Abstract
The configuration of the active site of E2 ligases, central enzymes in the ubiquitin/ubiquitin-like protein (Ub/Ubl) conjugation systems, has long puzzled researchers. Taking advantage of the wealth of newly available structures and sequences of E2s from diverse organisms, we performed a large-scale comparative analysis of these proteins. As a result we identified a previously under-appreciated diversity in the active site of these enzymes, in particular, the spatial location of the catalytic cysteine and a constellation of associated conserved residues that potentially contributes to catalysis. We observed structural innovations of differing magnitudes occurring in various families across the E2 fold that might correlate in part with differences in target interaction. A key finding was the independent emergence on multiple occasions of a polar residue, often a histidine, in the vicinity of the catalytic cysteine in different E2 families. We propose that these convergently emerging polar residues have a common function, such as in the stabilization of oxyanion holes during Ub/Ubl transfer and spatial localization of the Ub/Ubl tails in the active site. Thus, the E2 ligases represent a rare example in enzyme evolution of high structural diversity of the active site and position of the catalytic residue despite all characterized members catalyzing a similar reaction. Our studies also indicated certain evolutionarily conserved features in all active members of the E2 superfamily that stabilize the unusual flap-like structure in the fold. These features are likely to form a critical mechanical element of the fold required for catalysis. The results presented here could aid in new experiments to understand E2 catalysis.
INTRODUCTION
The E2 enzyme is the central component in the transfer of ubiquitin (Ub) and ubiquitin-like (Ubl) proteins to targets in diverse conjugation pathways. During the ubiquitination of proteins, an Ub/Ubl is first adenylated by the E1 enzyme, and then transferred to a conserved cysteine residue on the E1 protein through a thioester linkage formed with the terminal carboxyl group of the Ub/Ubl. A subsequent trans-thiolation reaction transfers the Ub/Ubl to a conserved cysteine residue on the E2 enzyme. In the final step of the cascade the Ub/Ubl is transferred from the E2 to the ε-amino group of lysine residues on protein substrates or the amine group of phosphatidylethanolamine in the case of the autophagy pathway (Ciechanover et al., 2000; Ichimura et al., 2000). This final step is usually mediated by E3 ligases that may function in one of two distinct ways: The HECT-like E3 ligases transfer the Ub/Ubl from E2 to an internal cysteine through a further trans-thiolation step before transferring it to the target, whereas the RING (U-Box) and A20 finger-type E3 ligases appear to only bridge the E2 enzyme and the substrate with the target lysine residues (Aravind et al., 2003; Ardley and Robinson, 2005; Wertz et al., 2004). The role of the E2 enzymes in the Ub/Ubl conjugation cascade, and the importance of the conserved cysteine shared by all catalytically active E2 enzymes in the trans-thiolation reaction are well-established (Berleth and Pickart, 1996; Dye and Schulman, 2007; Hershko et al., 1983; Pickart, 2001). The transfer of Ub/Ubl from E1 to E2 involves a nucleophilic attack by the conserved E2 cysteine on the carbonyl group of the Ub/Ubl-E1 thioester linkage (Pickart, 2001). Experiments have suggested that this reaction is primarily catalyzed by residues on the E1 protein in addition to the E2 cysteine (Wu et al., 2003).
Likewise, the conserved cysteine in the E2 protein is the only residue shown to be essential for the transfer of Ub/Ubl from E2 to the HECT ligases (Dye and Schulman, 2007; Wu et al., 2003). However, all other terminal Ub/Ubl transfer reactions involving E2 enzymes suggest a critical role for the target lysine. Studies on the ubiquitinating and sumoylating E2 enzymes, Ubc13 and Ubc9, indicate that the target lysine makes a nucleophilic attack on the carbonyl group of the Ub/Ubl-E2 thioester linkage upon deprotonation. Together, these studies also implicate three other residues (namely an asparagine, tyrosine and aspartate) in E2 function, which are proposed to mediate localization of the target lysine, and act to lower the effective pKa of the active site to allow the lysine’s deprotonation (Capili and Lima, 2007; Wu et al., 2003; Yunus and Lima, 2006). The asparagine is also proposed to stabilize an oxyanion formed in the reaction intermediate during the nucleophilic attack (Wu et al., 2003). These additional residues implicated in E2 action appear to be only required in the transfer of Ub/Ubl from E2 to the ε amino groups of target lysine residues in conjugations mediated by non-covalently interacting E3, like the RING superfamily (Knipscheer and Sixma, 2006; Melchior et al., 2003; Wu et al., 2003; Yunus and Lima, 2006; Zheng et al., 2000). However, the generality of this proposal for E2 function, and other aspects of the biochemical mechanism by which Ub transfer is effected, including the role of the E2s in target specificity, is poorly understood.
All eukaryotes possess several E2s ranging from about 8–11 in certain apicomplexans and Giardia to a little over 50 in certain multicellular plants and animals (Supplementary material). Most of them, barring those in the autophagy pathway (Apg3 and Apg10), are relatively close to each other in sequence. The entire diversity of eukaryotic E2s has not been systematically explored through the application of evolutionary principles to identify key functional residues. The extreme divergence of Apg3 and Apg10 families also did not allow for a direct comparison with the classical E2s (Ichimura et al., 2000; Yamada et al., 2007). More recently, we discovered the first prokaryotic homologs of the E2 enzymes proposed to function analogous to their eukaryotic counterparts in predicted Ub-conjugation like systems in bacteria (Iyer et al., 2006). A comparison of their conservation profiles with the eukaryotic E2s revealed that while the E2 catalytic cysteine is strictly conserved, they include representatives which are highly divergent in sequence. Importantly, comparisons of the prokaryotic and eukaryotic sequences suggested that the residues besides the catalytic cysteine proposed to be involved in E2 function might not be generally conserved. Concomitantly, several new crystal structures of E2 enzymes have been solved, including that of Apg3, which was also shown to lack the other residues involved in E2 function, despite conserving the catalytic cysteine (Arai et al., 2006; Mizushima et al., 2007; Yamada et al., 2007). Structures of catalytically inactive versions of the E2 fold such as the UEV1 proteins and the RWD domain have also been solved, providing structural information regarding the non-enzymatic adaptations of the fold (Eddins et al., 2006; Hau et al., 2006; Nameki et al., 2004).
These pieces of new data presented us with an opportunity to use the evolutionary information from bacterial E2 homologs in conjunction with the new crystal structures to explore previously unknown aspects of E2 catalysis and biochemical function. To this end, we performed a comprehensive comparative analysis of the E2 superfamily, identifying all structural and sequence variations. As a result we were able to uncover an unusually high and previously under-appreciated diversity in the sequence and structural features of the active sites of the E2 superfamily. In particular, it appears that the E2s might represent an infrequent example in enzyme evolution, where despite retention of the key catalytic residue and general biochemical function there is high active site diversity. We present evidence that the structural features identified here might provide new insights into the action of these enzymes, especially in terms of residues other than the catalytic cysteine.
RESULTS AND DISCUSSION
Comprehensive identification and classification of members of the E2 superfamily
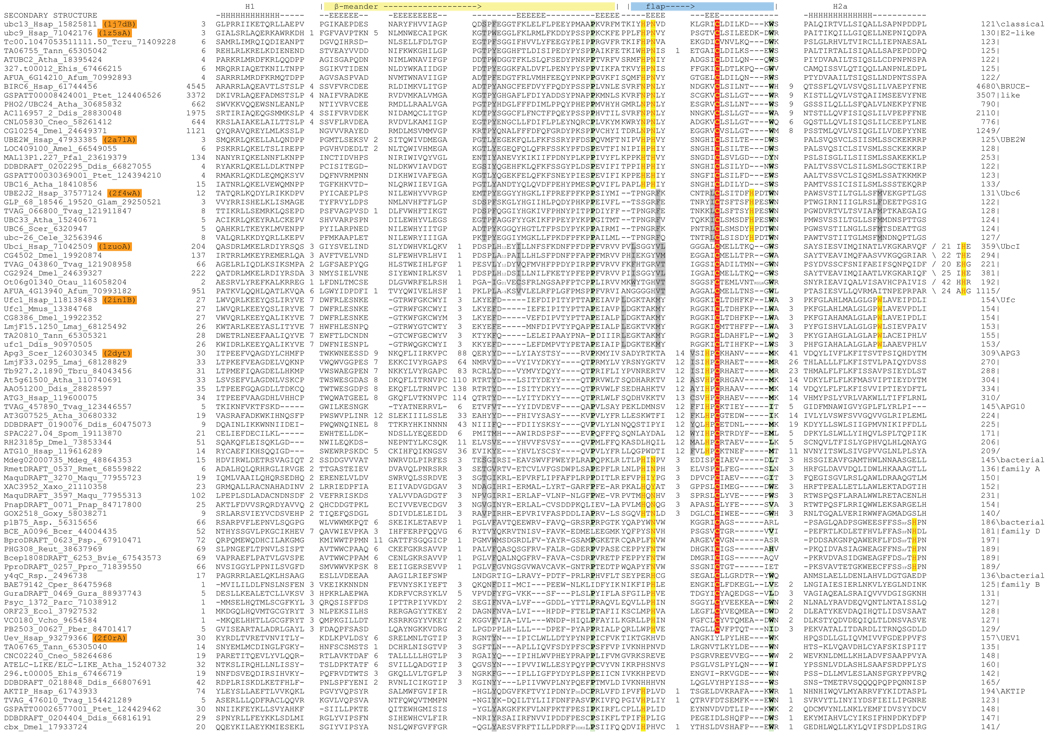
A multi-pronged search strategy utilizing sequence and structure similarity searches was used to comprehensively identify all members of the E2 fold. For structural searches we began with a non-redundant set of representatives of the E2 fold from the SCOP database ((Murzin et al., 1995); http://scop.mrc-lmb.cam.ac.uk/scop/) and iteratively searched the current version of the PDB database using the DALILITE program (Holm and Sander, 1995). In addition to previously described E2 proteins, these searches retrieved several E2 homologs (Z score > 9), such as UbcI (PDB: 2f4w) and Ufc1 (PDB: 2in1), crystallized as part of structural genomics initiatives. Sequences of all distinct structural representatives were then used as queries for sequence profile searches with the PSI-BLAST program (see Material and Methods for search strategy and threshold for significance). Alignments of all true positives were used for further HMM searches with the HMMER program (Eddy, 1998). The complete set of true positive sequences detected in these searches were clustered and classified into families based on uniquely shared sequence conservation patterns and distinct structural features (supplementary material). A structural multiple alignment of all available E2 fold structures was constructed using the MUSTANG program (Konagurthu et al., 2006). A sequence alignment including representatives of all detected families of the E2 domain was constructed using the MUSCLE and KALIGN programs and adjusted based on the above-mentioned structural alignment (Fig. 1; See Materials and Methods for details). As a result of our analysis, we identified 16 distinct families of E2 domains, 11 eukaryotic and 5 prokaryotic. Of these three families in eukaryotes and one family in prokaryotes were either predicted or have been experimentally shown to be inactive, and lacked the catalytic cysteine.
Fig. 1. Multiple alignment of selected E2 domains, representing active site variants.
Sequences are shown with their gene name, species abbreviations, and genbank ID (gi) numbers separated by underscores given to the left of the alignment. PDB ids of sequences with solved X-ray crystal structures are highlighted in orange. The top line of the alignment labels the major conserved features of the fold and the line below the alignment shows the consensus secondary structure; E and H denote β-strand and α-helix, respectively. Family names are listed to the right of the alignment. The bacterial families A–E correspond to representatives of rows 6A–6E in Table 2 of (Iyer et al., 2006). Columns in the alignment are colored based on the functional role of the residue: the catalytic cysteine is colored in yellow and shaded in red, flap histidine and asparagine residues and other residues predicted to perform analogous roles are colored in red and shaded in yellow, the proline and hydrophobic residue forming a conserved stabilizing contact are shaded in green, and the conserved aromatic residue and other residues predicted to form a chain of interacting residues are colored in gray. Species abbreviations are as follows: Afum, Aspergillus fumigatus; Amel, Apis mellifera; Asp., Azoarcus sp.; Atha, Arabidopsis thaliana; Bcer, Bacillus cereus; Bvie, Burkholderia vietnamiensis; Cele, Caenorhabditis elegans; Cneo, Cryptococcus neoformans; Cper, Clostridium perfringens; Ddis, Dictyostelium discoideum; Dmel, Drosophila melanogaster; Ecol, Escherichia coli; Ehis, Entamoeba histolytica; Glam, Giardia lamblia; Goxy, Gluconobacter oxydans; Gura, Geobacter uraniumreducens; Hsap, Homo sapiens; Lmaj, Leishmania major; Maqu, Marinobacter aquaeolei; Mdeg, Microbulbifer degradans; Mmus, Mus musculus; Otau, Ostreococcus tauri; Parc, Psychrobacter arcticus; Pber, Parvularcula bermudensis; Pfal, Plasmodium falciparum; Pnap, Polaromonas naphthalenivorans; Ppro, Pelobacter propionicus; Psp., Polaromonas sp.; Ptet, Paramecium tetraurelia; Reut, Ralstonia eutropha; Rmet, Ralstonia metallidurans; Rsp., Rhizobium sp.; Scer, Saccharomyces cerevisiae; Spom, Schizosaccharomyces pombe; Tann, Theileria annulata; Tbru, Trypanosoma brucei; Tcru, Trypanosoma cruzi; Tvag, Trichomonas vaginalis; Vcho, Vibrio cholerae; Xaxo, Xanthomonas axonopodis.
Among the eukaryotic E2 families, Ub ligase activity has been demonstrated in representatives across their entire spectrum of structural diversity (Fig. 1), ranging from the highly divergent families such as Apg3/Apg10 to the forms like UbcI and UBE2W-like families, which are closer to the classical E2 family (Bartke et al., 2004; Chen et al., 1993; Christensen et al., 2007; Hershko et al., 1983; Ichimura et al., 2000; Komatsu et al., 2004; Melner et al., 2006). Thus, the entire structure and sequence diversity of the eukaryotic members of the superfamily is bracketed by versions with Ub ligase activity, suggesting that this activity is a reasonable assumption for all representatives of the group that bear a conserved cysteine. The bacterial versions lack experimental evidence regarding catalytic activity. However, they show contextual associations in the form of domain fusions and conserved gene neighborhoods with homologs of other core components of the ubiquitin signaling system, such as E1, Ub and the JAB metalloprotease. Further, none of the contextual connections of the prokaryotic E2 proteins reveal associations suggesting participation in alternate enzymatic pathways. Hence it is highly probable that the prokaryotic E2s show similar enzymatic activity as their eukaryotic counterparts (Iyer et al., 2006).
A preliminary comparison of the residue conservation across the 12 catalytically active families revealed that other than the cysteine none of the other residues implicated in Ub/Ubl transfer based on the studies on UBC9 and UBC13 (Wu et al., 2003; Yunus and Lima, 2006) are widely conserved. The asparagine residue implicated in positioning the lysine and stabilizing the oxyanion (Wu et al., 2003) was conserved in 4 families (Fig. 1). Of the other residues implicated in providing the environment to reduce the pKa in the active site, the tyrosine residue was only detected in 3 eukaryotic families (although it is often a hydrophobic in others), whereas the aspartate residue was only detected in certain members of the classical E2 family (Fig. 1, Table 1). This immediately suggested that the proposed additional residues of the E2 active site are unlikely to be a general feature of E2 catalysis, and that there is a striking diversity in the active sites of the E2 superfamily. In order to understand better the role of this diversity in E2 function we used the above multiple sequence alignment, and the corresponding structural alignment for an exploration of the evolution of the active site of the superfamily. To do this we first identified the structure and sequence features conserved across the fold and used this as a baseline to systematically explore the innovations occurring in each distinctive family.
Table 1.
Distinguishing sequence and structural features of E2 families
| Family Namea |
Reference PDB/ Genbank index no. |
Catalytic cysteineb |
Flap asparagine |
Flap histidine |
Other polar residues |
Chain of conserved interacting residuesc |
|---|---|---|---|---|---|---|
| Classical E2-UBC13 | 1J7D-chain B | + | + | + | − | S45-F47-H77-N79-C87 |
| Classical E2-UBC9 | 1Z5S- chain A | + | + | + | Y87, D127 | T51-W53-H83-N85-C93 |
| Bruce-like E2 | 61744456 | + | + | N | − | 4591T-4593Y-4628N-4630N-C4638d |
| UBE2W | 2A7L | + | H | + | − | T47-Y49-H81-H83-C91 |
| Ubc6 | 2F4W | + | − | − | H101 | T54-Y56-M122-F87- C94-H101 |
| UbcI | 1ZUO | + | − | − | H362, E306 | L252-I268-L292-V297-L298-C304-H362 |
| Ufc | 2IN1 | + | K (backbone) | − | W145 | F80-L102-M109-C116-W145 |
| APG3 | 2DYT | + | − | − | H232 | Y169-M201-V229-T215-T213-A214-H232-C234 |
| APG10 | 119616289 | + | − | − | H164 | Y102-I128-F161-T149-T147-I146-H164-C166d |
| UEV1 | 2F0R | − | − | [H]e | − | − |
| AKTIP | 61743933 | − | H | − | − | − |
| Bacterial family A | 48864353 | + | + | + | − | S62-I64-H97-N99-C110d |
| Bacterial family D | 56315656 | + | + | − | H185 | − |
Only relevant families discussed in the text are shown. Specific members of a family that are being described are shown next to a hyphen
In columns, 3,4 and 5, + denotes presence and − denotes absence. Alternate conserved residues found in the same position are also shown
The chain is represented as a sequence of interacting amino acid residues, separated by hyphens. The number next to the residue is position of the residue in the reference sequence.
Chain of interacting residues predicted based on sequence conservation and comparison to known structures
Square brackets are enclose residues that are widely but not absolutely conserved
Core structure, active site configuration and surface features conserved in the E2 fold
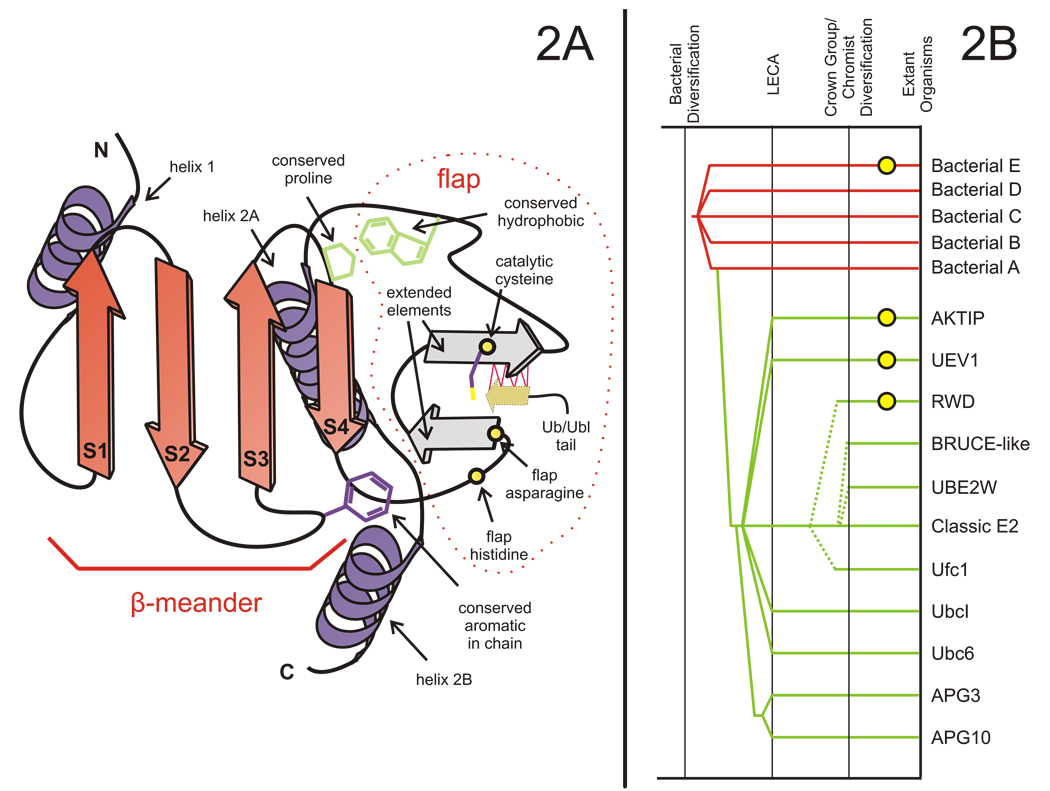
As previously described the core of the E2 fold is based on a four-stranded β-meander (henceforth labeled strands 1–4) that resembles a single “blade” of the β-propeller structures (Aravind et al., 2006). Beyond this basic core, the E2 fold displays certain distinctive structural accretions that distinguish it from all other comparable 4-stranded β-meanders. These include: 1) a C-terminal flap-like structure that contains within it two small elements in extended conformation forming a β-hairpin (hereinafter termed the “flap”; Fig. 2). 2) Flanking the meander and the flap, there is a helix at the N-terminus (helix 1) and 1–2 helices at the C-terminus (henceforth helix 2a and 2b, Fig. 2). The catalytic cysteine is usually located at the C-terminus of the flap, and in every available structure is in an exposed location (Capili and Lima, 2007; Mizushima et al., 2007; Moraes et al., 2001; Tong et al., 1997; Yamada et al., 2007). This flap appears to be an important feature for E2 catalysis—comparisons of structures with and without the Ub/Ubl substrate (Capili and Lima, 2007; Reverter and Lima, 2005; Yunus and Lima, 2006) show that the C-terminal extended segment of the flap undergoes a shift to an alternative conformation. It moves away from the N-terminal extended segment and pairs with the extended tail of the Ub/Ubl through hydrogen-bonding (Fig.2, see Supplementary material). This is critical for appropriate positioning the Ub/Ubl tail for attack by the target lysine (Reverter and Lima, 2005; Yunus and Lima, 2006).
Fig. 2. Topology diagram of generalized E2 domain and tree depicting general relationships between E2 domain families.
(A) Cartoon representation showing conserved elements of the E2 fold. The extended elements of the flap are colored in gray. Labels for all elements in the figure correspond to those in the text. Locations of the flap histidine and flap asparagine in the classic E2 active site are shown as circles colored in yellow, while other conserved residues mentioned in the text are depicted as line drawings. The C-terminal Ub/Ubl tail that forms hydrogen-bonding interactions with the C-terminal extended element is depicted as a brown colored arrow. The red lines represent hydrogen bonding occurring before attack by the target lysine. (B) Reconstructed evolutionary history of the E2 protein fold. Lineages in the fold are listed to the right, with inferred evolutionary depth traced by solid horizontal lines across the relative temporal epochs representing key evolutionary transition periods shown as vertical lines. Horizontal lines are color-coded according to observed phyletic distribution in a given lineage, red applied to bacteria-specific lineages and green applied to eukaryote-specific lineages. Dashed lines indicate uncertainty regarding the origins of a lineage. Yellow circles placed on a horizontal line indicate a lineage has lost catalytic activity.
The only conserved residue that is seen in a number of families in close proximity to the cysteine is the asparagine that was previously implicated in stabilizing the oxyanion hole ((Wu et al., 2003); hereafter termed “flap asparagine”). In structures with the co-crystallized substrates the terminal residue of the Ub/Ubl (glycine) is sandwiched between this asparagine and the cysteine (Reverter and Lima, 2005). In several families, including the classical E2 of Ub in eukaryotes and certain bacterial families the side-chain of the flap asparagine is flanked, on the side opposite to the catalytic cysteine and substrate Ub/Ubl, by a conserved histidine (termed flap histidine) that is present two residues upstream in the sequence (HxN signature, Fig. 1 and Fig. 2). As mentioned above, the tyrosine or an equivalent hydrophobic residue implicated in forming a part of the active site in studies on UBC9 is only conserved in a small number of families, and the co-crystal structures show it forming a part of the interface with the target peptide (Reverter and Lima, 2005; Yunus and Lima, 2006). Hence, its poor conservation is likely to be a consequence of the differences in target peptide recognized by different E2s, as well and alterations in the architecture of the active site seen in various families of the E2 fold (see below).
Examination of the sequence and structure alignments reveal several residues, besides the catalytic cysteine, that are highly conserved across most members of the fold, including the divergent Apg3/10 and bacterial families (Fig. 1). Amongst these are a proline N-terminal to strand 4 and its hydrophobic interacting partner (usually tryptophan or methionine) located between the flap and helix-2a (Fig. 1 and Fig. 2). This pair of residues is also conserved in inactive versions indicating a central role for them in stabilizing the E2 fold by tethering the flap to the core sheet of the β-meander (Fig. 1 and Fig. 2). Another well-conserved residue present in both active and inactive versions is a hydrophobic (mostly aromatic) residue in the loop between strand-2 and strand-3 (Fig. 1 and Fig. 2). This residue is part of a “chain” of interacting residues that, in some form, is seen in most members of the fold, and might play an important role both in connecting the flap to the core β-sheet as well as potentially allowing conformational changes during catalysis (see below for details).
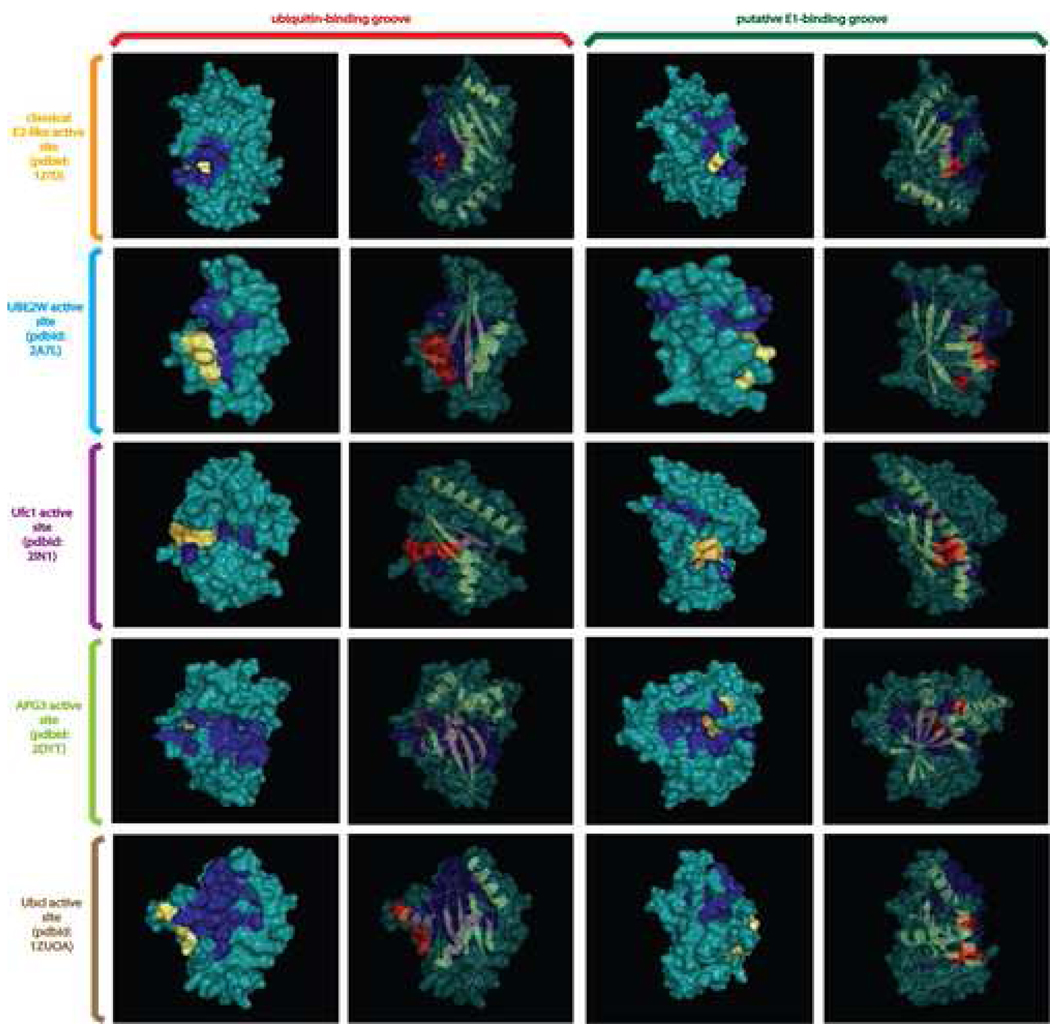
In order to determine if the different versions of the E2 fold might share a general mode of target interaction, we compared their protein surfaces and conservation of surface residues. We observed that all catalytically active versions including the divergent Apg3/Apg10 and the inactive UEV1 families shared the presence of two distinct surface grooves. These grooves are located on either face of the central β-sheet and are predominantly lined by hydrophobic residues that are conserved within individual families (Fig. 3, see supplementary material). Prior structural studies suggest that the groove on the face of the sheet which packs with the helices interacts with the C-terminal tail of Ub/Ubl (Reverter and Lima, 2005). The catalytic cysteine of the Ubc6 and APG3/APG10 families is located towards the center of the groove, while that of the other E2 families is closer to the end of the groove (Fig. 3). However, the invariant association of the catalytic cysteine with the groove suggests that, despite the differences in residues lining it in the various families, its role in binding the Ub/Ubl tail is an ancestral feature of the E2 superfamily. The role of the groove on the exposed face is less clear, but certain structures suggest that it might accommodate a part of the E1 during trans-thiolation (Wang et al., 2007). Its conservation across diverse E2 families suggests that an E1 interaction via this groove is a potentially ancestral feature of the E2 fold.
Fig. 3. Surface diagrams displaying conserved binding grooves.
Molecular surfaces are shown for the two conserved binding grooves in selected members of the E2 fold. The different grooves are labeled at the top of the figure, the ubiquitin-binding groove is on the left and the putative E1-binding groove is on the right. Two surface depictions in the same orientation are provided for each groove, the left surface provides an opaque view of the surface with the catalytic cysteine and flap asparagine/histidine residues colored in yellow, with family-specific conserved residues associated with the groove colored blue. The right surface provides cartoon depictions of the secondary structure of the domain colored in yellow framed by the transparent outline of the molecular surface. The catalytic cysteine and flap asparagine/histidine (or equivalent residues) are rendered as ball-and-sticks and colored in red. The surfaces of other conserved residues in a family are colored blue. Family names and PDB codes are given at the left.
Major structural variations in members of the E2 fold
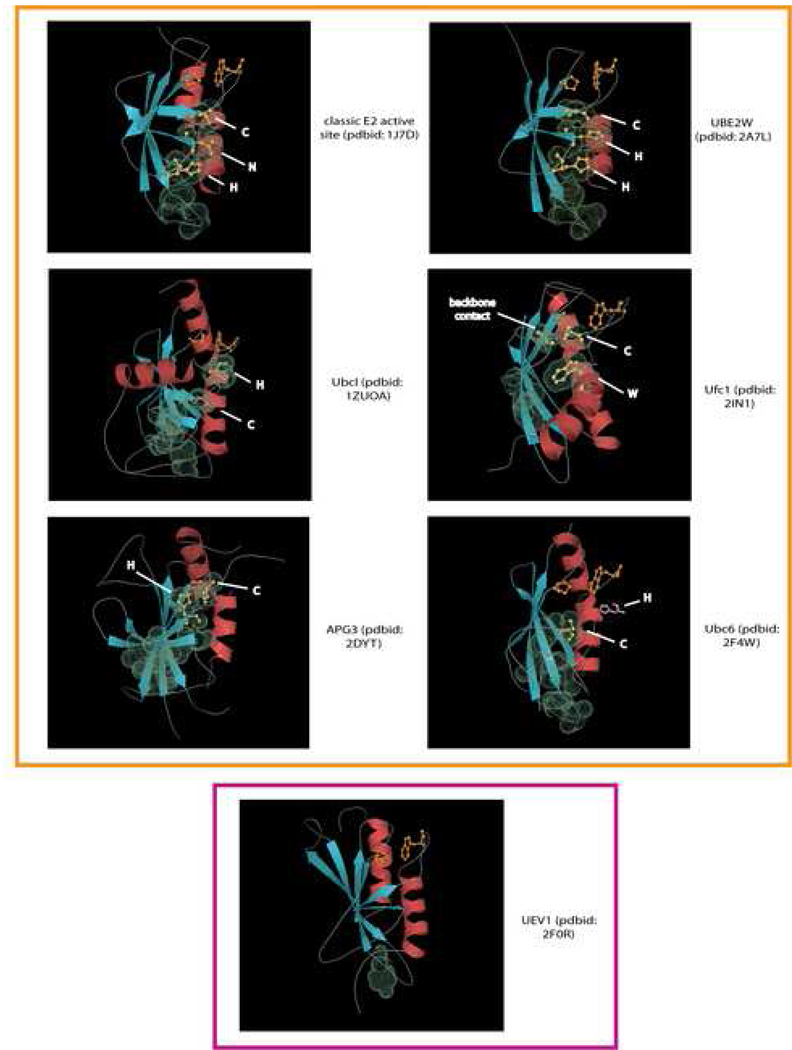
Although the core E2 topology is retained across the fold, and the catalytic cysteine shared by all active members, we observed rather dramatic structural variations in certain members that entirely altered the local architecture of the active site. The most striking of these was seen in the APG3 and APG10 families involved in direct E3-independent transfer of Ubls to protein or lipid substrates in autophagy (Ichimura et al., 2000; Yamada et al., 2007). In these two families there is a unique helix inserted into the N-terminal portion of the flap region, and the flap is also much longer than all other E2 families. Furthermore, instead of forming the usual β-hairpin that points away from the core β-meander, its extended regions stack as additional strands to the core β-meander (Fig. 2 and Fig. 4, PDB: 2dyt). Consequently, the catalytic cysteine is no longer in an equivalent location as that of the other E2 fold members, and is displaced further downstream. This unusual displacement precludes interactions with the position equivalent to the above-mentioned asparagine. Not surprisingly, members of the Apg3/Apg10 families lack the flap histidine and asparagine, instead possessing a strictly conserved histidine residue two residues N-terminal to the catalytic cysteine (Table 1). Strikingly, the spatial proximity of the side-chain of this conserved histidine vis-à-vis the catalytic cysteine is highly reminiscent of that of the flap asparagine vis-à-vis the cysteine of the classical E2 (Fig. 2 and Fig. 4, PDB: 2dyt). In addition, members of the Apg3/Apg10 families also contain large inserts between the 2nd and 3rd strands of the core β-meander.
Fig. 4. Cartoon structural diagrams showing E2 domain active site variants.
Cartoon representations of X-ray structures are shown with PDB codes and family names provided at the left. Strands are colored in blue and helices in red. The catalytic cysteine and the flap asparagine/histidine or predicted equivalent residues are labeled and rendered as ball-and-sticks colored in yellow. The molecular surfaces of these residues, as well as residues predicted to form the conserved interacting chain are rendered using their van der Waal radii. The histidine residue predicted to be important in the Ubc6 family, but not observed in the crystal structure due to structure disorder is placed in its approximate predicted spatial location and colored pink. The proline and hydrophobic resides forming a conserved hydrophobic contact are rendered as ball-and-sticks and are colored orange. The active versions of the domain are boxed in orange and the inactive UEV1 family boxed in purple.
A comparable structural variation is furnished by the Ubc6 family (PDB: 2F4W), which is unified by a distinctive three-residue insert in the flap after the catalytic cysteine (Fig. 1 and Fig. 2). The flap β-hairpin is reorganized due to hydrogen-bonding interaction between the first extended region and a neomorphic extended region between helices 2a and 2b (a loop in most other E2 families). This frees the second extended region of the β-hairpin that instead appears to hydrogen bond with the core β-meander with a consequent spatial displacement of the catalytic cysteine from its usual structural neighborhood. Consistent with this feature, the Ubc6 family lacks the flap histidine and asparagine, and the equivalent residues are not spatially positioned to interact with the active cysteine (Fig. 2 and Fig. 4). However, we uncovered a histidine, seven residues downstream of the catalytic cysteine in the family-specific insert, which is universally conserved in the Ubc6 family (Fig. 1 and Table 1). Although this region is not resolved in the available crystal structure, the location of the histidine suggests that it is likely to come close to the catalytic cysteine and flank it similar to the flap asparagine in the classical E2s, or the above-mentioned histidine of the Agp3/Apg10 families (Fig. 2 and Fig. 4). The third major structural variation is the UbcI family (PDB: 1ZUO) in which a constellation of interconnected modifications are observed. The N-terminal region of the flap upstream to the β-hairpin forms an extended region that packs with a neomorphic extended region between helices 2a and 2b (usually a loop in most other E2s). This not only appears to have displaced the flap hairpin with the catalytic cysteine “upwards” (Fig. 2 and Fig. 4), but also appears to have displaced helix 2b to pack against the flap hairpin. Keeping with these rearrangements, this family lacks the equivalent of the flap histidine and asparagine, instead bearing a family-specific conserved histidine at the C-terminus of helix 2b (Fig. 1 and Fig. 2, Table 1). This histidine occupies a spatial position equivalent to the flap asparagine or its structural analogs in the other variants discussed above (Fig. 4). An acidic residue (usually glutamate), universally conserved in this family and located two residues C-terminal to the catalytic cysteine, appears to tether the histidine in this position by forming a salt bridge.
The catalytically inactive RWD domain is the most structurally divergent version of the E2 fold. Here the flap appears to have assumed an entirely helical conformation. The retention of the hydrophobic residue, normally present between the flap and helix 2a, and its interacting partner, the conserved proline N-terminal to strand 4, suggests that the family-specific helix is derived from the flap and the structural transition may have been favored by the loss of catalytic activity.
Subtle sequence variations in E2 families affecting active site properties
Beyond the major structural variations in the fold and the corresponding changes in active site architecture, we detected several sequence variations specific to particular families that could alter the configuration of the active site. The most striking variation is seen in the Ufc1 family in which both the flap asparagine and histidine are absent (Table 1). In place of the former there is a family-specific conserved lysine whose backbone carbonyl is juxtaposed close to the catalytic cysteine rather than the side-chain. Additionally, this family contains a universally conserved tryptophan in the middle of helix 2A whose indole nitrogen also comes in close vicinity to the catalytic cysteine and backbone carbonyl of the lysine (Fig. 1 and Fig. 4). These together appear to form a constellation highly reminiscent of the flap asparagine side-chain of the classical E2s. Subtler changes are seen in the Bruce-like E2 family, which lack independent E3 ligases, and in which the E2 domain is always part of a larger multi-domain polypeptide ((Bartke et al., 2004) and AMB, LMI and LA unpublished observations). Here the flap histidine is substituted by an asparagine. Likewise, throughout the UBE2W family, the flap asparagine residue is replaced by a conserved histidine (Fig. 1 and Fig. 4), but its side chain occupies a position similar to the flap asparagine of the classical forms.
Four of the bacterial families (A–D) are predicted to be catalytically competent forms due to conservation of the active cysteine, with members of the family E likely to be inactive versions analogous to the eukaryotic UEV1 family. Though members of the bacterial families exhibit a high level of divergence amongst themselves, most members of families A, B and D conserve either an asparagine or a histidine in position equivalent to the flap asparagine of the classical E2s (Fig. 1). Family C currently has too few members to derive a meaningful conservation profile. Family A displays the same active site configuration as the classical E2 family with both a histidine and asparagine residue in the flap (i.e. with a HXN signature). All members of family B have a conserved histidine as the cognate of the flap asparagine and lack any other conserved polar residue that is likely to be in close proximity to the catalytic cysteine. Members of family D, which have a conventional flap asparagine, lack the flap histidine. Interestingly, they possess a highly conserved histidine between helices 2a and 2b. Based on alignments with known E2 structures this residue is predicted to occupy a spatial position equivalent to the flap histidine of the conventional forms ((Iyer et al., 2006); Fig. 1).
Members of the UEV1 family occasionally preserve the flap histidine and rarely the flap asparagine (Fig. 1, Table 1). The structure of the UEV1 protein suggests that the general architecture of the active site is retained despite lacking the active cysteine, and has been shown to function as a subunit of a heterodimeric E2 complex involved in polyubiquitination (Eddins et al., 2006). All members of the other inactive eukaryotic E2 family, namely the enigmatic AKTIP family, retain the flap histidine (Fig. 1, Table 1). Hence, these too might preserve the general active site architecture, and potentially function as an Ub-binding protein comparable to like UEV1.
Identification of a chain of interacting residues connecting the active site to the core β-meander
To understand better the role of the highly conserved hydrophobic (mostly aromatic) residue in the loop between strand-2 and strand-3 (Fig. 1) we systematically examined its interacting partners and their conservation. We found that this hydrophobic residue always interacted with the flap histidine when it was present. As the flap histidine (or cognate asparagine in the BRUCE family) in turn interacts with the flap asparagine, and that residue in turn with the catalytic cysteine, these residues formed an interacting chain connecting the active site to the core β-meander (Fig. 4). In four families (Fig. 1 and Fig. 4, Table 1), an alcoholic residue, also from the loop between strand-2 and strand-3 and two positions upstream of the highly conserved hydrophobic residue, is part of this interacting chain. The hydrophobic residue between strand-2 and strand-3 is also nearly absolutely conserved in families that have undergone major structural alterations, or those lacking the flap histidine and/or asparagine. An examination of these families suggested that even in these cases a comparable chain is constituted by the conserved hydrophobic residue along with other interacting residues conserved in family-specific manner (Table 1). For example, in the UbcI-like proteins the interaction chain has been elongated to include more residues, concomitant with the displacement of the active cysteine due to emergence of an additional extended region in the flap. The additional residues include conserved hydrophobic residues from strand-3, the neomorphic extended region in the flap and a couple from the first extended segment of the flap hairpin (Table 1).
In the Ubc6p family this chain, rather than directly leading to the cysteine, connects with the loop between helix-2a and helix-2b. This loop in turn furnishes a residue, which is usually hydrophobic, that appears to directly interact with the catalytic cysteine. Furthermore, the afore-stated residue might also convergently provide a hydrophobic environment for the cysteine as suggested for the tyrosine in experiments on Ubc9 (Fig. 1 and Fig. 4, Table 1). In line with the major structural modification seen in the APG3/APG10 families the interaction chain is longer and more diffuse than any other representatives of the fold. Here the conserved hydrophobic residue between strand -2 and strand-3 initiates the chain via a contact with a highly conserved methionine from the family-specific helix in the flap. This in turn interacts with a conserved hydrophobic residue and the Apg3/Apg10-specific ThTb motif (h: hydrophobic and b: big residues) respectively from the two extended regions of the exaggerated β-hairpin seen in these families. The ThTb motif then provides the link to the HPC motif at the active site (Fig. 1 and Fig. 4, Table 1).
Implications of the conserved and diversified features for catalytic mechanisms of the E2 fold
From the earliest studies on E2 mechanism it has been remarked that the “catalytic landscape of the E2 active site is remarkably sterile”(Johnson, 2004; Pickart, 2001; Wu et al., 2003). While subsequent studies have uncovered potentially important details regarding the E2 mechanism (Wu et al., 2003; Yunus and Lima, 2006), there has been no systematic attempt to unite these structure and mutagenesis studies with evolutionary conservation across the E2 fold and within individual families. The above observations emerging from our analysis based on a natural classification of the E2 domains and the wealth of new structures provides several new insights into E2 catalytic function:
In terms of mechanical properties of the E2 domain the presence of a conserved surface groove suggests a similar mode of Ub/Ubl-interaction throughout the fold (Fig. 3). Furthermore, conservation of the flap structure despite the variety in active site architecture suggests an important generalized role for it in E2 action (Fig.2 and Fig. 4). The opening and pairing of the second extended segment of the flap hairpin with the Ub/Ubl-tail suggests that this part of the flap is central to interaction with the Ub/Ubl tail and specific recognition of the cognate Ub/Ubl.
The conserved proline-hydrophobic side chain interaction at the “top” of the flap and the chain of interacting residues at the “bottom” of the flap may also be interpreted as contributing to the mechanical features of the E2-Ub/Ubl interaction (Fig. 2). These tether the C-terminal end of the flap and the first extended segment to the rest of the β-meander scaffold. Hence, they probably stabilize the rest of the structure while allowing the second segment of the hairpin to alternatively form hydrogen-bonds with the first segment (when there is no Ub/Ubl) or with the Ub/Ubl-tail (upon and after trans-thiolation) (See supplementary material; (Reverter and Lima, 2005; Yunus and Lima, 2006)).
In terms of reaction mechanism, the cysteine being the only universally conserved residue in all active forms is consistent with experimental evidence that it is the only required residue for most reactions involving E2s, except for the E3-dependent final transfer of Ub/Ubl to a target lysine. Our comparisons also show that the E2 fold represents an exceptional case in terms of enzyme evolution: While always being on the second extended segment of the flap and lying in an exposed position in a groove, the active cysteine occupies a remarkable diversity of spatial positions in different families with respect to the core scaffold of the fold (Fig. 4). This is no more strikingly illustrated than in a comparison of the Apg3 structure with that of a member of the classical E2 family (Fig. 4)(Yamada et al., 2007). This observation is again consistent with there being few constraints on the specific environment of the cysteine. It also means that there are no specific conserved residues across the E2 fold for creating the right environment for lowering the pKa of the attacking target lysine (Yunus and Lima, 2006). Instead this is likely to be achieved in a generic fashion due to the interface formed by target-E2 packing.
-
While the flap asparagine implicated in oxyanion hole stabilization (Wu et al., 2003; Yunus and Lima, 2006) is conserved only in certain families, we found that it is always substituted by a histidine from the cognate position, or other polar side chains from entirely different locations (Fig. 1 and Fig. 4, Table 1). In most instances, these alternative polar side chains are histidine, and in one case (Ufc1) appears to be replaced by a combination of a backbone carbonyl and the nitrogen of the tryptophan indole group. Thus, a polar group appears to have convergently emerged in an equivalent spatial location in the vicinity of the active cysteine in different families of the fold. The available structures suggest that this polar group is likely to interact with the C-terminus of the Ub/Ubl. Further, this residue being almost always an asparagine or a histidine is reminiscent of the fact that these two residues appear frequently in peptidase catalytic triads to stabilize oxyanion holes (Berg et al., 2002; Johnston et al., 1999; Rawlings and Barrett, 1994). Hence, it is possible that each of these represent independent innovations to stabilize the oxyanion hole and localize the Ub/Ubl tail for attack by lysine. However, in some cases like Apg3/Apg10 the conserved histidine could additionally provide a general base for activating the lysine. In a more general sense, this convergent evolution of a polar group in the vicinity of the cysteine in the E2s is reminiscent of the convergent evolution of the third (the cysteine and histidine being usually invariant and in their ancestral positions) residue of the catalytic triad in peptidases of the papain-like fold (Balakirev et al., 2003; Iyer et al., 2004).
The flap histidine which interacts with the flap asparagine and is part of the above-described interacting residue chain is fairly widespread. Moreover, in the bacterial family D, where it is absent it seems to have a functional analog in the form of another histidine from the loop between helices 2a and 2b. This suggests a potentially important role for this histidine with respect to the flap asparagine, by interacting with the oxo-group of the asparagine side-chain as suggested by some substrate-complexed structures (e.g. PDB: 1z5s; (Reverter and Lima, 2005)). Mutagenesis of the histidine in Ubc13 affected protein solubility/stability (Wu et al., 2003); hence, it is not clear if the experiments that failed to uncover a role for it in catalysis have objectively evaluated its function. Given the nature of the histidine imidazole ring it is possible that it acts as a switch that transiently regulates the asparagine, by alternately assuming acidic and basic states. As the substitutes for the flap asparagine are usually histidines, it is possible that in these cases the histidine directly shows such switch-like properties. The only multi-domain E2s are the BRUCE-like family, and these have an asparagine in place of the flap histidine (Fig. 1, Table 1). Here, the other globular domains fused to the E2 domain might help in providing an E3 like function. This raises the question as to whether the role of the flap histidine is mainly important in the context of E2s functioning with independent E3s of the non-HECT variety.
A corollary to the observation that there is a whole range of family-specific innovations in the region around the active cysteine is that these could play a role in target specificity. Extrapolating on the basis of the target-E2 complexes (e.g. PDB: 1z5s; (Reverter and Lima, 2005)), we propose that the neomorphic helices or strands in the flap region (for example in Agp3/Apg10, UbcI and Ubc6) and the region between helix2a and helix 2b and additional C-terminal helices would provide family-specific interfaces for target specificity. In other cases, apparently drastic changes in substrate specificity might also be achieved via relatively subtle changes. While Apg3 and Apg10 differ in respectively operating on a protein and lipid target they are closer to each other than any other E2 family. Autophagy or related processes are apparently universal to eukaryotes, but some eukaryotes lack Apg10, while possessing Apg3 (Supplementary material). This suggests that Apg3 might perform both roles in these organisms without any major changes in its active site. Nevertheless, we identified a few positions that might contribute towards the differences in target specificities of the two enzymes. Through systematic comparison of relative entropy the cognate alignment positions of the two families we identified one strongly differentially conserved position two residues upstream of the catalytic cysteine (Fig. 1). It is respectively a highly conserved threonine in Apg10 and a histidine in Apg3. The structure of Apg3 (Yamada et al., 2007) suggests that it is likely to line the groove containing the catalytic cysteine and control access to it. Taken together, these observations suggest that the residue at this position might play a role in target specificity of the two families.
Evolutionary history of E2 enzymes
The availability of complete genome sequences of early-branching eukaryotes like Trichomonas and Giardia, and ancient branches like the kinetoplastid-heterolobosan clade allows an objective reconstruction of the E2 superfamily in eukaryotes. Phyletic patterns of different E2 families suggest that the Last Eukaryotic Common Ancestor (LECA) possessed members of the large classical E2 family, along with other catalytically active families such as Ubc6p, APG10, APG3 and UbcI, and catalytically inactive versions such as UEV1 and AKTIP. Thus, the major structural variants in the E2 family seen in extant eukaryotes were already present in LECA (Anantharaman et al., 2007) and adapted to diverse niches such as degradation of proteins in the cytosol and endoplasmic reticulum (classical E2 and Ubc6p families), autophagy that involves transfer of Ubls to proteins (APG10) and lipids (APG3), and transfer of polyubiquitin to proteins (UEV1 family) (Fig. 2). The Ufc1 and the inactive RWD families are apparently absent in the basal eukaryotic branches and appear to have emerged slightly later, prior to the diversification of the heterolobosea-kinetoplastid clade (Fig. 2, Supplementary material). The UBE2W and Bruce-like E2 families emerged in the common ancestor of the crown-group eukaryotes (plants, slime molds, fungi and animals) and the chromalveolate clades. These diversifications involved 4 independent innovations of polar residues in the vicinity of the catalytic cysteine and loss of catalytic activity on at least 3 different occasions. This evolutionary reconstruction of the E2 families is consistent with the corresponding reconstructions of the cognate Ub/Ubl (Burroughs et al., 2007) and E1 families (MB, LMI, LA manuscript in preparation) that interact with the different E2 proteins. Thus, there appears to have been a major concomitant radiation of these three components resulting in multiple parallel Ub-conjugation systems prior to LECA itself, which were recruited to diverse physiological processes. In contrast, the E3-ligases show major lineage-specific expansions in different eukaryotic lineages (Lespinet et al., 2002) suggesting that they were the main players in recruiting the ancient core of the conjugation systems to various new targets.
In our earlier studies we had reported that the bacterial members of the E2 superfamily are encoded by predicted mobile operons that also code for E1-like, Ub-like and Jab-peptidase components (Iyer et al., 2006). Based on this we proposed that they constitute potential modification systems that were likely to have been precursors of the eukaryotic E2s. Of the 5 distinct bacterial E2 families, family A most closely resembles the large classical E2 family of eukaryotes with a flap histidine and flap asparagine. We suggest this is likely to represent the family from which the eukaryotic ancestral version emerged, most probably during the primary endosymbiotic event that spawned the eukaryotes.
GENERAL CONCLUSIONS
The catalytic mechanism of the E2 enzymes has been an enduring problem of interest in the enzymology of ubiquitin conjugation. Taking advantage of the wealth of recent structural data, sensitive sequence comparisons and an objective natural classification of the E2 superfamily we identify many under-appreciated features of the E2 fold. We present hypotheses for the roles of these features in mechanical and biochemical aspects of E2 catalysis. Crucially, we identify the major unifying themes of E2 catalysis ranging from the bacterial versions, to the divergent Agp3/Apg10 and the classical eukaryotic E2 proteins and their variants. Our studies show that the E2 superfamily has undergone considerable sequence and structure diversification in the region of its active site since the time of its emergence in the bacteria. Our analysis suggests that the sequence and structural diversity in the E2 active sites was not driven to a major extent by changes in the mode of E1 or Ub/Ubl binding. Instead, it is appears that the E2 diversification reflects the presence of relatively few constraints on the catalytic cysteine, and adaptations that facilitate target peptide (i.e. context of the target lysine) or lipid choice. A common theme is presence of a polar residue (mostly histidine or asparagine) that has convergently emerged in various families in the vicinity of the catalytic cysteine. This is likely to both position the Ub-tail as well as possibly stabilize the oxyanion hole during transfer of Ub/Ubl to lysine. We hope that the analysis presented here spurs further experiments in the form of site directed mutagenesis and structure determination to elucidate E2 catalysis.
MATERIAL AND METHODS
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, MD) was searched with the BLASTP program (Altschul et al., 1997). Profile searches were conducted using the PSI-BLAST program (Altschul et al., 1997) with either single sequences or multiple alignments as queries, with a profile inclusion expectation (e) value threshold of 0.01; searched were iterated until convergence. Hidden Markov models (HMMs) built from alignments using the hmmbuild program were also employed in searches carried out using the hmmsearch program from the HMMER package (Eddy, 1998). For queries and searches containing compositionally biased segments, the statistical correction option built into the BLAST program was used (Schaffer et al., 2001). Multiple alignments were constructed using the MUSCLE (Edgar, 2004) and/or KALIGN programs (Lassmann and Sonnhammer, 2005), followed by manual adjustment based on PSI-BLAST hsps and information provided by solved three-dimensional structures. Protein structures were visualized using the Swiss-PDB viewer (Guex and Peitsch, 1997) and cartoons were constructed with the PyMOL program (http://www.pymol.org). Protein secondary structure predictions were made with the JPRED program, which computes a consensus predicted structure using the information from the conservation profile, a PSI-BLAST position-specific score matrix and a hidden Markov model (Cuff and Barton, 2000).
Structure similarity searches were conducted using the standalone version of the DALI program with the query structures scanned against a local current version of the PDB that has all chains as separate entries (Holm and Sander, 1995). The structural hits for each query were collected, even if the DALI Z-score for the match was less than 2.0 and then parsed for topological congruence to the E2 structural template (Fig. 2) using a custom PERL script. To assess topological congruence, coordinates of the matching regions detected by DALI searches using known E2 domains as queries were extracted and analyzed for secondary structure using the DSSP program (Kabsch and Sander, 1983). These secondary structure elements were then represented as a string (corresponding to a row in table 1) along with the polarity of the secondary structure element determined from the DALI match to the query structure. These strings were then matched with the equivalent secondary structure pattern strings constructed of bona fide E2 domains. If a complete match was obtained these structures were tagged as congruent, while those which were not were ranked in descending order of elements that did not match. This discrimination of the potential candidates was further confirmed by visual examination of each structure. Structural multiple alignments were carried out using the MUSTANG program (Konagurthu et al., 2006). The interacting residues in various members of the E2 fold were deduced using a PERL scripts from the TASS package (V. Anantharaman, S. Balaji and LA, unpublished). The scripts encode interacting distance cut-off values of 5.0 Å and 3.5 Å between appropriate atoms in 3-D for deducing the hydrophobic and polar interactions respectively. These inferred interactions were confirmed via manually examination using Swiss-PDB viewer (Guex and Peitsch, 1997).
Preliminary clustering of the E2 superfamily was performed using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html) with a range of BLAST bit score density cutoffs (1-0.4) and length ratios for the alignments (0.8-0.6) in order to establish the core of potential families. These were further extended using reciprocal BLAST hits. The phyletic profiles of individual families were determined using scripts of the TASS package that query into the NCBI taxonomy database. Higher order relationship between families was inferred using shared conserved motifs in structure (when available) and sequence. Phylogenetic analysis within families was carried out using a variety of methods including maximum-likelihood, neighbor-joining, and minimum evolution (least squares) methods with the MEGA, PHYLIP and MOLPHY packages (Adachi and Hasegawa, 1992; Felsenstein, 1996; Kumar et al., 2004). All large-scale sequence and structure analysis procedures were carried out with the TASS software package, a successor to the SEALS package (Walker and Koonin, 1997).
Supplementary Material
Comprehensive supplementary material is also available at: http://www.ncbi.nlm.nih.gov/CBBresearch/Lakshmin/E2/
ACKNOWLEDGMENTS
The authors gratefully acknowledge the Intramural research program of the National Library Of Medicine, National Institutes of Health, USA for funding their research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adachi J, Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics Institute of Statistical Mathematics. Tokyo. 1992 [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation (*) Annu Rev Microbiol. 2007;61:453–475. doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- Arai R, Yoshikawa S, Murayama K, Imai Y, Takahashi R, Shirouzu M, Yokoyama S. Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:330–334. doi: 10.1107/S1744309106009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Koonin EV. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle. 2003;2:123–126. doi: 10.4161/cc.2.2.335. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Koonin EV. Comparative genomics and structural biology of the molecular innovations of eukaryotes. Curr Opin Struct Biol. 2006;16:409–419. doi: 10.1016/j.sbi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14:801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. Fifth ed. New York: W.H. Freeman and Co.; 2002. [Google Scholar]
- Berleth ES, Pickart CM. Mechanism of ubiquitin conjugating enzyme E2-230K: catalysis involving a thiol relay? Biochemistry. 1996;35:1664–1671. doi: 10.1021/bi952105y. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007 doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hau DD, Lewis MJ, Saltibus LF, Pastushok L, Xiao W, Spyracopoulos L. Structure and interactions of the ubiquitin-conjugating enzyme variant human Uev1a: implications for enzymatic synthesis of polyubiquitin chains. Biochemistry. 2006;45:9866–9877. doi: 10.1021/bi060631r. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3:1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. Embo J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Sixma TK. Divide and conquer: the E2 active site. Nat Struct Mol Biol. 2006;13:474–476. doi: 10.1038/nsmb0606-474. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. Embo J. 2004;23:1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign--an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Melner MH, Haas AL, Klein JM, Brash AR, Boeglin WE, Nagdas SK, Winfrey VP, Olson GE. Demonstration of ubiquitin thiolester formation of UBE2Q2 (UBCi), a novel ubiquitin-conjugating enzyme with implantation site-specific expression. Biol Reprod. 2006;75:395–406. doi: 10.1095/biolreprod.106.051458. [DOI] [PubMed] [Google Scholar]
- Mizushima T, Tatsumi K, Ozaki Y, Kawakami T, Suzuki A, Ogasahara K, Komatsu M, Kominami E, Tanaka K, Yamane T. Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem Biophys Res Commun. 2007;362:1079–1084. doi: 10.1016/j.bbrc.2007.08.129. [DOI] [PubMed] [Google Scholar]
- Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JN, Ellison MJ. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat Struct Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nameki N, Yoneyama M, Koshiba S, Tochio N, Inoue M, Seki E, Matsuda T, Tomo Y, Harada T, Saito K, Kobayashi N, Yabuki T, Aoki M, Nunokawa E, Matsuda N, Sakagami N, Terada T, Shirouzu M, Yoshida M, Hirota H, Osanai T, Tanaka A, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kinoshita K, Guntert P, Kigawa T, Yokoyama S. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 2004;13:2089–2100. doi: 10.1110/ps.04751804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Families of cysteine peptidases. Methods Enzymol. 1994;244:461–486. doi: 10.1016/0076-6879(94)44034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Hateboer G, Perrakis A, Bernards R, Sixma TK. Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J Biol Chem. 1997;272:21381–21387. doi: 10.1074/jbc.272.34.21381. [DOI] [PubMed] [Google Scholar]
- Walker DR, Koonin EV. SEALS: a system for easy analysis of lots of sequences. Proc Int Conf Intell Syst Mol Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- Wang J, Hu W, Cai S, Lee B, Song J, Chen Y. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol Cell. 2007;27:228–237. doi: 10.1016/j.molcel.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. Embo J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comprehensive supplementary material is also available at: http://www.ncbi.nlm.nih.gov/CBBresearch/Lakshmin/E2/