SUMMARY
Regulation of eukaryotic cell cycle progression requires sequential activation and inactivation of cyclin-dependent kinases. Previous RNA interference (RNAi) experiments in Trypanosoma brucei indicated that cyclin E1, cdc2-related kinase (CRK)1 and CRK2 are involved in regulating G1/S transition, whereas cyclin B2 and CRK3 play a pivotal role in controlling the G2/M checkpoint. To search for potential interactions between the other cyclins and CRKs that may not have been revealed by the RNAi assays, we used the yeast two-hybrid system and an in vitro GST pulldown assay and observed interactions between cyclin E1 and CRK1, CRK2 and CRK3. Cyclins E1-E4 are homologues of yeast Pho80 cyclin. But yeast complementation assays indicated that none of them possesses a Pho80-like function. Analysis of cyclin E1+CRK1 and cyclin E1+CRK2 double knockdowns in the procyclic form of T. brucei indicated that the cells were arrested more extensively in the G1 phase beyond the cumulative effect of individual knockdowns. But BrdU incorporation was significantly impaired only in cyclin E1+CRK1 depleted cells, whereas a higher percentage of cyclin E1+CRK2 knockdown cells assumed a grossly elongated posterior end morphology. A double knockdown of cyclin E1 and CRK3 arrested cells in G2/M much more efficiently than if CRK3 was depleted alone. Taken together, these data suggest multiple functions of cyclin E1. It forms a complex with CRK1 in promoting G1/S phase transition, with CRK2 in controlling the posterior morphogenesis during G1/S transition and with CRK3 in promoting passage across the G2/M checkpoint in the trypanosome.
Keywords: Trypanosoma brucei, cell cycle, RNAi, morphology, cyclin-dependent kinases
Introduction
The eukaryotic cell cycle is tightly regulated to ensure proper replication and segregation of genome into the two daughter cells. The primary and well-characterized regulation of cell cycle events is mediated by the cyclins and their partners, the cyclin-dependent kinases (CDK)1; 2; 3; 4; 5; 6. Since the CDKs require binding of cyclin for activity, they are regulated by the timely synthesis and degradation of cyclins7; 8. In the budding yeast Saccharomyces cerevisiae, a single CDK, Cdc28, regulates all phases of the cell cycle9 by binding to different cyclins10; 11. Thus a single CDK activity fluctuating via cyclin bindings is capable of regulating the progression of yeast cell cycle. In animal cells, the regulation of cell cycle is somewhat more complex with several families of cyclins and CDKs regulating S-phase and mitosis (cyclin A with Cdk2 or Cdk1), mitosis only (cyclin B and Cdk1), G1 progression (cyclin D and Cdk4 or Cdk6) and S-phase progression (cyclin E and Cdk2)12. Some cyclins have the capacity to bind multiple CDKs. For instance, cyclin A can bind both CDK1 and CDK2 to promote mitosis and S-phase progression, respectively13; 14; 15. Thus, the same cyclin has the capacity of regulating different phases of the cell cycle depending on its CDK binding partner.
Trypanosoma brucei is a parasitic protozoan and the causative agent of sleeping sickness in human and nagana in cattle in Sub-Saharan Africa. It is also among the most ancient and evolutionarily divergent eukaryotes with many unique biological features. Perhaps the most interesting feature of T. brucei cells is their single mitochondrion and the arrangement of the mitochondrial DNA into a disc-like structure, the kinetoplast. Equally interesting is the replication of the kinetoplast, which has discrete G1, S, G2 and D phases analogous to the four phases of the nuclear cell cycle16. Both cycles are coordinated with each other, suggesting close communication between the two17, yet both cycles are also autonomous as they can be uncoupled18; 19. Significant progress has been achieved recently in understanding the cell cycle of T. brucei. Four Pho80 homologues (cyclins E1–E4), three mitotic cyclin homologues (cyclins B1–B3) and five cdc-2 related kinases (CRK) 1–4 and 6 were identified in T. brucei from mining the trypanosome genome database and extensively characterized20; 21; 22. More recently, two additional cyclin homologues (CYC10, CYC11) and 6 more CRK (CRK7–12) homologues were identified from the genome database but not further characterized23; 24.
Recent RNA interference (RNAi) experiments have been effectively utilized to elucidate the mechanisms regulating the cell cycle progression in T. brucei. The outcome depicts that cyclin E1 and CRK1 are involved in controlling the G1/S checkpoint transition whereas cyclin B2 and CRK3 play specific pivotal roles in regulating the passage across G2/M checkpoint20; 22; 25; 26; 27. When expression of CRK1 and CRK2 were both knocked down in the procyclic-form T. brucei cells arrested in the G1 cell cycle phase and demonstrated strangely elongated and sometimes branched posterior ends, suggesting a coupling of posterior morphogenesis with G1/S transition in these cells28; 29.
The RNAi study has left several Pho80 and cyclin B homologues and CRKs without an apparent function in regulating the cell cycle progression in T. brucei. In order to verify if they still play a regulatory function, cyclin and CRK homologues were screened by yeast two-hybrid assays and in vitro pull down experiments for potential interactions. The four Pho80 homologues were tested for complementation in yeast and found devoid of the function of Pho80. The interactions between cyclin E1 and CRK1, CRK2 and CRK3 were further investigated by knocking them down in pairs via RNAi in T. brucei and screening for phenotypes. The outcome shows multiple functions of cyclin E1 via complexing with different CRKs.
Results
Interactions between cyclins and CRKs
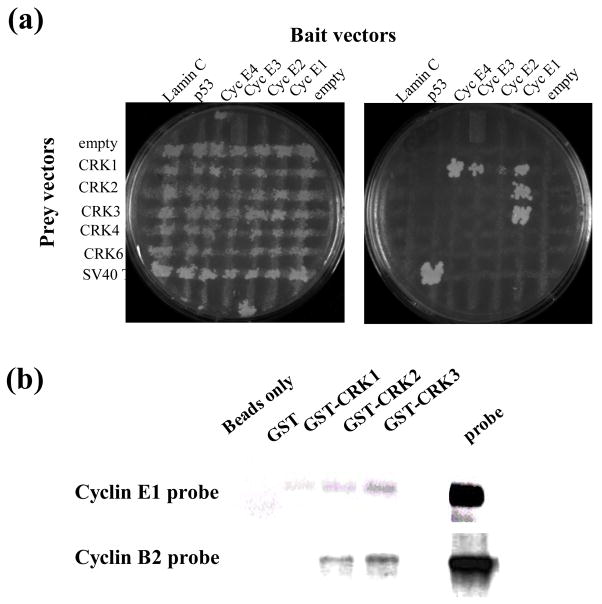
Physical interactions between T. brucei cyclins and CRKs had been previously demonstrated when both cyclin E1 and cyclin B2 were found to interact with CRK3 by yeast two-hybrid and immunoprecipitation20; 27. Our further investigation into the potential roles of CRK1 and CRK2 in cell cycle regulation25; 26; 28 prompted us to screen for possible interactions among the five CRKs (CRK1–CRK4, CRK6) and the seven cyclins (cyclins E1–E4, cyclins B1–B3) using yeast two-hybrid assays. The remaining newly discovered genes CYC10–11 and CRK7–12 do not appear to influence cell growth in any way, except for CRK9, which appears to function, however, by mechanisms other than directly regulating cell cycle progression (Gourguechon and Wang, manuscript in preparation). The complete ORFs for cyclins E1–E4 and B1–B3 and five CRKs (CRK1–4, 6) were cloned individually into the yeast two hybrid bait vector pGBKT7 and prey vector pGADT7 and transformed into the α and a mating type yeast, respectively. Mating between the two yeast types generated diploid yeast carrying both plasmids capable of growing on SD-LEU-TRP plates. (Figure 1(a), left panel). Interaction between the bait and prey proteins conferred to yeast the ability to grow on SD-ADE-HIS-LEU-TRP plates (Figure 1(a), right panel). A cross assay based on yeast mating allowed for simultaneous screening of interactions between cyclins and CRKs (Figure 1(a)) and enabled us to confirm the previously indicated interaction between cyclin E1 and CRK327, which is actually a little surprising, because cyclin E1 is involved in controlling G1/S transition22 whereas CRK3 is known to regulate the G2/M passage25. But cyclin E1 also interacted with CRK1 and CRK2 (Figure 1A) as would have been predicted from the previous RNAi data22; 25. No interaction was detected between cyclin E1 and CRK4, CRK6 or the unrelated protein lamin C or p53 (Figure 1(a)). The anticipated interaction between cyclin B2 and CRK320; 22; 25, which was shown previously in a two hybrid screen20, was, however, not demonstrated in our current assay with reasons remaining unclear to us.
Fig. 1. Novel cyclin-CRK interactions .
A, yeast two hybrid. Yeast AH109 were transformed with the indicated CRK in the pGADT7 vector (streaked horizontally) then mated to Y187 yeast containing the indicated cyclin cloned in pGBKT7 (streaked vertically) by replica plating on YPDA plates. After 24 hours mating, the cells were replica plated separately onto SD-LEU-TRP (left) and SD-ADE-HIS-LEU-TRP plates (right) and grown for three days. SD-LEU-TRP plates select for cells containing both bait and prey plasmids; SD-ADE-HIS-LEU-TRP plates select for cells that with bait and prey plasmids containing interacting pairs. B, in vitro interactions. Purified GST-CRK proteins (as indicated) immobilized on glutathione beads were incubated with each separate radiolabelled cyclin for two hours at 4 °C and washed extensively. Beads were then boiled in sample buffer and samples resolved by SDS PAGE followed by autoradiography. Beads only, no GST fusion protein included; probe, a sample of radiolabelled cyclin was loaded on the gel as control.
Verification of the postulated protein-protein interactions by an in vitro pull-down assay
The protein-protein interactions indicated in the yeast two-hybrid assay were re-examined in an in vitro pull-down assay. The GST-CRK fusion proteins expressed and purified from E. coli and immobilized on glutathione-Sepharose 4B beads were incubated with the radiolabeled cyclins synthesized in vitro. An analysis of the radiolabeled proteins pulled down by the beads indicated bindings between cyclin E1 and CRK1, 2 and 3 (Figure 1(b)), which are in good agreement with the results from the yeast two-hybrid assay. However, bindings between cyclin B2 and CRK3 and between cyclin B2 and CRK2, which were not demonstrated in the two-hybrid assay, showed up in the pull-down assay with strong signals (Figure 1(b)).
T. brucei Pho80 homologues do not complement S. cerevisiae Pho80
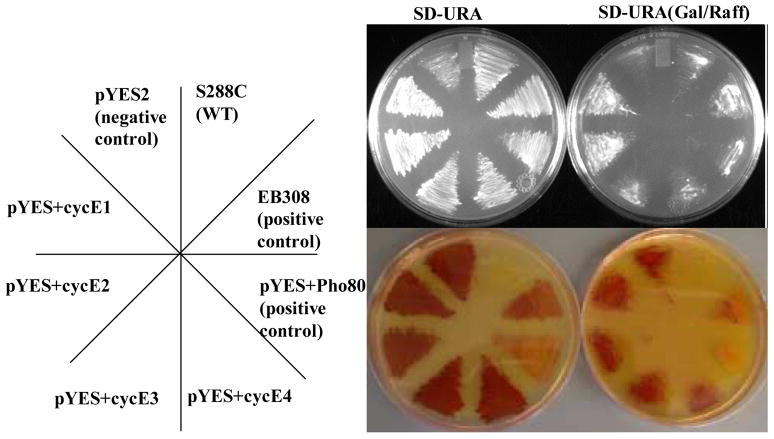
The observation that cyclin E1 interacted with CRKs 1, 2 and 3, whereas cyclins E2, E3 and E4 appeared not to bind any of the five CRKs in the yeast two hybrid assays (data not shown) raised the possibility that some of the cyclin E’s may not play a role in cell cycle regulation but possess a Pho80-like function instead. To see if these T. brucei Pho80 homologues can functionally replace Pho80 in yeast, we used a yeast complementation assay (see Experimental procedures). Briefly, by assaying the activity of Pho5, a downstream, secreted phosphatase, the activity of Pho80 can be rapidly assayed. The results show that cells transformed with an empty vector pYES2 were viable and turned red in the presence of the Pho5 substrates, showing an absence of Pho80 expression (Figure 2). In contrast, wild-type cells that retain the chromosomal copy of Pho80 remained white. Some Pho80 expression was detected in the absence of galactose for the pYES-Pho80 plasmid, as indicated by the less intense red color, possibly due to a leaky expression. However, upon addition of galactose, the red color was completely lost, showing a much enhanced expression of Pho80. Cells transformed with pYES-cyclins E1, E2, E3 and E4 all showed a strong red color after galactose induction, indicating the absence of Pho80-like function. Since the full-length cDNAs encoding yeast Pho80 and cyclins E1–E4 were all cloned into the same plasmid and tested under identical experimental conditions, their levels of expression in the transfected yeast cells were also most likely the same. Thus, the complementing action demonstrated by the yeast Pho80 but not by the others indicates that cyclins E1–E4 do not possess Pho80 function despite the structural homology. While cyclin E1 is clearly involved in controlling cell cycle progression, the function of cyclins E2, E3 and E4 remain unclear for the time being.
Figure 2. Complementation of Pho80 by T. brucei cyclins.
Wild-type yeast (S288C) and a yeast strain deleted for Pho80 (EY1251, courtesy of E. K. O’Shea) and transformed with plasmids (as indicated on the leftmost panel) were streaked as indicated on SD-URA plates containing glucose (left) or galactose (right) as sole carbon source. pYES vectors carry either a cyclin or Pho80 under the control of the galactose inducible promoter GAL1. EB308 is a plasmid containing Pho80 under its native promoter and serves as an additional positive control. After 24 hours growth cells were assayed for Pho5 phosphatase activity; this serves as a reporter assay for Pho80 activity. Plates were overlaid with molten agar containing the chromogenic substrates α-NP and FBSB41 and the color allowed to develop for 15 min. A red color indicates lack of Pho80 activity in these cells; normal white color indicates normal Pho80 activity. The top image shows growth of yeast cells; the bottom image is the photograph after 15 min showing conversion of the Pho5 substrate to a red product.
Combinatorial RNAi of cyclin E1 with CRK1 and CRK2
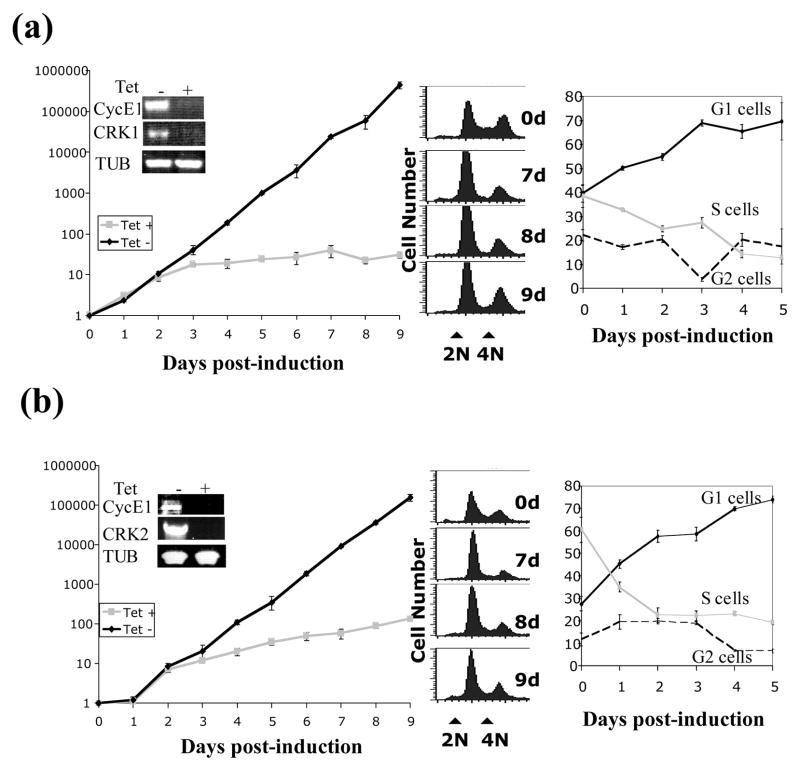
In view of the demonstrated physical interactions between cyclin E1 and CRK1 or CRK2, we carried out pairwise knockdowns of cyclin E1+CRK1 and cyclin E1+CRK2 with RNAi for potential additive and synergistic effects when comparing with those from the individual knockdowns performed in parallel. In both cases, inhibition of expression of both genes, shown by semi-quantitative RT-PCR (the insets from Figures 3(a) and 3(b), leftmost panels), was accompanied by significant growth inhibition that became apparent after 2 days. In the case of cyclin E1+CRK1 depletion, cells reached a density of 106 (Figure 3(a), left panel, Tet+ curve), compared to 1010 cells/ml in the un-induced control (Figure 3(a), left panel, Tet− curve) after 9 days. This extent of inhibited cell growth is much greater than that observed from knocking down either cyclin E1 (supplementary Figure 1(a), left panel) or CRK1 RNAi (supplementary Figure 1(b), left panel) alone where the cell densities reached 5×107 or 1×108 cells/ml after 9 days. When cyclin E1 and CRK2 were knocked down, there was a similar profile of growth inhibition reaching a density of 5×106 cells/ml after 9 days (Figure 3(b)). Again, it represented a more severe growth inhibition when compared with either cyclin E1 or CRK2 single knockdowns, which resulted in either reduced cell density (5×107 cells/ml after 9 days, supplementary Figure 1(a), left panel) or no inhibition at all (supplementary Figure 2(b) left panel, respectively). There is thus a synergistic inhibitory effect on cell growth when both cyclin E1 and CRK1 or cyclin E1 and CRK2 are depleted.
Figure 3. Growth analysis of CyclinE1/CRK1 and CyclinE1/CRK2 RNAi cells.
Cells with cyclin E1+CRK1 (A) or cyclin E1+CRK2 (B) knocked down by RNAi were grown for 5 days. Each day cell number was determined using a hemocytometer (left panels). RNA was extracted from cells after 5 days and the targeted genes were analyzed by RT-PCR to determine RNAi efficiency (inset). Cells were also stained with propidium iodide and analyzed by fluorescence-activated cell sorting (FACS). Histogram plots of frequency versus DNA content are shown in the two left panels (center panels). Cells were assigned G1, S or G2/M phase status using the ModFitLT software and their frequency plotted versus time (right panels).
We also stained the cyclin E1+CRK1 deficient cells with propidium iodide (PI) and examined the changes in cell cycle distribution over time by flow cytometry. Cell numbers were plotted versus DNA contents and the resulting histogram was analyzed using the ModFit software to determine the proportion of cells in the G1, S or G2/M phase. The number of cells in the G1 phase rose from 40 to 69% of the population accompanied by a concomitant decrease in the S phase cells from 40% to 12%, with no apparent change in the G2/M phase cells (Figure 3(a), right panel). When compared with the results from individual knockdowns, cyclin E1 depletion led to an increase of G1 cells from 40 to 63%, a decrease of S-phase cells from 40 to 22% and little change in the numbers of G2/M cells (supplementary Figure 1(a), right panel) while CRK1 deficiency alone resulted in a 20% increase of G1 cells accompanied with a corresponding 20% decrease in S-phase cells, whereas the percentage of G2/M cells remained relatively unchanged (supplementary Figure 1(b), right panel). Thus, the combined knockdowns have improved the extent of G1 arrest from knocking down CRK1 alone but have only modestly exceeded what was achieved from knocking down only cyclin E1.
Flow cytometry analysis of the cyclin E1+CRK2 depleted cells showed a 28 to 62% increase in G1 cells, accompanied by a corresponding decrease in S-phase cells from 60 to 25%, while the number of G2/M cells remained relatively unchanged (Figure 3(b), middle and right panels). Since CRK2 deficiency alone causes little shift in the distribution of cells (supplementary Figure 2(b), right panel), and the shift of cell distribution in cyclin E1+CRK2 deficient cells resembles that of cyclin E1 depleted cells, the G1 arrest caused by the double knockdown can be attributed to the missing cyclin E1, thus suggesting relatively little role for CRK2 in regulating the G1/S passage.
Morphological and BrdU incorporation analysis of the double knockdown cells
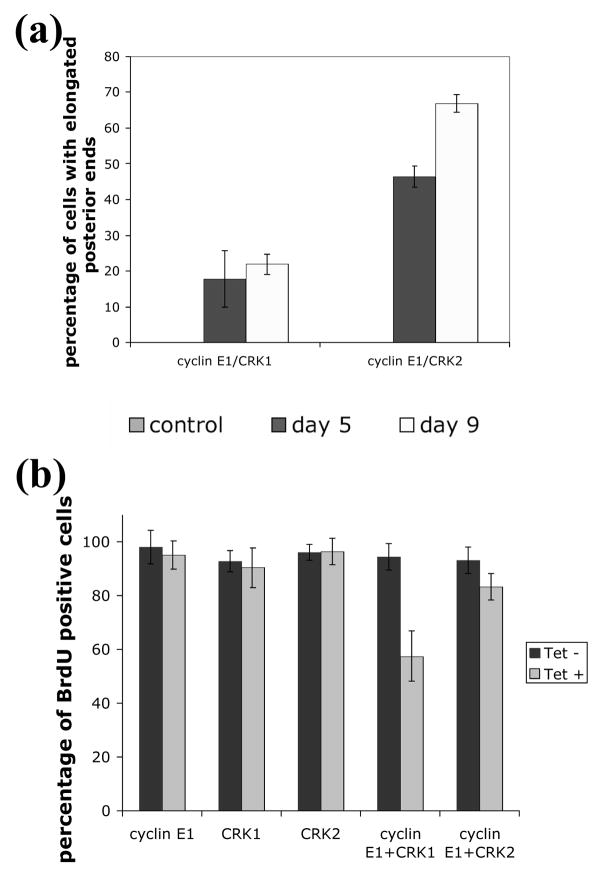
Arresting T. brucei procyclic-form cells in G1 by depleting either cyclin E1 alone22 or CRK1+CRK228 is known to lead to a strange phenotype where the cell’s posterior end is grossly elongated. This phenotype was apparently linked to the control cyclin E1 or CRK1 exerted on the G1/S phase transition28. Having demonstrated an interaction between cyclin E1 and both CRK1 and CRK2, we were interested to see if the cyclin+CRK double knockdowns resulted also in cells with elongated posterior ends. Cells were fixed and assessed by visual microscopy for their morphology. A notable difference between the cyclin E1+CRK1 and cyclin E1+CRK2 depleted cells is that whereas the former gave rise to cells with elongated posterior ends up to about 20% of the population, the latter gave rise to over 40% of cells showing an elongated posterior end after 5 days of RNAi (Figure 4(a)). These elongated cells became even more abundant to constitute 63% of the population after 9 days of cyclin E1+CRK2 RNAi induction (Figure 4(a)). It is noteworthy that RNAi of cyclin E1 alone causes about 20% of the cells to adopt elongated posterior end morphology after 5 days and 35% after 9 days. Neither CRK1 nor CRK2 single knockdowns resulted in appreciable (less than 3%) numbers of elongated cells (Figure 4(a)). It seems likely that the elongated cells observed in the cyclin E1+CRK1 RNAi experiment are solely attributed to the depletion of cyclin E1. CRK1 apparently does not participate per se in morphological maintenance, whereas CRK2 plays a major role in this aspect in a cyclin E1-dependent manner.
Figure 4. Morphology and BrdU incorporation in cyclin E1/CRK1 and cyclin E1/CRK2 depleted cells.
Cells depleted for cyclinE1+CRK1, cyclin E1+CRK2, cyclin E1, CRK1 or CRK2 were cultured in the presence of BrdU and stained for BrdU incorporation after 5 days. Cells were also counterstained with DAPI to visualize the nucleus and kinetoplast. Tet- cells, in which RNAi is not induced, were used as negative control. A, Cells inhibited for single genes or a combination of genes (as indicated) were examined and scored for abnormal elongated posterior morphology. B, Cells were counted and scored for BrdU incorporation, morphology and karyotype and plotted for frequency.
To further verify that the cyclin E1+CRK1 complex is mainly responsible for promoting passage from G1 to S-phase and that the cyclin E1+CRK2 complex regulates primarily cellular morphogenesis, BrdU incorporation experiments on the double knockdown cell lines were carried out to monitor nuclear DNA synthesis. Cells depleted of CRK1, CRK2 or cyclin E1 alone were shown to have little detectable defect in BrdU incorporation, since over 90% cells could be stained with BrdU (Figure 4(b)). But cells depleted of both cyclin E1 and CRK1 showed only 50% BrdU positive cells among those of normal morphology (Figure 4(b)) and only 20% BrdU positive cells among those with elongated posterior ends (data not shown). This reduction in BrdU incorporation does not vary among the 1N1K (cell with one nucleus and one kinetoplast), 1N2K and 2N2K cells, indicating a similarly repressed S-phase among them with an un-identified mechanism (data not shown). It is worth noticing that in no case was BrdU incorporation completely blocked by RNAi. This could be attributed to; (1) the incomplete protein depletion by RNAi; (2) RNAi depleted only a part of the large protein complex controlling G1/S transition. Thus, the experimental outcome represented most likely a slowed rather than a totally blocked passage across the G1/S boundary. This slow-down could be more vividly demonstrated by the enrichment of G1 cells in flow cytometry than an inhibited BrdU incorporation, which which is a less quantitative assay.
For the cyclin E1+CRK2 deficient cells, 83% of the population were BrdU positive despite the fact that over 40% of these cells were elongated (Figure 4(a) and 4(b)); thus BrdU incorporation and cell morphology are separately controlled. We thus conclude that the cyclin E1+CRK1 complex controls entry into S-phase, hence its depletion causes a defect in BrdU incorporation, whereas the cyclin E1+CRK2 complex controls the cell morphology during G1/S transition.
A double knockdown of cyclin E1 and CRK3 arrests cells at the G2/M boundary
The interaction detected between cyclin E1 and CRK3 raises the question whether a cyclin E1+CRK3 double knockdown will arrest the cells in G1, G2/M or both phases. We selected a procyclic-form T. brucei stable cell line with an integrated construct carrying RNAi fragments for both cyclin E1 and CRK3. An efficient reduction of cyclin E1 and CRK3 mRNAs, evidenced by semi-quantitative RT-PCR (Figure 5(a), inset), was achieved within 48 hours of RNAi induction. This double knockdown was accompanied by a rapid and essentially complete inhibition of cell growth 24 hr after the induction of RNAi (Figure 5(a)). Compared to the single RNAi experiments for cyclin E1, where cells reached 5×107 (supplementary Figure 1(a), left panel), or CRK3, where cells reached 108 cells/ml (supplementary Figure 2(a), left panel) after 9 days, the cyclin E1+CRK3 double knockdown causes apparently a much more rapid and severe growth inhibition, suggesting a synergistic inhibitory effect.
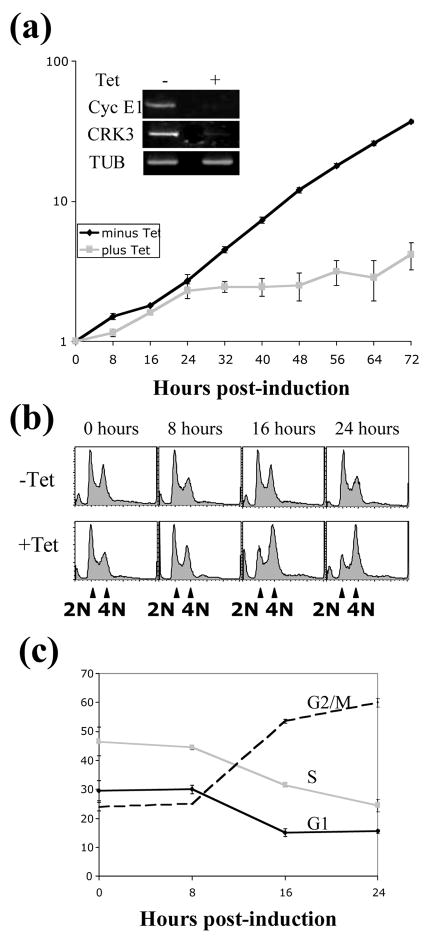
Figure 5. Growth analysis of CyclinE1/CRK3 depleted cells.
Top, cells carrying the integrated RNAi construct for both cyclin E1 and CRK3 were grown for three days. Cell number was determined using a hemocytometer every 8 hours. RNA was extracted from cells after 2 days and the targeted genes were analyzed by RT-PCR to determine RNAi efficiency (inset). Bottom, cells with cyclin E1 and CRK3 knocked down by RNAi were harvested and fixed over a three day period, stained with propidium iodide and analyzed by fluorescence-activated cell sorting (FACS). Bottom right, histogram plots of frequency versus DNA content. Control cells (−Tet) were not induced for RNAi. Bottom left, cells were assigned G1, S or G2/M phase status using the ModFitLT software and their proportion of total cell number plotted versus time.
To identify the specific stage(s) of cell cycle block from the double knockdowns, time samples of the RNAi experiment were examined by flow cytometry. The results showed that cells began to be significantly enriched in the G2/M phase after 16 hours, and reached maximum G2/M enrichment after 24 hours (Figure 5(b)). ModFit analysis of the FACScan data indicated that cells in G2/M phase increased from 21% to 60% within 24 hr, accompanied by a decrease in G1- and S-phase from 30% to 10% and 45% to 20%, respectively (Figure 5(c)). This outcome is completely different from that of depleting cyclin E1 alone, which led to a significant enrichment of G1 cells and depletion of S-phase and G2/M cells (supplementary Figure 1(a), right panel). For the CRK3 depleted cells, the G1 cells were decreased from 40 to 20%, whereas the G2/M cells were increased from 20 to 50% and the S-phase cells decreased from 40 to 30% after 5 days of RNAi (supplementary Figure 2(a), right panel). A synergistic effect in arresting cells at the G2/M is thus apparent from the combined knockdown of cyclin E1 and CRK3. The extraordinarily fast rate of this G2/M arrest is only comparable with that from knocking down the mitotic cyclin B222.
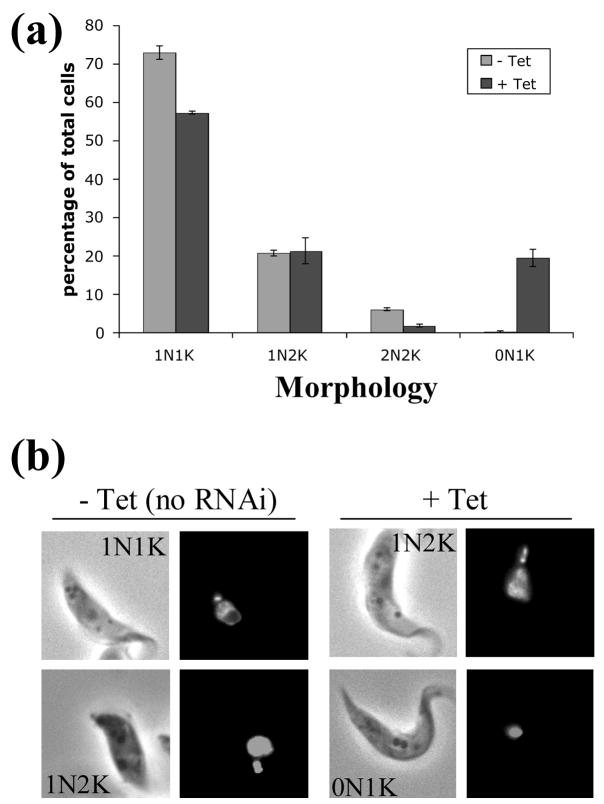
Depletion of cyclin E1 and CRK3 resulted in substantial emergence of anucleated cells (zoids)
Cells depleted of cyclin E1 and CRK3 were stained with 4′,6-diaminido-2-phenylindole (DAPI), examined by fluorescence microscopy, and the percentages of cells containing different numbers of nuclei and kinetoplasts were tabulated. The results demonstrated a decrease among the 1N1K cells from 74% down to 56% of the population (Figure 6(a)). The remaining 1N1K cells appeared to have an enlarged and elongated nucleus (Figure 6(b)), indicating the state of an arrested mitosis. No cells with elongated posterior ends, which were generated from knocking down cyclin E1 alone22, were detectable. Numerous zoids emerged from virtual nonexistence to 20% of the population after the double knockdowns (Figures 6(a) and (b)). This is a typical phenomenon whenever G2/M transition is blocked in the procyclic form of T. brucei22; 25. It provides yet another supporting evidence that the cyclin E1/CRK3 complex plays a major role in regulating the passage of procyclic-form cells across the G2/M checkpoint.
Figure 6. Karyotype analysis of the cyclin E1/CRK3 RNAi cell line.
A, cells were categorized into 1N1K, 1N2K, 2N2K, and 0N1K as described previously and the frequency of each category plotted as percentage of the total cell population. B, Cells with cyclin E1 and CRK3 knocked down by RNAi were fixed 24 hours after induction, stained with DAPI and examined by fluorescence microscopy. In Control cells no RNAi induced inhibition was performed.
Discussion
In our present study, we demonstrated that despite the homology observed between the cyclin boxes of yeast Pho80 cyclin and trypanosome cyclins E1–E4, the latter could not complement the function of Pho80 in yeast. This finding suggests that the cyclin E’s are probably not involved in regulating phosphate metabolism in the trypanosome. This conclusion, plus the previous data showing that cyclin E1 could rescue yeast cells arrested at the G1/S boundary27 and that cyclin E1 RNAi inhibited G1/S transition in procyclic-form T. brucei 22, reinforce the concept that cyclin E1 is a cyclin similar to cyclin A in animal cells contributing to the regulation of G1/S transition (in combination with CRK1) and G2/M transition (in combination with CRK3). The functions of cyclins E2, E3 and E4 remain to be further explored.
By performing yeast two hybrid tests and the subsequent pull-down experiments, we made an effort in examining the potential physical interactions between cyclin E1 and the five CRKs 1–4 and 6. The outcome indicated that cyclin E1 interacts with CRK1, CRK2 and CRK3. The interaction between cyclin E1 and CRK1 is supported by evidence from the previous studies when individual knockdowns of cyclin E1 or CRK1 by RNAi both resulted in similar enrichment of G1 cells22; 25. A single knockdown of CRK2 gave no detectable phenotype. But when it was depleted together with CRK1, the cells arrested in the G1 phase exhibited a grossly elongated and branched posterior end28, suggesting a potential function of CRK2 in controlling cellular morphogenesis during G1/S transition. Thus, it is likely that cyclin E1 and CRK2 could also form a complex during G1/S transition.
The outcome from the present study indicated that when cyclin E1 and CRK1 are both knocked down, the cells are enriched in G1 phase to a slightly greater degree than that observed with cyclin E1 RNAi. Depletion of cyclin E1 and CRK1 together led to a detectable reduction in BrdU incorporation into nuclear DNA, whereas depletion of cyclin E1, CRK1 or CRK2 alone had no apparent effect on BrdU incorporation. The percentage of cells with elongated posterior ends in cyclin E1+CRK1 depleted cells is similar to that from knocking down cyclin E1 alone. Depletion of cyclin E1+CRK2 enriched the G1 cells to the same extent as if cyclin E1 alone is depleted with insignificant reduction in BrdU incorporation in both cases. But a considerably higher percentage of the cells (47% after 5 days of RNAi) showed an elongated posterior end compared to cyclin E1 deficient cells (16% after 5 days of RNAi). We thus conclude that the cyclin E1/CRK1 complex is responsible for promoting transition through the G1/S phase while the cyclin E1/CRK2 complex’s primary function is to limit the posterior cytoskeleton growth during this transition. A concurrent G1 arrest is most likely a necessary condition to observe the elongated cell phenotype caused by loss of CRK2 activity.
Assuming that the cyclin E1 level begins to rise at the start of the G1 phase of the cell cycle, CRK2 could have a lower threshold for binding to cyclin E1, so the cyclin E1/CRK2 complex forms during early G1 and acts to repress any further cytoskeletal elongation. CRK1 could require a much higher threshold of cyclin E1 to form an active complex, so it will only become active during the late G1 phase or early S-phase when cyclin E1 levels have risen. In animal cells, the first cyclin/Cdk complex to be activated in G1 phase is cyclin D/Cdk4, which becomes active and removes the inhibition exerted by Retinoblastoma (Rb), p21/Cip, and p27/Kip on the cyclin E/Cdk2 complex30; 31, which is the main regulator of S-phase progression in animal cells11; 31; 32; 33; 34; 35. It is possible that the situation is similar in T. brucei cells with the cyclin E1/CRK2 complex causing downstream activation of the cyclin E1/CRK1 complex to guide the cells through S-phase. Like cyclin D/Cdk4, T. brucei cyclin E1/CRK2 could promote S-phase indirectly by relieving inhibition to T. brucei cyclin E1/CRK1 but cannot promote entry into S-phase directly30. The latter then promotes the passage through the G1/S phase boundary. After passage through the G1/S phase, the level of cyclin E1 drops, leading to reduced CRK1 and CRK2 activities, paving the way for another phase of cytoskeletal morphogenesis needed for basal body and kinetoplast segregation. One caveat of this model, however, is that no homologue of Rb, p21 or p27 has so far been identified in the T. brucei genome database. More effort needs to be channeled into identification of potential protein candidates associated with the cyclin E1/CRK1 and cyclin E1/CRK2 complexes.
A physical combination between cyclin E1 and CRK3, originally unanticipated from the previous RNAi data, turns out to exert a highly specific and powerful control of the G2/M transition. A simultaneous knockdown of cyclin E1 and CRK3 resulted in a swift G2/M arrest within 24 hrs (see Figure 5). There was no sign of inhibited G1/S transition in the cyclin E1+CRK3 double knockdown cells that had been repeatedly observed in cells depleted of cyclin E1, cyclin E1+CRK1 or cyclin E1+CRK2 after 5 days. This time difference may in fact result in accumulating most of the cells in the G2/M phase long before the G1 blockade takes effect due to cyclin E1 deficiency. In fact, all of the phenotypes caused by the double RNAi of cyclin E1 and CRK3 are strikingly similar, indeed almost identical, to those observed with the mitotic cyclin B2 RNAi22. These outcomes suggest two possible models. Firstly, both the cyclin E1/CRK3 and the cyclin B2/CRK3 complexes may play an essential role in promoting the entry into mitosis. Though attractive, this model raises the question of why the depletion of cyclin E1 alone does not lead to any G2/M arrest. It is possible that the primary function of cyclin E1 is to activate CRK1 and CRK2. A single knockdown of cyclin E1 may be insufficient to cause loss of activity of CRK3 because of the presence of cyclin B2. In mammalian cells, it has been reported that the Atype cyclins have dual activities very much like that of cyclin E1 in trypanosome36; 37, though no significant structural homology between the A-type cyclins and the E-type cyclins from T. brucei has been observed22. Alternatively, the cyclin E1/CRK3 complex may act upstream of the cyclin B2/CRK3 complex, possibly by promoting phosphorylation and activation of CRK3. Homology analysis of CRK3 has shown that it is similar to the S. cerevisiae Cdk activating kinase, a kinase responsible for activating CDK28 by phosphorylating Thr-161 in CDK2838; 39; 40. This raises the possibility that the cyclin E1/CRK3 complex may trigger a positive feedback loop at the end of either G1 or S-phase that results in full activation of CRK3 in preparation for entry into mitosis. A depletion of either cyclin E1 or CRK3 alone could be insufficient to deplete the entire cyclin E1/CRK3 complex to stop the positive feed back loop. But depletion of both cyclin E1 and CRK3 could bring the activity of the cyclin E1/CRK3 complex below a critical threshold to inhibit the activation of CRK3. This will in turn lead to a depletion of the cyclin B2/CRK3 complex due to the deficiency in CRK3.
In conclusion, we have obtained data demonstrating direct interactions between cyclin E1 and either CRK1, CRK2 or CRK3. The cyclin E1/CRK1 complex regulates the G1/S phase transition, whilst the cyclin E1/CRK2 complex appears to co-ordinate the cytoskeleton morphogenesis with the nuclear cycle in the procyclic form of T. brucei. The cyclin E1/CRK3 complex plays a pivotal role in regulating the G2/M transition without any involvement in regulating the G1/S transition. Further studies will be necessary to clarify the entire spectrum of cyclins and CRKs involved in regulating cell cycle progression in the procyclic form of T. brucei.
Materials and methods
Complementation of Pho80 function in yeast
To find out if the four T. brucei Pho80 homologs, cyclins E1–E4, can complement the function of Pho80 in S. cerevisiae, an assay originally developed by Dr. Erin O’Shea of UCSF41 was used. Pho80 and the kinase that is activated by it, Pho85, play a key role in the phosphate metabolism in yeast 42; 43 by inhibiting the activity of the transcription factor Pho4, which is required for production of Pho5, an extracellular phosphatase. The assay uses chromogenic substrates 44 which turn red in the presence of Pho5; yeast colonies with no Pho5 activity remain white, whereas colonies expressing Pho5 are colored red. Thus, an expression of Pho80 function gives white colored cell colonies. The T. brucei Pho80 homologs and yeast Pho80 were cloned into the pYES vector that places genes under the control of the galactose-inducible GAL1 promoter. A ΔPho80 yeast strain, EY1251, was transformed with the pYES constructs and streaked onto SD-URA and SD-URA-Galactose/Raffinose plates. After 24 hours of growth at 30° C, the plates were overlaid with molten agar containing the Pho5 chromogenic substrates and incubated at room temperature for 15 min and then photographed.
Plasmid construction and manipulations
Purification and manipulation of plasmid DNA were carried out by standard procedures 45 or using the manufacturer’s specific instructions (Qiagen, Promega). DNA sequencing (Elim Biopharmaceuticals) was used to verify correctness of plasmid constructs. Full length open-reading-frames for the seven cyclins (cyclins E1–E4, B1–B3) and the five CRKs (CRK1–4, 6) were amplified by RT-PCR using gene-specific primers (sequences available upon request). For the yeast two-hybrid screen, full-length genes were cloned into both the bait vector pGBKT7 and the prey vector pGADT7 to generate the GAL4 DNA binding domain fusions and activation domain fusions, respectively. For the production of recombinant GST fusion proteins in the pull down experiments, the five CRK genes were each cloned into the vector pGEX4T3 (Amersham Pharmacia). For RNAi, fragments of each cyclin and CRK gene (250–500 bp in length) previously used for RNAi of individual gene expression22; 25 were inserted in pairs into the pZJM vector. The newly formed recombinant sequences from conjugating the two fragments were checked against the BLAST database to ensure that no homology to any gene other than the RNAi targets existed. Recombinant pZJM vectors were linearized with NotI prior to transfection to allow for homologous recombination to the ribosomal DNA spacer locus in the procyclic-form T. brucei.
Yeast two hybrid
The Matchmaker™ system (BD Biosciences Clontech) was used according to the manufacturer’s instruction. S. cerevisiae was grown at 30°C. Vectors expressing bait-cyclin fusions were transformed into the Y187 strain (mating type a) whereas vectors expressing prey-CRK fusions were transformed into the AH109 strain (mating type α). Recombinant yeast clones were selected on SD-TRP plates for bait fusions and SD-LEU plates for prey fusions. Colonies were then further streaked horizontally on fresh plates and grown for 2 days. Separate SD-TRP and SD-LEU plates were then replica plated perpendicularly to each other onto YPDA plates and grown for 24 hr to allow for yeast mating. These plates were then replica plated onto SD-LEU-TRP plates and SD-ADE-HIS-LEU-TRP plates, grown for three days and photographed. Plating on SD-LEU-TRP selected for the presence of bait and prey plasmid; SD-ADE-HIS-LEU-TRP plates selected for the presence of both plasmids and interaction between the two encoded proteins.
In vitro pull-down assays
CRK proteins were expressed in BL21(DE3) Escherichia coli cells transformed with the pGEX-CRK plasmids. The transformed cells were grown to an OD of 0.6 at 37°C in 1 L of LB supplemented with 100 μg/ml ampicillin, then shifted to 20°C and induced with 0.1 mM isopropyl-D-thiogalactoside (IPTG) overnight. Cells were harvested by centrifugation at 5,000×g, resuspended in lysis buffer (20 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA), lysed by lysozyme treatment (1 mg/ml) and followed by sonication. Cellular debris was removed by centrifugation at 10,000×g for 10 min and the cleared lysate was applied to a glutathione-Sepharose 4B column (Amersham Pharmacia Biotech). Unbound proteins were washed off the column, and the bound protein was eluted with 10 mM reduced glutathione. This procedure typically yielded 1–5 mg of pure (as estimated by SDS PAGE analysis) CRK protein. Purified proteins were then dialyzed against phosphate-buffered-saline (PBS) to remove the glutathione.
Radiolabeled cyclin proteins were produced by in vitro transcription/translation using the TNT rabbit reticulocyte system (Promega) according to the manufacturer’s instruction. 35S-methionine (0.5–2 μCi, Amersham Pharmacia) was included in each reaction to radiolabel the protein to allow for direct detection.
For the in vitro binding reaction, the purified bait GST-CRK proteins were incubated at 400 nM with an equimolar amount of glutathione-Sepharose 4B beads (Pharmacia) for 60 min at 4°C, then incubated with the prey proteins (5–10 nM) for 2 hours and washed three times with a binding buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, 0.2 % Tween-20). The beads were recovered by centrifugation and boiled in sample buffer. The bead-bound proteins were separated by SDS-PAGE, which was followed by autoradiography using a phosphorimager screen
Cell culture, transfection and RNA interference (RNAi)
Procyclic-form T. brucei cell strain 29–13 was maintained in Cunningham’s medium supplemented with 10 % fetal bovine serum (Atlanta Biological) and grown at 26°C. G418 (15 μg/ml) and hygromycin B (50 μg/ml) were also added to the culture medium to maintain the T7 RNA polymerase and tetracycline repressor expressed in the cells.
Transfections were carried out as described previously22. Briefly, 1 × 108 log-phase cells were washed and resuspended in 0.5 ml cytomix buffer46 containing 10 μg of linearized DNA for electroporation. The transfected cells were selected using phleomycin (2.5 μg/ml) and single cells were cloned on soft agar followed by limiting dilution25. RNAi was induced by adding 1 μg/ml tetracycline to the culture medium. Cells were counted with a microscope using a hemocytometer to monitor the effect of RNAi on cell growth.
Semi-quantitative RT-PCR
Total RNA was treated with DNase I to remove contaminating DNA and used to generate the first-strand cDNA by reverse transcription. PCR was performed using the first-strand cDNA (200 ng) and gene-specific primers that were different from the primer pair used in generating the RNAi constructs. The PCR cycling program was set for 28 cycles at 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s followed by a final elongation time at 72 °C for 5 min.
Fluorescence activated cell sorting (FACS)
The experiment was performed by a previously published procedure22; 25. Time samples of T. brucei cells were collected, before and after induction of RNAi, by centrifuging the cells at 1,000×g. The cells were washed twice with phosphate-buffered saline (PBS) and suspended in 100 μl of PBS and mixed sequentially with 200 μl PBS, 10% ethanol, 5% glycerol, followed by 200 μl PBS, 50 % ethanol, 5% glycerol and finally 1 ml PBS, 70 % ethanol, 5% glycerol. The mixture was then incubated at 4°C overnight. Propidium iodide (PI) (20 μg/ml) and DNase-free RNase (5 μg/ml) were added to each cell sample and the DNA content of cells was analyzed by fluorescence activated cell sorting (FACS) using a Facscalibur machine with the Cellquest software (BD Biosciences). The percentage of cells in G1, S, or G2/M phases of the cell cycle was determined using the Modfit software (BD Biosciences).
Morphological analysis
Cells were fixed using 4% paraformaldehyde at 4°C for 20 minutes, washed, incubated with 1 μg/ml 4′,6-diaminido-2-phenylindole (DAPI) and mounted on a coverslip, then examined by fluorescence microscopy and scored for the number of nuclei (N) and kinetoplasts (K) per cell and their morphology (normal or with elongated posterior end)28; 29. The data were tabulated from a population of 200–500 cells, performed in triplicate.
Assay of 5-bromo-2′-deoxyuridine (BrdU) incorporation into DNA
Cells with induced RNAi were cultured for two days. BrdU was then added to a final concentration of 0.3 mM and the cells were grown for another 3 days. They were then harvested and fixed as described previously for morphological analysis, overlaid onto poly-L lysine coated coverslips and allowed to adhere for 20 min. The cellular DNA was denatured by adding 2 M HCl to the coverslip, incubated for 20 min, washed twice with PBS and followed by blocking for 1 hr at room temperature with PBS/0.5 % BSA. Anti-BrdU antibody (Sigma), diluted 1:100 with PBS/0.5 % BSA, was then added and incubated at room temperature for 1 hour, washed twice with PBS and incubated with the secondary anti-mouse FITC conjugated antibody (Sigma) diluted 1:40 in PBS/0.5 % BSA. The cells were washed twice in PBS, incubated for 5 min in 1μg/ml 4, 6-diamino-2-phenylindole (DAPI) and mounted in mounting medium (Vectashield). Coverslips were sealed with wax and examined by fluorescence microscopy as described earlier.
Supplementary Material
Supplementary figure 1. RNAi of CRK1 and cyclin E1. Cell lines carrying constructs for RNAi of cyclin E1 (a) and CRK1 (b) were selected and cloned. RNAi was induced and cell growth monitored daily over the course of 9 days using a hemocytometer (left panels). RNA was extracted three days after induction and used as template for semi-quantitative RT-PCR to ascertain efficiency of RNAi against the targeted genes (left panels, insets). Cells were also harvested and fixed over a five day period, stained with propidium iodide, analyzed by fluorescence-activated cell sorting (FACS) and assigned G1, S or G2/M phase status using the ModFitLT software and their proportion of total cell number plotted versus time (right panels).
Supplementary figure 2. RNAi of CRK2 and CRK3. T. brucei cell lines carrying the pZJM plasmid for RNAi of CRK3 (a) or CRK2 (b) were selected and cloned. RNAi was induced and cell growth monitored daily over the course of 9 days using a hemocytometer (left panels). RNA was extracted three days after induction and used as template for semi-quantitative RT-PCR to ascertain efficiency of RNAi against the targeted genes (left panels, insets). Cells were also harvested and fixed over a five day period, stained with propidium iodide, analyzed by fluorescence-activated cell sorting (FACS) and assigned G1, S or G2/M phase status using the ModFitLT software and their proportion of total cell number plotted versus time (right panels).
Acknowledgments
We thank Noah Dephoure and Dr Erin K O’Shea of the UCSF for the generous gift of plasmid EB308 and yeast strains S288C and EY1251. We also wish to thank Dr Paul T. Englund of the John Hopkins University School of Medicine for the RNAi vector pZJM and Dr George Cross of the Rockefeller University for the T. brucei procyclic 29-13 strain. We are also grateful to our colleagues for technical help and to Drs W. Zacheus Cande and Lee Douglas for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murray A. Cell cycle checkpoints. Curr Opin Cell Biol. 1994;6:872–6. doi: 10.1016/0955-0674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 2.Murray AW. Cyclin-dependent kinases: regulators of the cell cycle and more. Chem Biol. 1994;1:191–5. doi: 10.1016/1074-5521(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 3.Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–96. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 4.Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–80. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 5.Pines J. Cell cycle. Checkpoint on the nuclear frontier. Nature. 1999;397:104–5. doi: 10.1038/16344. [DOI] [PubMed] [Google Scholar]
- 6.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–4. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- 8.King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–9. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 9.Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–79. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 11.Miller ME, Cross FR. Cyclin specificity: how many wheels do you need on a unicycle? J Cell Sci. 2001;114:1811–20. doi: 10.1242/jcs.114.10.1811. [DOI] [PubMed] [Google Scholar]
- 12.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–64. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 13.Girard F, Strausfeld U, Fernandez A, Lamb NJ. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–79. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 14.Pagano M, Durst M, Joswig S, Draetta G, Jansen-Durr P. Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene. 1992;7:1681–6. [PubMed] [Google Scholar]
- 15.Walker DH, Maller JL. Role for cyclin A in the dependence of mitosis on completion of DNA replication. Nature. 1991;354:314–7. doi: 10.1038/354314a0. [DOI] [PubMed] [Google Scholar]
- 16.Matthews KR, Gull K. Cycles within cycles: the interplay between differentiation and cell division in Trypanosoma brucei. Parasitol Today. 1994;10:473–6. doi: 10.1016/0169-4758(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 17.McKean PG. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr Opin Microbiol. 2003;6:600–7. doi: 10.1016/j.mib.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J Cell Sci. 1999;112 ( Pt 24):4641–50. doi: 10.1242/jcs.112.24.4641. [DOI] [PubMed] [Google Scholar]
- 19.Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol. 1995;128:1163–72. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J Biol Chem. 2003;278:22877–86. doi: 10.1074/jbc.M300813200. [DOI] [PubMed] [Google Scholar]
- 21.Mottram JC, Smith G. A family of trypanosome cdc2-related protein kinases. Gene. 1995;162:147–52. doi: 10.1016/0378-1119(95)00350-f. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Wang CC. A PHO80-like cyclin and a B-type cyclin control the cell cycle of the procyclic form of Trypanosoma brucei. J Biol Chem. 2003;278:20652–8. doi: 10.1074/jbc.M301635200. [DOI] [PubMed] [Google Scholar]
- 23.Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naula C, Parsons M, Mottram JC. Protein kinases as drug targets in trypanosomes and Leishmania. Biochim Biophys Acta. 2005;1754:151–9. doi: 10.1016/j.bbapap.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu X, Wang CC. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J Biol Chem. 2004;279:20519–28. doi: 10.1074/jbc.M312862200. [DOI] [PubMed] [Google Scholar]
- 26.Tu X, Wang CC. Pairwise knockdowns of cdc2-related kinases (CRKs) in Trypanosoma brucei identified the CRKs for G1/S and G2/M transitions and demonstrated distinctive cytokinetic regulations between two developmental stages of the organism. Eukaryot Cell. 2005;4:755–64. doi: 10.1128/EC.4.4.755-764.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Hellemond JJ, Neuville P, Schwarz RT, Matthews KR, Mottram JC. Isolation of Trypanosoma brucei CYC2 and CYC3 cyclin genes by rescue of a yeast G(1) cyclin mutant. Functional characterization of CYC2. J Biol Chem. 2000;275:8315–23. doi: 10.1074/jbc.275.12.8315. [DOI] [PubMed] [Google Scholar]
- 28.Tu X, Wang CC. Coupling of posterior cytoskeletal morphogenesis to the G1/S transition in the Trypanosoma brucei cell cycle. Mol Biol Cell. 2005;16:97–105. doi: 10.1091/mbc.E04-05-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu X, Mancuso J, Cande WZ, Wang CC. Distinct cytoskeletal modulation and regulation of G1-S transition in the two life stages of Trypanosoma brucei. J Cell Sci. 2005;118:4353–64. doi: 10.1242/jcs.02567. [DOI] [PubMed] [Google Scholar]
- 30.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–90. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 31.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 32.Boonstra J. Progression through the G1-phase of the on-going cell cycle. J Cell Biochem. 2003;90:244–52. doi: 10.1002/jcb.10617. [DOI] [PubMed] [Google Scholar]
- 33.Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–34. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 34.Pagano M, Jackson PK. Wagging the dogma; tissue-specific cell cycle control in the mouse embryo. Cell. 2004;118:535–8. doi: 10.1016/j.cell.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Kornbluth S. All aboard the cyclin train: subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol. 1999;9:207–10. doi: 10.1016/s0962-8924(99)01577-9. [DOI] [PubMed] [Google Scholar]
- 36.Brown NR, Noble ME, Endicott JA, Garman EF, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson LN. The crystal structure of cyclin A. Structure. 1995;3:1235–47. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 37.Murray A. Cyclin ubiquitination: the destructive end of mitosis. Cell. 1995;81:149–52. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 38.Espinoza FH, Farrell A, Nourse JL, Chamberlin HM, Gileadi O, Morgan DO. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–73. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross KE, Kaldis P, Solomon MJ. Activating phosphorylation of the Saccharomyces cerevisiae cyclin-dependent kinase, cdc28p, precedes cyclin binding. Mol Biol Cell. 2000;11:1597–609. doi: 10.1091/mbc.11.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsakraklides V, Solomon MJ. Comparison of Cak1p-like cyclin-dependent kinase-activating kinases. J Biol Chem. 2002;277:33482–9. doi: 10.1074/jbc.M205537200. [DOI] [PubMed] [Google Scholar]
- 41.Komeili A, O’Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–80. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 42.Kaffman A, Herskowitz I, Tjian R, O’Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–6. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 43.Madden SL, Johnson DL, Bergman LW. Molecular and expression analysis of the negative regulators involved in the transcriptional regulation of acid phosphatase production in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5950–7. doi: 10.1128/mcb.10.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenburg ME, O’Shea EK. Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics. 2001;157:39–51. doi: 10.1093/genetics/157.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook JaRDW. Molecular cloning: a laboratory manual 2001 [Google Scholar]
- 46.Mutomba MC, To WY, Hyun WC, Wang CC. Inhibition of proteasome activity blocks cell cycle progression at specific phase boundaries in African trypanosomes. Mol Biochem Parasitol. 1997;90:491–504. doi: 10.1016/s0166-6851(97)00197-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. RNAi of CRK1 and cyclin E1. Cell lines carrying constructs for RNAi of cyclin E1 (a) and CRK1 (b) were selected and cloned. RNAi was induced and cell growth monitored daily over the course of 9 days using a hemocytometer (left panels). RNA was extracted three days after induction and used as template for semi-quantitative RT-PCR to ascertain efficiency of RNAi against the targeted genes (left panels, insets). Cells were also harvested and fixed over a five day period, stained with propidium iodide, analyzed by fluorescence-activated cell sorting (FACS) and assigned G1, S or G2/M phase status using the ModFitLT software and their proportion of total cell number plotted versus time (right panels).
Supplementary figure 2. RNAi of CRK2 and CRK3. T. brucei cell lines carrying the pZJM plasmid for RNAi of CRK3 (a) or CRK2 (b) were selected and cloned. RNAi was induced and cell growth monitored daily over the course of 9 days using a hemocytometer (left panels). RNA was extracted three days after induction and used as template for semi-quantitative RT-PCR to ascertain efficiency of RNAi against the targeted genes (left panels, insets). Cells were also harvested and fixed over a five day period, stained with propidium iodide, analyzed by fluorescence-activated cell sorting (FACS) and assigned G1, S or G2/M phase status using the ModFitLT software and their proportion of total cell number plotted versus time (right panels).