Abstract
Co-transcriptional RNA processing not only permits temporal RNA processing before the completion of transcription, but also allows sequential recognition of RNA processing signals on nascent transcripts threading out from the elongating RNAPII complex. Rapid progress in recent years has established multiple contacts that physically connect the transcription and RNA processing machineries, which centers on the C-terminal domain (CTD) of the largest subunit of RNAPII. While co-transcriptional RNA processing has been substantiated, the evidence for “reciprocal” coupling starts to emerge, which emphasizes functional integration of transcription and RNA processing machineries in a mutually beneficial manner for efficient and regulated gene expression.
Introduction
Although separately dissected at the molecular level, various machineries for RNA polymerase II (RNAPII)-mediated gene expression are intimately coupled in vivo from transcription to RNA processing to RNA export, which has been the subject of intensive studies in recent years (for comprehensive reviews, see [1-4]). Temporal coupling not only allows efficient gene expression to meet the demand for growth and proliferation, but also permits rapid response to diverse signaling events.
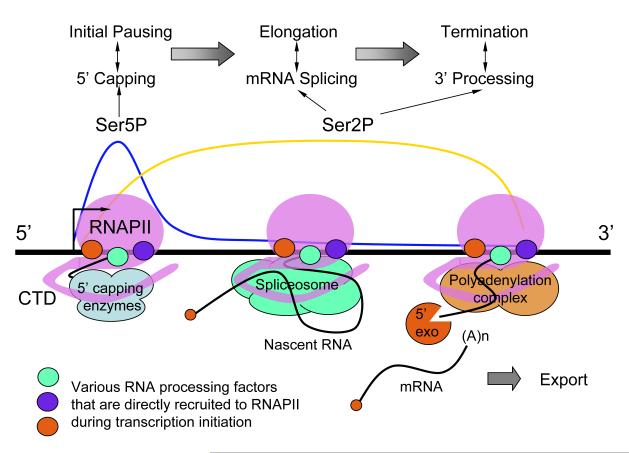
RNAPII-mediated transcription is known to take place in three phases, promoter clearance, productive elongation, and transcriptional termination [5], which is coupled, respectively, with 5′ capping, intron removal, and 3′ end formation [6,7]. These processes are functionally dependent on phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNAPII with Ser5 phosphorylation peaking at the beginning of the gene and Ser2 phosphorylation occurring in an incremental fashion along the gene [3]. As illustrated in Fig. 1, this dynamic change in CTD phosphorylation suggests that the RNAPII complex is rearranging its content during transcription to allow sequential action of distinct machineries for co-transcriptional RNA processing. However, sequential action does not necessarily mean sequential recruitment of RNA processing factors to the RNAPII complex because many “downstream” factors seem to be recruited at the very beginning of transcription [8,9].
Figure 1.
Coupling between transcription and pre-mRNA processing. The RNA polymerase II (RNAPII) is modified on its CTD with Ser5 phosphorylation predominately at the beginning of the gene (blue line) and Ser2 phosphorylation in the middle and end of the gene (yellow line). 5′ capping enzymes are recruited through direct interactions with Ser5 phosphorylated CTD to catalyze the co-transcriptional capping reaction. Various splicing factors are recruited during the elongation phase of transcription, most of which in a CTD Ser2 phosphorylation dependent manner, to facilitate co-transcriptional splicing. The 3′ end formation is functionally tied to transcription termination. Importantly, increasing evidence now suggests that the transcription and RNA processing machineries are functionally integrated in a reciprocal fashion such that individual co-transcriptional processing events also influence transcription at different phases.
While most studies focus on understanding how RNA processing takes advantage of the transcriptional machinery to execute co-transcriptional processing for efficient gene expression, increasing evidence suggests that transcription may also benefit from and/or depend on specific RNA processing reactions. The most compelling case is the critical role of 3′ end formation in transcriptional termination, which is triggered by RNA cleavage at the polyadenylation site [6]. This review concentrates on our recent understanding of molecular links between transcription and RNA processing machineries, with a particular emphasis on “reciprocal coupling”, i.e. the interdependence of transcription and co-transcriptional RNA processing.
Multiple molecular contacts to facilitate functional coupling
Proteomic analysis of affinity purified RNAPII complex demonstrates the association of a large number of RNA processing and transport factors with the complex, suggesting that RNA processing factors are physically integrated with the transcriptional machinery in eukaryotic cells [10]. Consequently, many RNA processing factors are tethered to the transcribed genes, which can be detected by chromatin immuniprecipitation (ChIP) [11-15]. However, it is also clear that purified RNAPII does not contain all RNA processing factors, indicating that only a subset of stable interactions survived biochemical fractionation. It is currently unclear whether all RNA processing machineries are fully loaded to the RNAPII complex at the beginning of transcription to execute sequential actions during transcription or distinct sets of RNA processing factors are dynamically recruited at different phases of transcription.
Table 1 lists documented physical interactions between transcription and RNA processing machineries. The CTD serves as a general loading pad for RNA processing enzymes. Except for the polypyrimidine tract binding splicing regulator PSF [16] and related p54nrb [17], which appear to bind to CTD independently of its phosphorylation state [18], most recruitment events require CTD phosphorylation. The core transcription factor TFIIH catalyzes Ser5 phosphorylation, which is essential for the recruitment of 5′ capping enzymes at the beginning of the gene [19,20]. This step coincides with the initial RNAPII pausing event ∼30 nt downstream the transcriptional start. The initial pausing is also thought to be a checkpoint for the recruitment of CTD Ser2 kinases, such as P-TEFb, for RNAPII to enter the productive mode of transcriptional elongation [5,21]. In contrast to the capping enzymes, a large of number of RNA proteins (Table 1) implicated in RNA splicing and 3′ end formation bind to CTD in a Ser2 or Ser5/Ser2 phosphorylation dependent manner [22-27].
Table 1.
Factors or domains involved in physical interactions between transcription and RNA processing Machineries
| Transcription factors |
RNA processing factors |
Key domain for binding |
Coupled functions |
References |
|---|---|---|---|---|
| Ser5P CTD | Gtase, MT | GTase domain | Txn reinitiation/5′ capping | 19,20 |
| Ser2P CTD | SR proteins | RS domain | Txn/Splicing commitment | 23 |
| Prp40 (U1 snRNP) | FF domain | Txn/Splicing reaction | 27 | |
| SCAFs | CID | Txn/3′ end formation | 22,25,26 | |
| Pcf11 | ? | Txn/3′ end formation | 24 | |
| CTD | PSF | FF domain | Txn/Alternative splicing | 16,18 |
| p54nrb | FF domain | Txn/Alternative splicing | 17 | |
| CHD | SF3a (U2 snRNP) | ? | TriMeH3K4 recognition/ Txn elongation/splicing |
32 |
| ASC-2 | CAPERα, β | RRM1 | Txn initiation/ Alternative splicing |
33,34 |
| PC4 | CstF64 | ? | Txn/3′ end formation | 29 |
| TFIID | CPSF | ? | Txn/3′ end formation | 28 |
| TAT-SF1/Cus2 | dual role? | Txn elongation/ Spliceosome assembly |
38,39.40 | |
| SKIP/Prp45 | dual role? | Txn elongation/ Splicing reaction |
14,41 | |
| CA150 | SF1 | WW domain Proline-rich motif |
Txn elongation/ Spliceosome assembly |
35,36 |
| THO and Spt6 | UAP56, Aly | ? | Txn elongation/ RNA export |
42,44,45 |
Besides CTD-mediated interactions, multiple RNA processing factors interact directly with specific transcriptional factors. The polyadenylation factors CPSF and CstF64 bind to the general transcription factor TFIID and coactivator PC4, respectively [28,29]. These and likely other molecular interactions may underline physical tethering between both ends of the gene observed in yeast and mammalian cells [11,30,31]. More recently, the chromodomain protein CHD capable of recognizing trimethylated H3K4, a generally mark for active genes at gene promoters, was found to directly interact with SF3a, a component of the U2 snRNP particle [32]. This joins a growing list of RNA processing factors that directly interact with specific transcription factors and cofactors (www.nursa.org), such as the two U2AF65 homologues called HCC1.3/CAPERαand β, which bind to the coactivator ASC2 in nuclear hormone-mediated transcription and alternative splicing [33,34]. These findings suggest that different promoters may nucleate distinct sets of RNA processing factors, which may travel with the elongating RNAPII complex to affect downstream RNA processing events [8].
Interestingly, several transcriptional elongation factors appear to mediate molecular interactions between transcription and RNA splicing machineries or even have a dual role in these processes. In particular, the transcriptional elongation factor CA150 is able to bridge the splicing factor SF1 via its WW domain and phosphorylated CTD through its FF domain [35,36]; and interestingly, the interaction between CA150 and several splicing factors seems to subject to regulation by arginine methylation [37]. The transcriptional elongation factor TAT-SF1 forms a stable complex with the spliceosome [38] and its homologue in yeast (Cus2) has been shown to function as a critical proofreading enzyme during early spliceosome assembly [39,40], suggesting that TAT-SF1/Cus2 may be a dual function protein in transcription and mRNA splicing. Similarly, SKIP/Prp45 has been characterized as a dual functional factor in both transcriptional elongation and splicing [14,41]. These findings indicate that the transcriptional machinery is structurally and functionally intertwined with specific components of the splicing machinery.
Finally, the THO transcriptional elongation complex co-transcriptionally recruits critical mediators for RNA export, including Sub2/UAP56 and Yra1/Aly in yeast [42,43]. However, mammalian cells seem to use a different strategy to couple transcription with RNA export: The transcriptional elongation factor Sp6 was recently found to interact with an adaptor to mediate the Aly recruitment [44], which seems to be unloaded onto nascent transcripts in a splicing dependent manner [45,46]. Together, these data demonstrate a crucial role of transcriptional elongation factors in coupling transcription with mRNA splicing and export.
Functional consequences of sequential recognition of cis-acting elements
Given the extensive molecular interaction between transcription and RNA processing machineries, what is the functional benefit for mRNA splicing to take place co-transcriptionally, instead of post-transcriptionally? One obvious possibility is that mRNA splicing may occur more efficiently co-transcriptionally than post-transcriptionally. Purified RNAPII has been shown to stimulate both splicing and polyadenylation in vitro, suggesting that the polymerase may facilitate the assembly of RNA processing machineries [47,48]. The existence of functional coupling has been extensively investigated by the development of in vitro coupled transcription and splicing systems [49-53]. Although the data generated so far are not entirely consistent with one another, at least some evidence points to the importance of RNAPII, rather than transcription per se, in facilitating efficient mRNA splicing by comparing RNAPII with a phage polymerase [53].
Further exploration of the coupled transcription/splicing revealed a critical role of co-transcriptional commitment of pre-mRNA to the splicing pathway in achieving a rapid kinetics in mRNA splicing [10], comparable to that observed in vivo [54]. This is accomplished by a family of splicing commitment factors known as SR proteins, which are essential for multiple steps of spliceosome assembly in mammalian cells [55]. It is likely that, during transcriptional elongation, SR proteins and other spliceosomal components may be able to recognize emerging splicing signals in a timely manner in competition with other RNA binding proteins, such as hnRNP proteins, to nucleate the efficient assembly of functional spliceosomes on nascent transcripts [53].
Another benefit of co-transcriptional RNA splicing is the recognition of splicing signals in a sequential manner, which may affect splice site selection in a distinct mechanism from the situation when all splice sites are exposed simultaneously to the splicing machinery in nuclear extracts [56]. Indeed, it has been documented that the rate of transcriptional elongation is able to dictate splice site selection in vivo [57,58]. According to the current kinetic model, weak splice site may be selected during transcriptional elongation if the elongating RNAPII is slowed down to leave sufficient time for the recognition of the weak splice site before the appearance of a stronger competing splice site [59]. Consequently, transcriptional elongation was found to have a profound impact on splicing commitment [60] and alternative splicing in vivo [61]. Since multiple RNA processing factors are linked to the transcriptional machinery via physical interaction with various transcriptional elongation factors, it is conceivable that many transcription elongation factors may also regulate alternative splicing in mammalian cells.
Transcription/RNA processing coupling: A mutually beneficial arrangement
If the coupling with transcription facilitates RNA processing, is the converse also true? As illustrated in Fig. 1, this is clearly the case with transcriptional termination, a process that has been fully integrated with the 3′ end formation of transcribed genes. Unlikely genes in prokaryotes or intronless histone genes or other classes of transcriptional units mediated by RNAPI or RNAPIII in eukaryotes, the vast majority of RNAPII-mediated transcription does not seem to terminate at a defined location at the end of the gene. Nuclear run-on assay suggests that RNAPII may continue to transcribe ∼1.5kb after the polyadenylation site [62]. An allosteric model proposes that the nascent RNA is cleaved downstream the polyadenylation site, which may trigger some sort of conformational change in the elongating RNAPII complex, thereby inducing pausing and release of the elongating RNAPII complex from template DNA. Alternatively, the torpedo model suggests that the exposed 5′ end becomes attackable by the nuclear 5′-3′ exonuclease Xrn2 and rapid trimming of the free 5′ end ultimately causes the fall off of the elongating RNAPII complex at the end of the gene [6]. Perhaps, both models reflect specific aspects of the process as suggested by a recent study [63]. The important message here is that 3′ end formation is part of the transcription termination process for most protein-coding genes.
Similar to the coupling events at the 3′ end of the gene, the 5′ capping enzymes also play a major role as a checkpoint in transcription reinitiation at the beginning of the gene. It was observed earlier that an excess amount of the methylation donor SAM inhibited transcription initiation [64] and that the 5′ capping enzymes were able to repress transcription [65], suggesting that the 5′ capping reaction and the subsequent release of the capping enzymes are both critical for the transition from transcriptional initiation to elongation. Indeed, further studies revealed that both Ser5 phosphorylated CTD and the transcription elongation factor Spt5 help recruit the 5′ capping enzymes, which then counteract the inhibitory NELF complex, thereby facilitating promoter clearance and enhancing the entrance of the initially paused RNAPII to the mode of productive transcription elongation [66-68]. After this transition, the CTD phosphatase FCP1 seems responsible for removing Ser5 phosphates and dismantling the capping enzymes [20,68].
If co-transcriptional RNA processing at both the 5′ and 3′ of the gene benefits transcription, does co-transcriptional splicing also play a role in transcription? The first piece of evidence for this “reciprocal” coupling came from the demonstration that the TAT-SF1 associated splicing complex was able to stimulate transcriptional elongation [38]. However, it has been unclear which component(s) in the complex provided such function. A hint to this important question came from a striking observation that in vivo depletion of the splicing commitment factor ASF/SF2 (a prototypic SR protein) caused widespread double-strand DNA breaks [69]. Multiple SR proteins seem to have a similar role in maintaining genome stability [70,71]. Further studies revealed that ASF/SF2 plays a direct role in resolving transcription bubbles that are transiently formed behind the elongating RNAPII complex in which a portion of nascent RNA remains base-paired with the template DNA, a configuration known as R-loop [69]. Because the resolution of transcriptional R-loops is likely critical for transcriptional elongation, it is conceivable that SR proteins and other single-stand RNA binding proteins may directly facilitate transcriptional elongation, which may be intimately coupled with splicing commitment. Therefore, SR proteins may play a bigger role in gene expression, explaining the indispensable requirement of SR proteins for cell proliferation and animal development [71,72]. It is interesting that nature has evolved distinct strategies to prevent R-loops by coupling transcription with translation in bacteria, with RNA export in yeast, and with splicing in metazoans [4].
Conclusions
Increasing evidence has now established that transcription and co-transcriptional RNA processing are an integrated process for gene expression in eukaryotic cells. Such coupling seems to be mutually beneficial. The elucidated molecular connections between transcription and RNA processing machineries may be just a tip of iceberg, which may reflect a series of recruitment and/or stabilization events at different phases of transcription. The challenge is to further dissect the mechanics of transcription-RNA processing coupling, and more importantly, understand how various coupling events may be regulated and how one event may influence the other to give rise to final outcomes of gene expression in a cell-type specific manner or in response to diverse internal and external signals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 2.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 3.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 5.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 10.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036.SR proteins were found to co-transcriptionally commit nascent RNA to the splicing pathway in in vitro coupled transcription-splicing system. The work showed that co-transcriptional splicing is much more efficient than post-transcriptional splicing, demonstrating the functional coupling between transcription and splicing.
- 11.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 14.Bres V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swinburne IA, Meyer CA, Liu XS, Silver PA, Brodsky AS. Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 2006;16:912–921. doi: 10.1101/gr.5211806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kameoka S, Duque P, Konarska MM. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. Embo J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. Rna. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 24.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 25.Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci U S A. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 27.Morris DP, Greenleaf AL. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 28.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 29.Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 30.O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 31.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030.This work indicates that gene looping seems to be a general phenomenon in both yeast and mammalian cells, which is mediated by RNA processing factors bound at both ends of the gene.
- 32.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010.A novel link was established between a chromodomain-containing transcription factor (CHD) and a specific U2 snRNP component (SF3a) and the interaction was shown to be critical for mRNA splicing both in vitro and in vivo. CHD is part of complex recognizing the TriMeH3K4 mark and the CHD-containing complex is also implicated in transcriptional elongation, suggesting a new connection between transcriptional initiation, elongation, and co-transcriptional RNA splicing.
- 33.Jung DJ, Na SY, Na DS, Lee JW. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem. 2002;277:1229–1234. doi: 10.1074/jbc.M110417200. [DOI] [PubMed] [Google Scholar]
- 34.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Alvarez M, Goldstrohm AC, Garcia-Blanco MA, Sune C. Human transcription elongation factor CA150 localizes to splicing factor-rich nuclear speckles and assembles transcription and splicing components into complexes through its amino and carboxyl regions. Mol Cell Biol. 2006;26:4998–5014. doi: 10.1128/MCB.01991-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MJ, Kulkarni S, Pawson T. FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol Cell Biol. 2004;24:9274–9285. doi: 10.1128/MCB.24.21.9274-9285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019.The authors show that the arginine methyltransferase CARM1 recognizes the transcriptional elongation factor CA150 and a number of spliceosome components, thereby regulating the coupling between transcription and splicing.
- 38.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 39.Perriman R, Ares M., Jr. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- 40.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albers M, Diment A, Muraru M, Russell CS, Beggs JD. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. Rna. 2003;9:138–150. doi: 10.1261/rna.2119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 43.Reed R. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 44.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107.The elongation factor Spt6 was found to recruit the mRNA export factor Aly via a novel adaptor called Iws1. RNAi-knockdown of IwsI impaired poly(A)+ mRNA export in HeLa cells. The study reveals a new coupling mechanism for transcriptional elongation and mRNA export.
- 45.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 46.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 48.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarev D, Manley JL. Concurrent splicing and transcription are not sufficient to enhance splicing efficiency. Rna. 2007;13:1546–1557. doi: 10.1261/rna.595907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Natalizio BJ, Garcia-Blanco MA. In vitro coupled transcription splicing. Methods. 2005;37:314–322. doi: 10.1016/j.ymeth.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Hicks MJ, Lam BJ, Hertel KJ. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods. 2005;37:306–313. doi: 10.1016/j.ymeth.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Audibert A, Weil D, Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol Cell Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin S, Fu X-D. SR Proteins and related factors in alternative splicing. In: Blencowe B, Graveley B, editors. Alternative splicing in postgenomic era. Vol. 623. 2007. pp. 107–122. (Eurekah Bioscience Series). [DOI] [PubMed] [Google Scholar]
- 56.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 57.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Howe KJ, Kane CM, Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. Rna. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. Rna. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 62.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 63.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jove R, Manley JL. Transcription initiation by RNA polymerase II is inhibited by S-adenosylhomocysteine. Proc Natl Acad Sci U S A. 1982;79:5842–5846. doi: 10.1073/pnas.79.19.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P. The yeast capping enzyme represses RNA polymerase II transcription. Mol Cell. 2002;10:883–894. doi: 10.1016/s1097-2765(02)00644-5. [DOI] [PubMed] [Google Scholar]
- 66.Kim HJ, Jeong SH, Heo JH, Jeong SJ, Kim ST, Youn HD, Han JW, Lee HW, Cho EJ. mRNA capping enzyme activity is coupled to an early transcription elongation. Mol Cell Biol. 2004;24:6184–6193. doi: 10.1128/MCB.24.14.6184-6193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 68.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Li X, Niu T, Manley JL. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. Rna. 2007;13:2108–2115. doi: 10.1261/rna.734407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing Regulator SC35 Is Essential for Genomic Stability and Cell Proliferation during Mammalian Organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07.This work revealed that in vivo depletion of SR proteins causes widespread double-strand DNA breaks in MEFs, which in turns triggers the activation of the ATM pathway and cell cycle arrest. This work demonstrates elucidates a molecular pathway that ties the functional requirement of SR proteins to cell proliferation and regulation of cell cycle progression.
- 72.Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao RP, Ross J, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. Embo J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]