused with the appropriate repressor protein. Simultaneously, fluorescence imaging techniques significantly evolved during the last years allowing the observation of the time evolution of processes in living specimens. In this context, the motion of the tagged locus was observed and analyzed to extract quantitative information regarding its dynamics. This review focuses on recent advances in our understanding of chromatin dynamics in interphase with the emphasis placed on the information obtained from single particle tracking (SPT) experiments. We introduce the basis of SPT methods and trajectories analysis, and summarize what has been learnt by using this new technology in the context of chromatin dynamics. Finally, we briefly describe a method of SPT in a two-photon excitation microscope that has several advantages over methods based on conventional microscopy and review the information obtained by using this novel approach to study chromatin dynamics.
Keywords: single particle tracking, two photon microscopy, chromatin dynamics, review
Chromatin Organization in the Interphase Nucleus
Chromatin organization in interphase nuclei has been a subject of great interest over the last decade (Heard & Bickmore 2007).
First results pointing toward a compartmentalization of the interphase nucleus were intuitively striking because the nucleus does not contain compartments limited by membranes as organelles in the cell cytoplasm do. It is now widely accepted that nuclear substructures constitute compartments in which specific functions are performed [reviewed in (Dundr & Misteli 2001; Handwerger & Gall 2006)].
Similarly, the idea that chromatin was organized in defined regions later known as chromosomes territories was unexpected. Even current textbooks teach us that chromatin is a polymer randomly entangled in the interphase nucleus [see references in (Cremer & Cremer 2006)]. Despite that a model of territorial organization of chromatin in interphase was initially proposed by Rabl and Boveri in the early 20th century [for a recent review see (Cremer & Cremer 2006)] several years had pass before new tools in microscopy and cell biology allowed the experimental demonstration of the existence of chromosome territories [see for example, (Branco & Pombo 2006; Cremer et al. 1982; Lichter et al. 1988)].
These studies demonstrated that chromosomes in interphase cells occupy well-defined volumes in the nucleus; moreover, they have preferred positions relative to the nuclear membrane and with respect to each other.
The internal architecture of these territories is also not random: some studies showed that gene-rich and gene-poor sequences are mostly localized in the periphery and in the core of the chromosome territory, respectively (Kurz et al. 1996). These results leaded to a model proposing that the transcription machinery is located in interchromatin domains, i.e. regions between chromosomes territories. According to this model, active genes are mainly located in the surface of these territories to access the transcription and splicing machineries. This model was further supported by studies showing that genes may move to the interchromatin region in response to transcription (Chambeyron et al. 2005; Volpi et al. 2000).
However, the architecture of chromatin in interphase seems to be more complex and there are no general rules that can be applied to every chromosome territory in every cell line. Contrary to the proposed models, coding sequences were found in the interior of chromatin territories which do not change their positions during gene expression (Mahy et al. 2002). Also, transcription sites were found to be located all over chromatin territories (Abranches et al. 1998; Branco & Pombo 2006; Verschure et al. 1999) in contrast to the proposition of transcription taking place only at interchromatin regions. Moreover, the nuclear periphery contains silencing compartments (Andrulis et al. 1998) as well as activating compartments. Contrary to the observations in mammalian cells, nuclear pores in Saccharomyces cerevisiae have been identified as important sites of transcription (Brickner & Walter 2004; Menon et al. 2005; Schmid et al. 2006) and, in some instances, transcription activation is accompanied by motion of the sequence toward the nuclear periphery (Brickner & Walter 2004).
A significant amount of evidences has been accumulated in the last years pointing toward a model in which chromatin territories are interspersed with interchromatin domains. To activate transcription, in some cases specific local reorganization of regions of these territories occurs (Branco & Pombo 2006; Branco & Pombo 2007) while in others, the transcription machinery is physically excluded from particular chromatin territories providing a mechanism to silence gene expression (Chaumeil et al. 2006).
Two novel technologies, chromosome conformation capture (Dekker et al. 2002) and chromosome conformation capture-on-chip (Simonis et al. 2006) allowed studying intra and interchromosomal interactions for the first time and showed the relevance of these interactions to gene activity regulation. In these methods isolated nuclei are treated with formaldehyde producing crosslinking of nearby chromatin segments. The crosslinked products are digested with a restriction enzyme and analyzed to quantify the relative frequencies with which different sites of chromatin were crosslinked. By using this technology, Simonis et al (Simonis et al. 2006) showed that the actively transcribed β-globin locus in fetal liver interacts with a completely different set of loci from its transcriptionally silent counterpart in brain. While the majority of the β-globin interacting loci in fetal liver contained actively transcribed genes, most of the interacting loci in brain did not show gene activity.
In conclusion, in spite that we do not have a complete knowledge of the three-dimensional architecture of chromatin in the nucleus, factors such as the relative positioning of genes, regulatory sequences and transcription factors seems to be very important for transcription. It is therefore a priority investigating chromatin organization during interphase to fully understand the mechanisms involved in gene expression regulation.
Studying Intracellular Dynamics by Single Particle Tracking
In recent years, single particle tracking techniques (SPT) have been increasingly used to study the dynamics of cellular compounds ranging from small lipids and proteins to organelles [for recent reviews see (Cheezum et al. 2001; Levi & Gratton 2007)].
These techniques were developed to follow the position of individual particles in time. Provided the spatial and temporal resolution of the method is adequate, these trajectories can be analyzed to extract quantitative information about the mechanism involved in the motion of the particle. Since the properties of the particles are not averaged as in bulk measurements, SPT can easily distinguish populations of particles with different motion properties and recognize changes between different mechanisms of motion thus allowing the study of complex processes [see for example, (Dahan et al. 2003; Lakadamyali et al. 2003)]. These characteristics make SPT an appealing technique to achieve the ultimate goal of understanding dynamics in cells.
The most common methods used to track fluorescence particles are based on recording images of the cells in a widefield or confocal fluorescence microscope as a function of time and afterwards locating the particle of interest in every frame of the stack.
A point-like particle observed through an optical microscope forms a diffraction-limited image of width approximately equal to λ/(2·NA), where λ is the wavelength of the light and NA is the numerical aperture of the objective (Abramowitz 2003). This diffraction limit implies that the image of the particle would appear to have a diameter of ∼200 nm for visible light, making it impossible to achieve optical resolution below this limit.
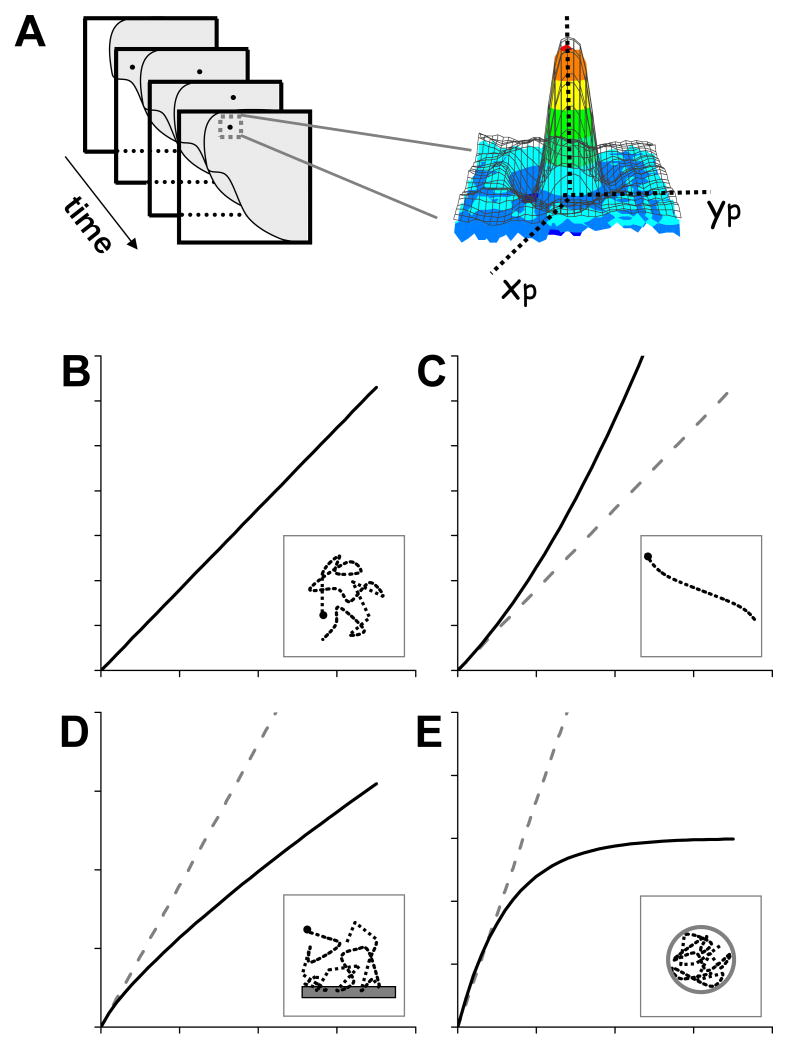
Several techniques have been proposed to locate particles with higher precision than that given by the diffraction limit. A common approach is based on the fact that the intensity of the image of a point-like particle follows a diffraction-limited distribution whose center corresponds to the position of the particle. Then, this position can be determined with high precision using an algorithm that finds the center of the distribution. The most common algorithm used to track particles is based on fitting a Gaussian distribution function to the intensity profile of the image (Figure 1A). By using this method, the precision of the position determination can be as high as 1.5 nm (Yildiz et al. 2003) depending on experimental factors such as the number of collected photons per particle, pixel size and background noise (Thompson et al. 2002).
FIGURE 1. Single particle tracking.
A. Determination of a particle trajectory by analysis of an image stack. A theoretical distribution function (black grid) is fitted to the image of a punctual particle in every image (colored in intensity-code scale). The center of the distribution function corresponds to the position of the particle. (B-E). Expected MSD curves for random diffusion (B), active transport (C), anomalous subdiffusion (D) and confined diffusion (E). Dashed lines in each plot represent the behavior expected for a free, randomly diffusing particle. The insets to the plots schematically show trajectories expected for particles moving under these regimes and possible interactions or restrictions that accounts for these trajectories.
Once the position of the particle is obtained in every frame of the movie, its trajectory is analyzed to extract quantitative information regarding the mechanism underlying the motion [for a general review see (Saxton & Jacobson 1997)]. The most common approach to analyze single particle trajectories is to calculate the mean square displacement (MSD) as follows:
| [1] |
where x, y and z are the particle coordinates and τ is a lag time.
MSD values at longer τ are calculated with a lower number of data. Thus, these values have lower statistical significance. A common criterion is to restrict the analysis to τ values lower than 1/4 of the total time of the trajectory (Saxton & Jacobson 1997).
MSD indicates how far a particle traveled after a time lag. Thus; its dependence with τ is related to the motion properties of the particle. Consequently, a possible mechanism for the particle motion can be obtained by comparing the experimental MSD plot with predictions from different motion models. Figure 1B-D shows MSD plots expected for simple mechanisms of motion such as random diffusion, corralled diffusion, active transport and anomalous subdiffusion (Qian et al. 1991; Saxton 1993; Saxton 1994; Saxton 1994; Saxton 1995; Saxton 1996; Saxton 1997; Saxton & Jacobson 1997). This last behavior can be the consequence of the binding of the particle to a given fixed target or due to the presence of obstacles to the free motion.
A common concern when doing tracking experiments in living cells is that regions of the cell may move during the experiment and thus the trajectory of the studied particle will reflect its intrinsic motion as well as the motion of the region where the particle is included. In the case of tracking experiments in the nucleus, different strategies have been followed to uncouple the motion of the fluorescent particle from the motion of the whole nucleus. Most of them are based on introducing an internal reference e.g. labeling the nuclear envelope, to study the motion of the particle relative to the reference [see for example, (Heun et al. 2001; Marshall et al. 1997)] Another possibility is to use cells whose nuclei are fluorescent and then to align the nucleus in every image before the tracking analysis [see for example (Chuang et al. 2006)]; this approach has the disadvantage that nuclear rotations may not be detected.
Chromatin Dynamics in the Interphase Nucleus
As we mentioned above, the spatial organization of chromatin is intimately related to gene expression. Important and not completely answered questions are how the organization of chromatin is achieved and how specific regions change their position in the nucleus.
Most of the techniques used to study chromatin architecture cannot reveal details regarding the dynamics of chromatin providing only a static view of the nucleus in a given condition. To observe chromatin motion, it is required to use labeling techniques compatible with living cell observation.
Initial works made use of DNA-specific fluorescent dyes such as 4,6-diamidino-2-phenylindole or DAPI (Crissman & Hirons 1994) and fluorescent nucleotides (Zink et al. 1998). With the emergence of green fluorescent protein (GFP) technology, histones fused to fluorescent protein variants (Kanda et al. 1998) were used to label chromatin.
A significant breakthrough in the field was the development of a novel approach to label specific DNA sequences in live cells which consist in the insertion of lac operator repeats at specific chromosome locations. This locus can be detected through the binding of the lac repressor protein fused to GFP and engineered to contain a nuclear localization signal (Belmont et al. 1999; Robinett et al. 1996). The fluorescent labeled sequence is seen as a bright dot in the nucleus which is dimly fluorescent due to unbound EGFP-Lac repressor. The motion of this bright dot can be followed and the recovered trajectory analyzed to obtain information about the mechanism by which the tagged sequence is moving.
Studies on Saccharomyces cerevisiae (Marshall et al. 1997), mammalian cells (Chubb et al. 2002) and Drosophila spermatocyte (Vazquez et al. 2001) have shown that GFP tagged chromatin loci undergo Brownian motion limited to a sub-region of the nucleus during interphase.
However, the motion seems to be more complex than constrained, passive diffusion. Chubb et al. (Chubb et al. 2002) demonstrated that chromatin associated with nucleolus or localized in the nuclear periphery is more restricted in its movements than other more nucleoplasmic genomic regions. Rosa et al (Rosa et al. 2006) showed that two chromosomal sites exhibiting preferential association with the nuclear membrane, are confined to regions of different size, with the site with higher levels of transcription exploring larger regions. Heun et al. (Heun et al. 2001) reported that early and late origins of replication in yeast are more mobile in G1 phase than in S phase. Also, the movement in G1 phase was highly sensitive to ATP depletion and to changes in metabolic status.
On the other hand, Tumbar and Belmont (Tumbar & Belmont 2001) showed that a specific DNA region in CHO cells changes its position from the nucleus periphery to the center in response to VP16 transcriptional activator.
An important factor that limits the temporal resolution and observation time window in studies of chromatin dynamics is that the motion of the labeled sequence is highly sensitive to photodamage, as was shown in yeast and mammals cells (Chuang et al. 2006; Hediger et al. 2004). For example Chuang et al (Chuang et al. 2006) showed that the motion of a tagged chromatin sequence significantly changes after taking as little as 10 images when using a mercury lamp as excitation source in regular imaging conditions. Thus special care has to be taken with respect to the illumination conditions, exposure time and maximum number of frames that can be acquired.
Two Photon Microscopy
In 1990, Denk, Stricler and Webb invented two-photon excitation microscopy (Denk et al. 1990). This relatively new microscopy is an attractive alternative when dealing with photobleaching and photodamage in widefield and confocal fluorescence microscopes. Excellent reviews on this technique can be found in the literature [see, for example, (So et al. 2000; Svoboda & Yasuda 2006)].
Two-photon excitation is a non-linear process involving the almost simultaneous absorption of two photons(∼10-15 s) that in the simplest case will have half of the energy required for the transition to the excited state (Figure 2A). The probability of this simultaneous absorption increases with the square of the photon flux since two photons have to encounter the fluorophore almost at the same time. Thus, very high photon fluxes are necessary to achieve significant levels of two-photon excitation. This condition is fulfilled by using, for example, femtosecond-pulse lasers that provide pulses of high intensity during brief periods of time but have low average excitation power therefore limiting the photodamage to the sample for at least two reasons. Firstly, the excitation of the fluorophore only occurs at the focal point of the laser beam and secondly because the near-IR laser light has relatively low absorption in biological samples.
FIGURE 2. Two-photon microscopy.
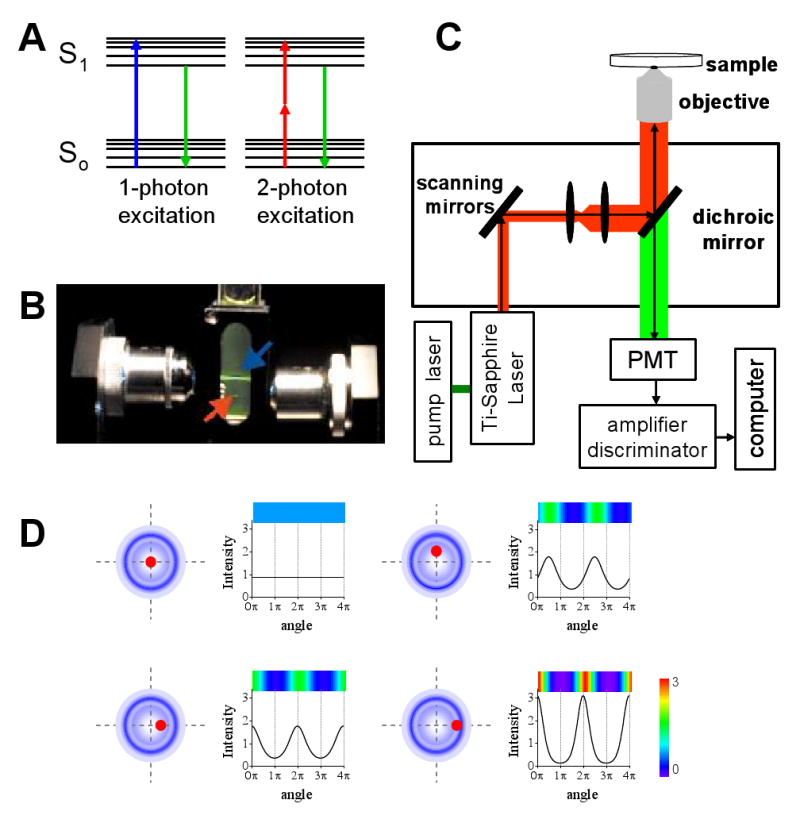
A. Energy diagrams showing the electronic transitions occurring during one-photon (blue) and two-photon (red) excitation. B Emission volumes obtained in a fluorescein solution excited under one-photon (blue arrow) and two-photon (red arrow) conditions. The numerical apertures of the objectives used in this experiment were the same. C. Scheme of the two-photon excitation microscope used for single particle tracking experiments. D. Intensity profile determined along two cycles of the tracking routine represented as function of the angle of rotation of the laser, for different relative positions of the particle (black dot) with respect to the center of scanning (right panels). The laser orbits are represented with blue circles.
Figure 2B shows the emission volume obtained in a sample of fluorescein under two-photon or one-photon excitation. While two-photon excitation occurs only in the femtoliter volume in which the laser was focused, one-photon excitation is also observed in out-of-focus regions.
The key feature of two-photon microscopy is that emission occurs in a femtoliter focal volume. This small volume defines several advantages of this technique over one-photon excitation microscopy. It provides improved axial depth discrimination and image contrast and less out of focus photobleaching and photodamage since no excitation occurs in out of focus regions. On the other hand, the background fluorescence in two-photon microscopy is usually lower because the excitation wavelength is significantly red-shifted with respect to emission. Thus, it is easier to separate excitation from emission lights than in a one-photon experiment where excitation and emission are spectrally close to each other. Another advantage of two-photon microscopy is that it is better suited for imaging thick specimens because the near-IR light used for the excitation is less scattered and absorbed than the UV or visible excitation light required for the excitation of common fluorophores in one-photon microscopy.
Figure 2D shows the two-photon microscope setup used for the tracking experiments described below. In this setup, a mode-locked titanium-sapphire laser is used as excitation source. These lasers are ideal for two-photon microscopy because they provide femtosecond pulses with a repetition rate of ∼100 megahertz, and can be tuned in the range 700-1000 nm covering the excitation of common fluorescent probes including most of the variants of fluorescent proteins. There are several papers describing the determination of two-photon excitation cross-section for fluorescent probes [see for example, (Larson et al. 2003; Wokosin et al. 2004; Xu & Webb 1996; Xu et al. 1996)].
The laser is then directed into the microscope by two galvomotor-driven scanning mirrors moved by voltage generated in a computer card. In a conventional two-photon microscope, the mirrors are programmed to raster scan the laser on a squared region of the sample.
The laser light is reflected in a low-pass dichroic mirror and focused on the sample with the objective. Fluorescence emission is collected by the objective and passes through the dichroic mirror and a short-pass filter to eliminate any reflected excitation light. It then exits the microscope to the photomultiplier detector; which output is further amplified, passed through a photon counting discriminator and finally, photons are counted with a data acquisition card.
Single Particle Tracking In A Two Photon Microscope
Motivated by the results mentioned above regarding the high level of photodamage observed in chromatin studies using standard fluorescence microscopes, we designed a routine to do particle tracking in a two-photon excitation setup which, as we mentioned above, provides improved photostability of the sample (Levi et al. 2005).
The main limitation of scanning microscopes is that acquiring an image usually takes in the order of a second depending on the size of the image to scan, making any tracking routine based on imaging very slow. Thus, a completely different approach to imaging-based methods was followed by our group (Kis-Petikova & Gratton 2004; Levi et al. 2005; Levi et al. 2005) based in the early theoretical work of Enderlein (Enderlein 2000).
The tracking routine starts with a fast raster scan of an area of the sample that includes the particle of interest. Then, the user selects the particle to be tracked by clicking on top of its image. This directs the laser beam to the chosen particle and the computer saves these coordinates as the initial position for the tracking routine. During each cycle of tracking, the excitation beam traces a given number of circular orbits surrounding the particle of interest and the fluorescence intensity is integrated at different points along the orbit. Figure 2D represents the fluorescence intensity profile expected during these circular scans for different relative positions of the particle to the center of scanning. The dependence on the intensity profile with the particle position during circular scans was mathematically derived providing a way to determine the coordinates of the particle. (Kis-Petikova & Gratton 2004). The spatial resolution depends on the diameter of the orbit and can be as high as 1 nm when the diameter of the circle is equal to the radial waist of the point spread function (Kis-Petikova & Gratton 2004).
During the tracking routine, the determination of the particle position is done on the fly by analyzing the fast Fourier transform of the intensity signal (Kis-Petikova & Gratton 2004). Before the next cycle of tracking, the center of scanning is moved to the position determined for the particle in the previous cycle. Thus, the laser follows the particle in real time; in contrast to imaging-based methods in which the trajectory is recovered from recorded data.
The method can also be used to do simultaneous two-particle tracking. The tracking routine starts on top of one of the particles; after a given number of cycles, the laser jumps to the position of the second particle where it performs the same number of tracking cycles. Then, the center of scanning is moved to the new position determined previously for the first particle. Thus the positions of the particles are recovered alternately. It should be noticed that by doing so the temporal resolution of the method is reduced to one half.
Experimentally, we determined that the spatial and temporal resolutions of the method are 20 nm and 32 ms, respectively. Also, we showed that the error of the position determination is inversely proportional to the square of the S/N ratio, as it was predicted (Kis-Petikova & Gratton 2004) being approximately constant for S/N >2 (Levi et al. 2005). Most methods based in images analysis only work for S/N ratios higher than 4 (Cheezum et al. 2001).
The background noise –due to the detection of other compounds different from the particle of interest- is usually a limit to the accuracy of the tracking when doing experiments in living cells. The theoretical accuracy of the particle position obtained by the scanning method does not change in the presence of a locally homogeneous background representing up to 10 % of the total photons emitted by the particle. In contrast, the theoretical accuracy on the particle position obtained by Gaussian deconvolution abruptly decreases by adding a similar background (Levi & Gratton 2007).
Tracking Chromatin in a Two-Photon Excitation Microscope
We re-inspected the motion of EGFP-tagged chromatin sequences in interphase cells by using the two-photon microscopy tracking technique described above (Levi et al. 2005). As we mentioned previously, two-photon excitation usually causes less out-of-focus photobleaching and photodamage than one photon excitation. In addition, the excitation laser moves in a very small volume of the cell during the tracking routine, introducing no damage in regions far from the tagged sequence.
For this study, we used the C6-14 CHO cell line which has 10–20 copies of a plasmid containing lac operator repeats and a DHFR cDNA transgene driven by a viral promoter (Tumbar & Belmont 2001). These copies are integrated at a single, internal chromosome site and can be observed in the two-photon excitation microscope as a bright dot (Figure 3A).
FIGURE 3. Dynamics EGFP-tagged sequence in living nuclei.
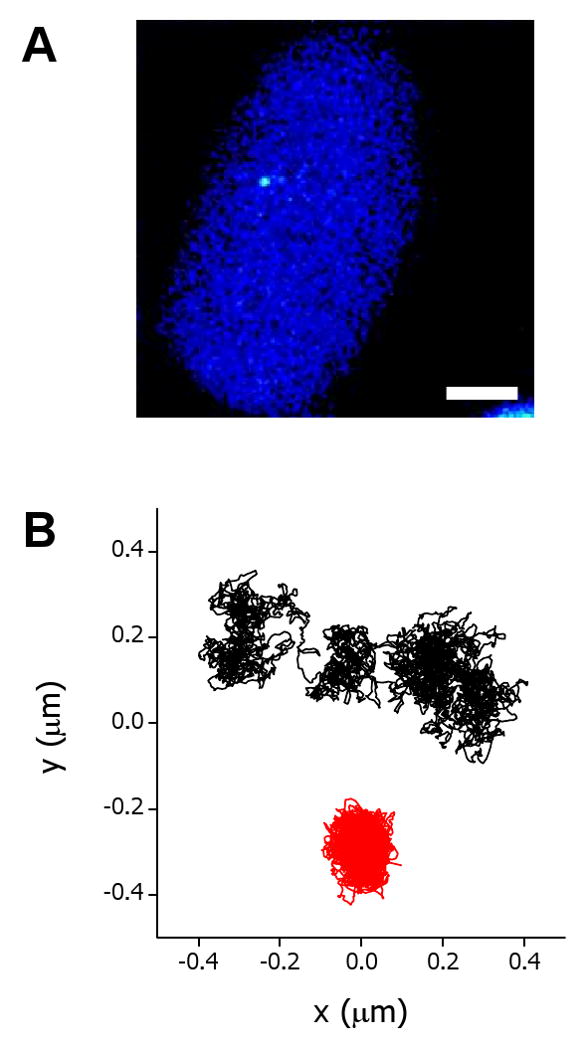
A. Two photon excitation fluorescence image of a representative C6-14 cell. Bar, 5 μm. B Trajectories of the sequence in a control (black) and azide-treated (red) cells.
The trajectories obtained for this EGFP-tagged sequence showed long periods of confinement in regions of sizes similar to the 30 nm fiber of chromatin -probably reflecting local, thermal fluctuations- interspersed by short periods in which the sequence moves ∼ 150 nm following a curvilinear path (Figure 3B).
We were puzzled by the presence of these jumps in position since; as far as we know, they were not observed in previous studies on chromatin motion. However, jumps occurred in the time range 0.3-2 s, thus they could not be observed using a tracking method with low temporal resolution. Moreover, the characteristic jump distance was 150 nm, similar to the spatial resolution of previous methods.
We ruled out that jumps were due to the motion of the nucleus using the DHFR-BAC cell line which has multiple lac operator-repeats insertions and also expressed the EGFP-lac repressor protein. Each of the insertions is visualized in a fluorescence microscope as a bright dot separated from the neighbor dots by less than 1-2 μm. The trajectories of these labeled DNA sequences also presented jumps but they occurred independently from each other showing that the jumps reflect local, short-distance motion of the chromatin sequence (Figure 4).
FIGURE 4. Simultaneous tracking of EFGP-tagged sequences in DHFR-BAC cells.
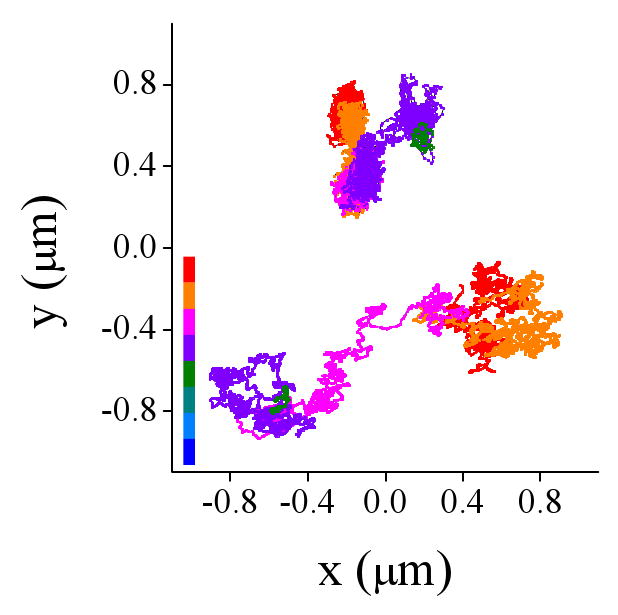
The trajectories were registered by simultaneous tracking two labeled sequences in a DHFR-BAC nucleus following the procedure described in the text. The color code represents the time evolution of the trajectories.
The way the sequence moves during jumps may remind us of the motion of a particle during active transport. However, in order to interpret the data properly, it is extremely important to do a careful statistical analysis of the data. A visual examination of isolated segments of any trajectory would suggest different mechanisms of motion since there is always a non-zero probability of observing a given path in any trajectory. Unfortunately, most of the tools designed for trajectories analysis are based on MSD analysis, in which processes that occur only during short periods of time are averaged out.
Thus we have designed new, statistical tools to get insight in the mechanism underlying the motion of the sequence during jumps and complemented this analysis with experiments to test the hypothesis derived from the statistical analysis. The statistical analyses were designed to compare the motion of the sequence during jumps with the predictions obtained considering a passive diffusion process. All our analyses indicated that the sequence moves an average of four times faster than in the periods between jumps and in paths more rectilinear than predicted for random diffusion motion suggesting that an active process is responsible for the transport of the sequence during short periods of time. This hypothesis was supported by experiments showing, for example, that there are no jumps in the trajectories after ATP depletion (Figure 3B) suggesting that jumps likely reflect energy-dependent chromatin movements.
Final Remarks
The view of the internal structure and function of the nucleus has drastically changed in the last years. It is now accepted that the nucleus is a well organized and highly compartmentalized organelle. Chromatin has also been shown to be structurally organized in interphase and its organization was found to be very important to gene regulation. Significant advances in our understanding of chromatin dynamics during interphase were consequences of technological developments in the fields of fluorescence microscopy and Cell Biology. SPT constituted an important new tool to reveal new aspects of chromatin dynamics.
The emerging model coming from SPT experiments is that chromatin diffuses within a constrained volume of the nucleus during most of the time but can also move through an energy dependent mechanism during brief periods. Important new data from Chuang et al. (Chuang et al. 2006) gave some clues regarding chromatin active motion. They observed a fast, directional motion of an interphase chromosome site from the nuclear periphery to the interior after targeting a transcriptional activator to this site. The directional motion was suppressed in actin and myosin mutants suggesting that they constitute the transport system required for this repositioning. More work will be needed to fully understand the mechanism involved in the fast, active motion of chromatin and more important, what is the relevance of this motion to gene regulation.
References
- Abramowitz M. Microscope: basics and beyond. Olympus America Inc.; New York: 2003. [Google Scholar]
- Abranches R, Beven AF, Aragon-Alcaide L, Shaw PJ. Transcription sites are not correlated with chromosome territories in wheat nuclei. J Cell Biol. 1998;143:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Li G, Sudlow G, Robinett C. Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system. Methods Cell Biol. 1999;58:203–222. doi: 10.1016/s0091-679x(08)61957-3. [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Chromosome organization: new facts, new models. Trends Cell Biol. 2007;17:127–134. doi: 10.1016/j.tcb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur J Histochem. 2006;50:161–176. [PubMed] [Google Scholar]
- Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem. 2006;50:223–272. [PubMed] [Google Scholar]
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet. 1982;62:201–209. doi: 10.1007/BF00333519. [DOI] [PubMed] [Google Scholar]
- Crissman HA, Hirons GT. Staining of DNA in live and fixed cells. Methods Cell Biol. 1994;41:195–209. doi: 10.1016/s0091-679x(08)61718-5. [DOI] [PubMed] [Google Scholar]
- Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderlein J. Tracking of fluorescent molecules diffusing within membranes. Appl Phys B-Lasers O. 2000;71:773–777. [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Hediger F, Taddei A, Neumann FR, Gasser SM. Methods for visualizing chromatin dynamics in living yeast. Methods Enzymol. 2004;375:345–365. doi: 10.1016/s0076-6879(03)75022-8. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Kis-Petikova K, Gratton E. Distance measurement by circular scanning of the excitation beam in the two-photon microscope. Microsc Res Tech. 2004;63:34–49. doi: 10.1002/jemt.10417. [DOI] [PubMed] [Google Scholar]
- Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, Zirbel RM, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, Webb WW. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300:1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- Levi V, Gratton E. Exploring dynamics in living cells by tracking single particles. Cell Biochem Biophys. 2007;48:1–15. doi: 10.1007/s12013-007-0010-0. [DOI] [PubMed] [Google Scholar]
- Levi V, Ruan Q, Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Sheetz MP, Elson EL. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys J. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Maddocks JH, Neumann FR, Gasser SM, Stasiak A. Measuring limits of telomere movement on nuclear envelope. Biophys J. 2006;90:L24–26. doi: 10.1529/biophysj.105.077974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Lateral diffusion in an archipelago. Single-particle diffusion. Biophys J. 1993;64:1766–1780. doi: 10.1016/S0006-3495(93)81548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Anomalous diffusion due to obstacles: a Monte Carlo study. Biophys J. 1994;66:394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Single-particle tracking: models of directed transport. Biophys J. 1994;67:2110–2119. doi: 10.1016/S0006-3495(94)80694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Single-particle tracking: effects of corrals. Biophys J. 1995;69:389–398. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Anomalous diffusion due to binding: a Monte Carlo study. Biophys J. 1996;70:1250–1262. doi: 10.1016/S0006-3495(96)79682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Single-particle tracking: the distribution of diffusion coefficients. Biophys J. 1997;72:1744–1753. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- So PT, Dong CY, Masters BR, Berland KM. Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr Biol. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Spatial relationship between transcription sites and chromosome territories. J Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wokosin DL, Loughrey CM, Smith GL. Characterization of a range of fura dyes with two-photon excitation. Biophys J. 2004;86:1726–1738. doi: 10.1016/S0006-3495(04)74241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Webb WW. Measurement of two-photon excitaton cross sections of molecular fluorophores with data from 690 nm to 1050 nm. J Opt Soc B. 1996;13:481–491. [Google Scholar]
- Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: new spectral windows for biological nonlinear microscopy. Proc Natl Acad Sci U S A. 1996;93:10763–10768. doi: 10.1073/pnas.93.20.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EH. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]