Abstract
Biomaterials play a pivotal role in regenerative medicine, which aims to regenerate and replace lost/dysfunctional tissues or organs. Biomaterials (scaffolds) serve as temporary 3D substrates to guide neo tissue formation and organization. It is often beneficial for a scaffolding material to mimic the characteristics of extracellular matrix (ECM) at the nanometer scale and to induce certain natural developmental or/and wound healing processes for tissue regeneration applications. This article reviews the fabrication and modification technologies for nanofibrous, nanocomposite, and nanostructured drug-delivering scaffolds. ECM-mimicking nanostructured biomaterials have been shown to actively regulate cellular responses including attachment, proliferation, differentiation and matrix deposition. Nano-scaled drug delivery systems can be successfully incorporated into a porous 3D scaffold to enhance the tissue regeneration capacity. In conclusion, nano-structured biomateials are a very exciting and rapidly expanding research area, and are providing new enabling technologies for regenerative medicine.
Keywords: Biomaterials, Nanofiber, Drug Delivery, Tissue Engineering, Regenerative Medicine
1. Introduction
Regenerative medicine aims to repair and replace lost or damaged tissues by initiating the natural regeneration process. Most approaches currently pursued or contemplated within the framework of regenerative medicine, including cell-based therapies and living tissue engineering, rely greatly on the ability to synthesize or otherwise generate novel biomaterials, to fabricate or assemble biomaterials into appropriate two-dimensional (2D) and three-dimensional (3D) forms, and to tailor physical, chemical, structural and biological properties to achieve desired clinical efficacy. Biomaterials, including natural macromolecules, synthetic polymers, ceramics, biological factors, and various combinations of these material types, play important roles in regenerative medicine.[1-5]
In a regeneration strategy, biomaterials promote new tissue formation by providing adequate space (porosity) and appropriate surface to foster and direct cellular attachment, migration, proliferation, desired differentiation of specific cell phenotypes throughout the scaffold where new tissue formation is needed.[6, 7] Consequently, the 3D architecture plays a pivotal role. It refers to the way in which a bulk material is distributed in space from the macro, micro to nano scales (corresponding to tissue, cellular and molecular scales in a specific tissue, respectively).[8] Such hierarchical porous architectures not only define the mechanical properties of the scaffold, but also the initial void space that is available for regenerating cells to form new tissues (including new blood vessels) as well as the pathways for mass transport via diffusion and/or convection. While interconnected macroporosity of a biomaterial is important to provide sufficient space for cellular activity,[9-12] interactions between cells and biomaterials occur at the interface, i.e. the entire internal pore walls of a 3D scaffold. The surface morphology or topography directly and significantly affects cell-scaffold interactions and ultimately tissue formation and function.[13]
Nanotechnology has made great strides forward in the creation of new materials, new surfaces, and new 3D architectures, which find numerous applications in the biomedical area.[13-15] Nanostructured biomaterials such as nanoparticles, nanofibers, nanosurfaces, nanocomposites have gained increasing interest in regenerative medicine[16, 17] because these materials often mimic the physical features of natural extracellular matrix (ECM) at the nano scale. The present review summarizes recent advances in the development of porous 3D nanostructured biomaterials and their applications in tissue regeneration. Special attention is paid to nanofibers, nanocomposites, and nanosphere-immobilized biomaterials. The effects of nanofibers on the biological activities of cells and drug-delivering nanofibrous scaffolds for tissue regeneration are highlighted. Two-dimensional nanostructured biomaterials (films, non-porous bulk materials) are beyond the scope of this review, but their advances have been discussed in several recent reviews.[18-20] To consistently discuss biological responses, osteoblastic cells and bone regeneration are frequently used as examples in this article. However, these nanostructured biomaterials are not limited to bone regeneration, but rather are often similarly advantageous in a variety of other tissue regeneration applications.
2. Nanofibrous Biomaterials
An important class of nanostructured biomaterials on which intensive research has been carried out is nano-fibrous materials, especially biodegradable polymer nanofibers. This class of biomaterials mimics the nanofibrillar structure of extracellular matrix (ECM). Collagen is the major ECM component of many tissues and has been investigated as a substrate or scaffold for cell attachment, proliferation and differentiation.[21, 22] Importantly, the nanoscaled collagen fibrillar structure (50-500 nm in diameter) has been found to enhance cell/matrix interactions.[23, 24] To serve as a temporary ECM for regenerative cells, it may be beneficial for a biomaterial to emulate such nanofibrous features of collagen (and other natural ECM components such as fibronectin, proteoglycans, and so forth). Biomaterials that mimic collagen fiber bundles in nanometer size have been developed using a few different techniques: electrospinning, phase separation, and self-assembly. These technologies generate nanofibrous biomaterials with varying degrees of success in tissue regeneration applications.
2.1. Electrospinning
Electrospinning is a well-established process that has been used to produce ultrafine fibers including microfibers (>1 μm) or nanofibers (<1000 nm).[25] In electrospinning, a high voltage is applied to a polymer solution or melt, which overcomes the surface tension to form a charged jet. The charged polymer solution or melt is ejected, dried and solidified onto a grounded substrate. The ejected polymer solutions repel each other during the travel to the grounded collector, which forms thin fibers after solvent evaporation. By controlling the spinning conditions, the resulting fibers can range from about 0.02 μm to about 20 μm. When a stationary collector is used, the resulting fibers deposit randomly due to the random motion of the electrospinning polymer solution. However, preferentially oriented fibers can be obtained by using an electrically grounded rotating drum as the collector.[26]
Many electrospun nanofibrous biomaterials have been investigated as tissue regeneration scaffolds.[27, 28] Materials used in electrospinning can be natural macromolecules such as collagen, chitosan, silk fibroin; synthetic biodegradable polymers such as PGA, PLGA, PLLA, PCL; and combinations of these natural and synthetic polymers.[26, 29-33] In addition, various substances (proteins, growth factors, and hydroxyapatite) can be incorporated into nanofibrous materials during electrospinning.[33, 34] Therefore, electrospun nanofibrous biomaterials have been explored to engineer various tissues. The main advantage of electrospinning process is the relative quick and simple way to fabricate a variety of materials into nanofibrous structure. However, significant challenges still exist in using this technique to fabricate complex 3D scaffold shapes or to generate designed internal pore structures, limiting its potential for many tissue engineering applications.
2.2. Phase Separation
Phase separation techniques have been used to prepare porous polymer membranes for purification and separation purposes.[35] The phase separation process can be induced either thermally or by using a non-solvent. Phase separation induced by a non-solvent results in heterogeneous pore formation, which are generally not suitable for uniform tissue engineering scaffold fabrication. In thermally induced phase separation (TIPS), a homogeneous polymer solution becomes thermodynamically unstable under certain temperature conditions and tends to separate into a multi-phase system (e.g. a polymer-rich phase and a polymer-lean phase) to lower the free energy. After removal of the solvent, the polymer-rich phase solidifies to form the structure while polymer-lean phase becomes pores. A variety of biodegradable polymers have been fabricated into three-dimensional porous scaffolds using phase separation techniques.[36, 37] Depending on the polymer system and phase separation conditions, various pore structures and morphologies have been reported and these scaffolds have been investigated for tissue regeneration applications.
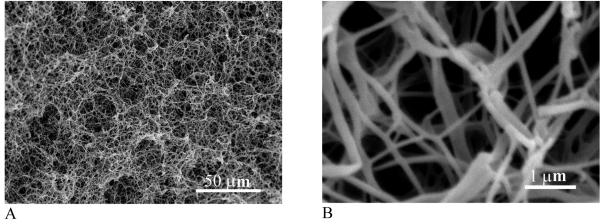
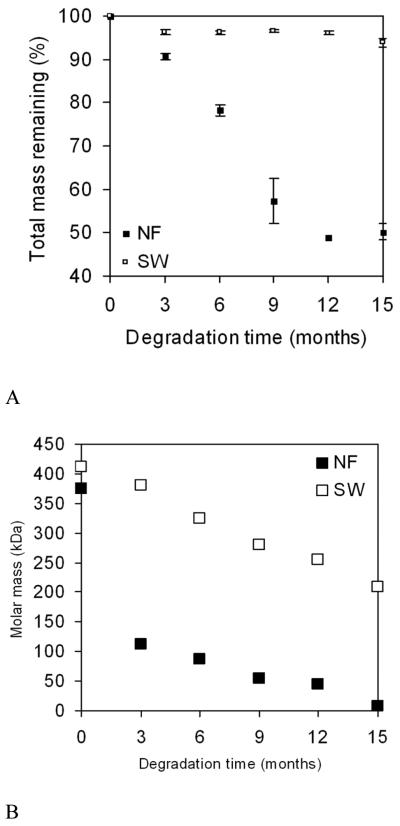
A novel phase separation technique has been developed in our laboratory to generate nanofibrous structures by manipulating the phase separation process.[38-41] For example, a PLLA solution is thermally induced to separate through a spinodal liquid-liquid phase separation and subsequent self assembly into nanofibers. After the removal of solvent a porous structure with 3D continuous nanofibrous network is formed (Figure 1).[42] The fibrous scaffold can have a porosity of 98% and contains nanofibers ranging from 50-500 nm in diameter, which is similar to natural collagen fibers in size. PLLA nanofibrous scaffolds have surface area about two orders of magnitude higher than non-fibrous solid-walled scaffolds.[43] Due to the substantial surface area difference, degradation is much more rapid in such nanofibrous scaffolds (Fig. 2), in which the overall mass loss is 51% while mass loss in solid-walled non-fibrous foams is only 6% after 15 months. During degradation, both molar mass and surface area of nanofibrous scaffolds decrease more rapidly than those of solid-walled scaffolds, indicating that nanofibrous characteristics accelerate the rate of hydrolytic degradation of the polymer scaffolds.[43]
Fig. 1.
Scanning electron micrographs (SEM) of a PLLA nanofibrous scaffold prepared from 2.5% PLLA/THF solution at a phase separation temperature of 8 °C: (A) 500x; (B) 20,000x. From Ma et al.[42] Copyright © 1999 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons.
Fig. 2.
Normalized change in average mass (A) and molar mass (B) of PLLA nanofibrous (NF) and solid-walled (SW) scaffolds after in vitro degradation in PBS. From Chen et al.[43] Copyright © 2001 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons.
One limitation of the early nanofibrous materials generated using the phase-separation technique is the lack of interconnected macropores, which are critical for cell seeding and recruiting, mass transfer, vascularization and tissue organization. To overcome this problem, phase separation techniques are used in combination with other scaffold fabrication techniques such as porogen leaching and solid freeform fabrication (SFF). The combined technique provides broader control over porous architectures from macro, micro to nano scales.[39, 41, 44] The hierarchical pore structure has been shown to substantially promote tissue regeneration.[39]
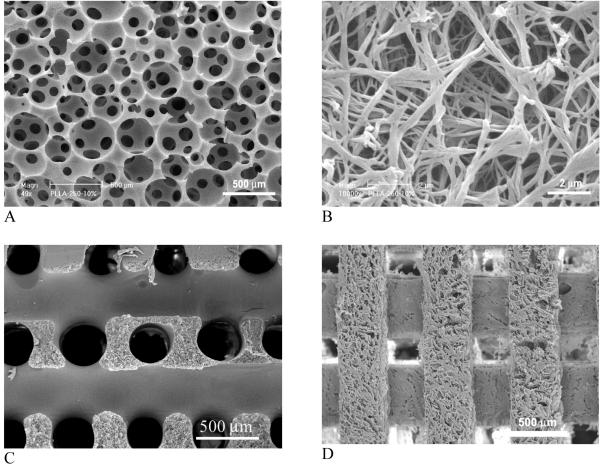
A combined technique of phase separation and sugar sphere template leaching has been developed recently. Water-soluble sugar spheres are prepared and bound to form a template. Phase separation occurs when a polymer solution is cast on to a sugar template and the temperature is lowered. Removal of sugar template leads to the formation of macroporous and nanofibrous scaffolds[44] (Fig. 3 A, B). The technique advantageously controls macropore shape and size by sugar spheres, interpore opening size by assembly conditions (time and temperature of heat treatment for sugar spheres), and pore wall morphologies by phase separation parameters. Alternatively, sugar fiber template or solid freeform fabricated wax mold are used to prepare nanofibrous scaffolds with interconnected macro-tubular structures or channels, respectively (Fig. 3, C, D).[38, 39] Using a computer-assisted-design (CAD) technique, external scaffold shape can be generated from computed-tomography scans or histological sections. For example, a nanofibrous polymer scaffold with the shape of a human ear is precisely created using the phase-separation technique and a mold reconstructed from its histological sections (Fig. 4). Despite the different external scaffold shapes and macroporous structures (spherical, channeled), all scaffolds have nanofibrous architectures on the pore walls (Fig. 3B and Fig. 4C). The nanofibrous scaffolds also have high porosity (>90%) and high surface area (around 100 m2/g).
Fig. 3.
SEM micrographs of 3D macroporous and nanofibrous PLLA scaffolds. (A, B) prepared from sugar sphere template leaching and phase separation; (C) prepared from sugar fiber template leaching and phase separation; (D) prepared from solid freeform fabrication and phase separation. (A, B) From Wei and Ma[44] Copyright © 2006 by John Wiley & Sons; (C) From Zhang and Ma[38] Copyright © 2000 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons. (D) From Chen et al.[39] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
Fig. 4.
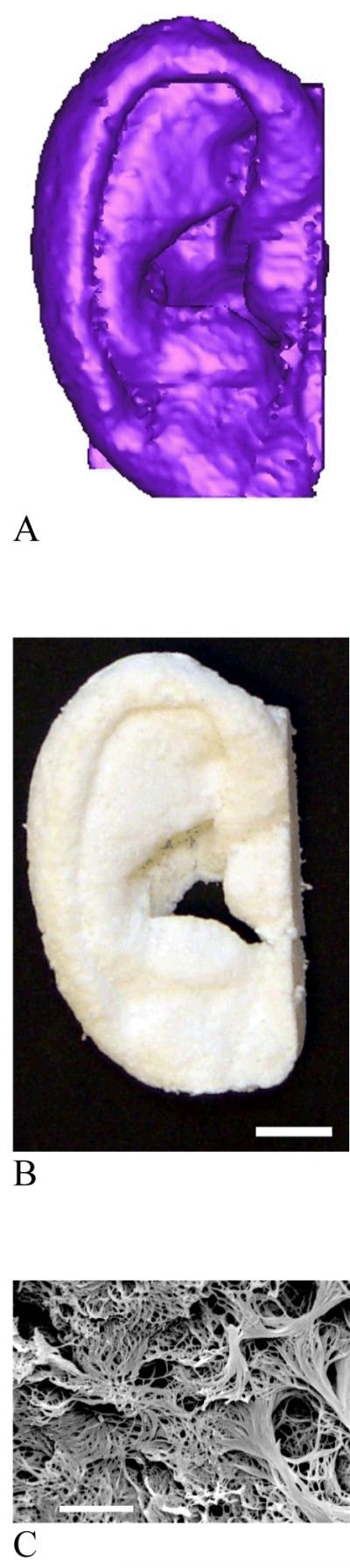
Nanofibrous scaffolds created from 3D medical images and a phase-separation technique. (A) human ear template reconstructed from histological sections; (B) resulting nanofibrous scaffold of the human ear (scale bar: 10 mm); (C) the nanofibrous pore wall morphology (scale bar: 5 μm). From Chen et al.[39] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
2.3. Self-assembly
Nanofibrous matrices can be created by a molecular self-assembly approach which involves the spontaneous organization of individual molecules into a well-defined and stable hierarchical structure with preprogrammed non-covalent interactions.[45-47] As a natural process for several essential biological components including nucleic acid or protein synthesis, self-assembly technology usually incorporates some specific biological components of the extracellular matrix (ECM) and mimics ECM assembly process.
Self-assembling molecules require some specific configurations to be assembled into nanofibers. For example, nanofiber forming peptide-amphiphiles (PAs) are designed to have several critical structural features: a long alkyl tail that conveys hydrophobic characteristics to drive self-assembly; four consecutive cysteine residues that create disulfide bonds to polymerize the structure; a linker region of three glycine residues to provide the flexibility to the hydrophilic head group; a phosphorylated serine residue that interacts strongly with calcium ions intended to enhance mineralization; and an Arg-Gly-Asp (RGD) peptide to aid in cell adhesion.[48] These PAs when subject to an acid are induced to self-assemble to form nanofibers with 5-8 nm in diameter and 1 μm in length. This involves treating PA solutions with dithiothreitol at a pH of 8 and then reducing the pH to 4. As the solution is acidified the PAs rapidly become insoluble and form nanofibers of a gel. Besides pH-driven, self-assembly of PAs can also be achieved by drying and introductions with divalent ions such as calcium.[49] Some of the self-assembled hydrogels are being explored for cell entrapment utilizing polyvalent metal ions in the media, and one of them has been shown to allow for osteoblastic cells (MC3T3-E1) to survive for 3 weeks.[50]
Zhang and coworkers have synthesized ionic self-complementary oligopeptides that consist of regular repeats of alternating ionic hydrophilic and hydrophobic amino acids. In water, these oligopeptides form a β-sheet and both a polar surface (with charged ionic chains) and a non-polar surface (with alanines) are present. Upon exposure to monovalent alkaline cations or under physiological conditions, the oligopeptides spontaneously assemble into hydrogels of various shapes.[51] These hydrogel matrices are interwoven nano-fibers approximately 10-20 nm in diameter with pores about 50-200 nm in diameter.[52, 53] The nanofibrous matrices support mammalian cell attachment, proliferation and differentiation.[53]
In addition to PAs and oligopeptides, synthetic diblock/triblock copolymers[54, 55] and dendrimers[56, 57] can be self-assembled into nanofibrous structures. However, as regenerative biomaterials, self-assembled nanofibrous scaffolds are currently limited to biological molecules such as peptides in the form of hydrogels.[45, 58] One of the limitations is their inability to controllably form mechanically stable 3D geometry. The peptide nanofibers can be fragmented and may be susceptible to endocytosis.[50] In addition, their degradation has not been systematically addressed. The peptide backbone structure can be degraded by enzymes, making it difficult to control their in vivo degradation behavior at will.
2.4. Surface Modification of Nanofibrous Scaffolds
Although a variety of biomaterials have been fabricated into nanofibrous scaffolds using various techniques discussed above, many of them have low biological compatibility with cells, which has limited their regeneration applications. Surface modification is an effective way to improve cellular interactions with the nanofibrous scaffolds. Different from bulk modification which usually changes the chemical and mechanical properties of the biomaterials, surface modification has the advantage of not altering the scaffold architectures significantly. This is especially desirable for the modification of ultrafine structures such as nanofibers.
Plasma exposure has been used to introduce desired functional groups and molecular chains onto the nanofibrous surface of an electrospun matrix.[59, 60] Non-woven poly(epsilon-caprolactone) (PCL) nanofibers were prepared by electrospinning and type-I collagen was then immobilized on the nanofibers after surface modification by plasma treatment. Collagen immobilization enhanced the attachment, spreading and proliferation of human dermal fibroblasts.[59] Grafting gelatin on to plasma treated PCL nanofibers improved endothelial cell spreading and proliferation as well as cell orientation.[60] Because of the limited depth of plasma penetration, this technique can be used for 2D films or very thin 3D structures but is not adequate for large 3D scaffolds with complex architectures.
To mimic both the nanofibrous structure and chemical composition of collagen fibers, 3D nanofibrous scaffolds have been prepared using a phase separation technique and surface modified with gelatin, a biomacromolecule derived from ECM collagen.[61-63] An entrapment method was developed to effectively incorporate gelatin onto nanofibrous wall surfaces of both interior and exterior pores.[61] Compared to a simple coating of gelatin onto the scaffold, entrapped gelatin molecular chains entangle with the molecular chains of scaffold materials and are physically locked on the surface permanently. The entrapment modification method can be used for various geometries, morphologies, and thicknesses of 3D polymer scaffolds without interfering bulk properties and architectures of a scaffold. Surface modification can also be carried out during scaffold fabrication using a porogen-induced modification method.[62] For example, gelatin spheres can act as both the porogen for scaffold fabrication and the agent of surface-modification, with which scaffold formation and surface modification are completed in a simple one-step process.[62] The gelatin-modified porous nanofibrous scaffold has been demonstrated to significantly improve osteoblast cell adhesion and proliferation throughout the scaffold.[61, 62]
Since most biomacromolecules are charged cationic or anionic polyelectrolytes, the immobilization of these biomacromolecules onto 3D nanofibrous scaffolds can also be achieved by a layer-by-layer self-assembly process.[63-65] For example, a macroporous and nanofibrous PLLA scaffold was activated in an aqueous poly(diallyldimethylammonium chloride) (PDAC) solution to obtain positively charged pore wall surface. The scaffold was subsequently immersed in a solution of negatively charged biomacromolecules (e.g. gelatin). Surface layers with positive or negative charges are developed by alternative immersion in the two different solutions. Self-assembly approach is carried out in aqueous solutions under mild conditions and offers a controlled way to regulate the surface charge type and the thickness of the surface modification layer.[63] Growth factors and DNAs (as a gene-delivery strategy) may also be self-assembled on to nanofibrous scaffolds to regulate cellular response and gene expression.
2.5. Biological Effects of Nanofibers
Nanofibers have special properties arising from their dimensions and high surface area to volume ratio. Nanofibrous scaffolds serve as promising tissue engineering scaffolds because nanofibrous surfaces elicit special biological responses of cells during cultivation and after implantation. The nanofibers regulate protein adsorption, cell attachment, proliferation, and differentiation as well as extracellular matrix deposition.
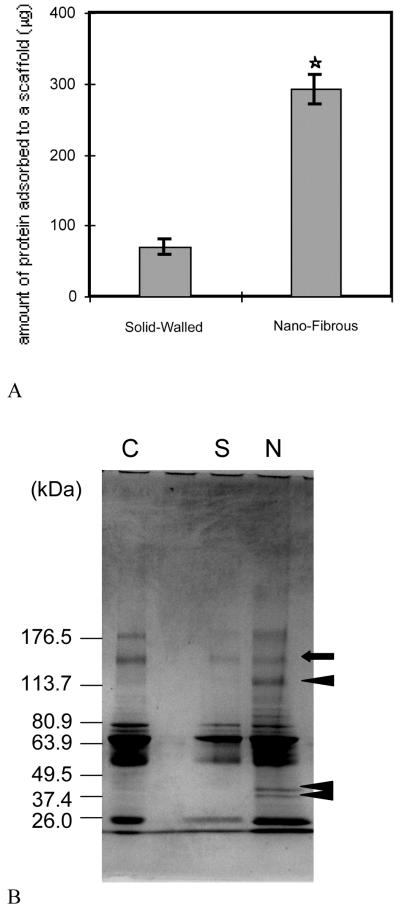
The nanofibrous architectures built in a macroporous scaffold enhance protein adsorption in vitro.[39, 40] Compared to solid-walled scaffolds without nanofibrous structures, nanofibrous scaffolds were found to adsorb 4 times more human serum proteins (Fig. 5A). More interestingly, the profile of serum proteins adsorbed onto the nanofibrous scaffolds was different from that adsorbed onto the solid-walled scaffolds. With SDS-PAGE analysis, the protein with an approximate size of 150 kDa (arrow) adsorbed to both scaffolds similarly while the proteins with approximate sizes of 120, 45, or 40 kDa (arrowheads) exclusively adsorbed onto the nanofibrous scaffold (Fig. 5B). Based on the measurements of band intensities of proteins, the amounts of proteins adsorbed to nanofibrous scaffolds were 0.57–13.35 times those of corresponding proteins (bands) in control serum, whereas the amounts of proteins adsorbed to solid-walled scaffolds were 0.24–0.79 times those of corresponding proteins in the control serum. Furthermore, Western blot analysis showed that nanofibrous scaffolds adsorbed large amounts of fibronectin and vitronectin from serum, while these cell-adhesion proteins were barely detectable on the solid-walled scaffolds. The findings suggest that nanofibers alone may have certain features enhancing protein-affinity and binding strength.[40]
Fig. 5.
(A) Amounts of adsorbed serum proteins onto the scaffolds. *Significantly different from solid-walled scaffolds, p< 0.05, n = 4. (B) Protein adsorption profile on nanofibrous and solid-walled scaffolds. Polyacrylamide gels stained with Coomassie blue. Lane C, bovine serum proteins; lane S, adsorbed bovine serum proteins to the solid-walled scaffold; lane N, adsorbed bovine serum proteins to the nanofibrous scaffold. From Woo et al.[40] Copyright © 2003 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons.
Cells behave differently on nanofibrous scaffolds from those on solid-walled scaffolds, both pretreated with fetal bovine serum. About 70% more osteoblastic progenitor cells (MC3T3-E1) were attached on a nanofibrous scaffold than on a solid-walled scaffold (24 h after seeding).[40] Cell filopodia were reported to direct to certain particulate structures.[66] The nanofibrous architecture may allow filopodia to anchor more tightly, and this mode of anchorage could also contribute to the stronger cell adhesion to the nanofibrous scaffolds. Osteoprogenitor cells also proliferated more rapidly on the nanofibrous scaffolds compared with solid-walled scaffolds. Since both types of substrates (3D nanofibrous vs. solid-walled scaffolds or 2D nanofibrous vs. solid films) were made from the same polymer and had the same macro-structures for cell accommodation, it appears that the nanofibrous architecture provides a more favorable environment for cell attachment and proliferation. Similar results were reported when comparing smooth muscle cells (SMCs) on electrospun nanofibrous films and plain polymer films. The adhesion and proliferation of SMCs on the nanofibrous films were significantly improved over those on the plain polymer films.[27]
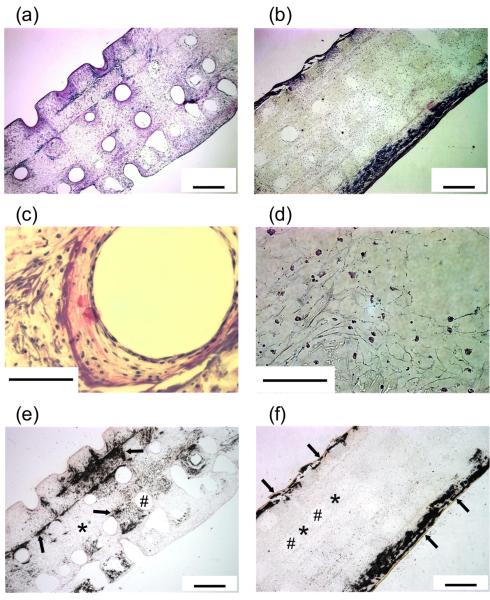
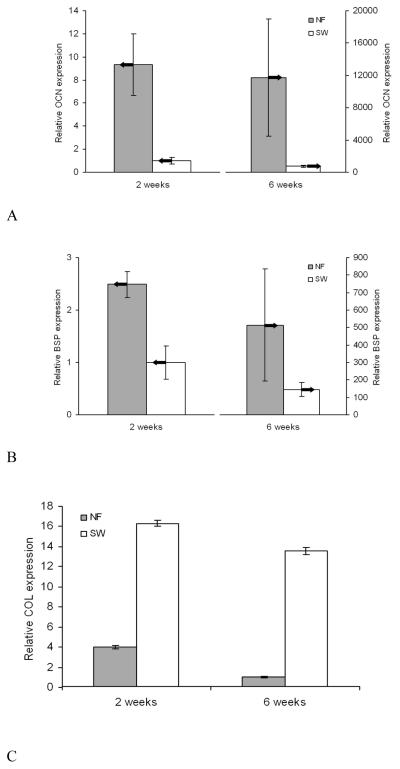
When bone tissue is engineered, cells and mineralized extracellular matrix (ECM) are distributed throughout nanofibrous scaffolds while cells and mineralized ECM are primarily deposited in the outer regions of the solid-walled scaffolds (Fig. 6).[39] Fast biomineralization is also demonstrated using a calcium assay, which shows 13-fold greater amount of mineral deposition on nanofibrous scaffolds than on solid-walled scaffolds.[67] At the molecular level, cells on nanofibrous scaffolds express significantly higher levels of bone markers such as osteocalcin (OCN) and bone sialoprotein (BSP) (Fig. 7A, B). In contrast, cells on solid-walled scaffolds express a substantially higher level of collagen (COL) mRNA (Fig. 7C).[39] The results suggest that bone cells differentiate more quickly on nanofibrous scaffolds than on solid-walled scaffolds, resulting in decreased COL mRNA expression and the increased late-stage bone marker expression (OCN and BSP) on nanofibrous scaffolds. The enhanced osteoblastic differentiation by the nanofibrous scaffolds has recently been found to be associated with the RhoA-ROCK signaling pathway.[68] In addition, our unpublished data show that nanofibrous architectures also facilitate the differentiation of both adult and embryonic stem cells. In a rat calvarial bone defect regeneration model, the amount of new bone formation was about two times greater in the nanofibrous scaffolds than in the solid-walled scaffolds (data to be published). These advantageous properties of the nano-fibrous scaffolds are also corroborated by data of cartilage tissue engineering using electrospun nanofibrous materials.[69]
Fig. 6.
Responses of MC3T3-E1 cells on nanofibrous (A, C, E) and solid-walled (B, D, F) PLLA scaffolds. Scaffolds were prepared using a combined solid freeform fabrication and phase separation technique, having the same macropore structures. Cells were cultured in vitro for 6 weeks. (A-D): H&E staining, (E, F): von Kossa staining. * denotes the PLLA scaffold, # a scaffold pore. Arrows in (E, F) denote mineralization. From Chen et al.[39] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
Fig. 7.
Relative levels of bone marker expression in nanofibrous and solid-walled scaffolds after 2 and 6 weeks of culture under differentiation conditions. (A) osteocalcin (OCN) expression; (B) bone sialoprotein (BSP) expression; and (C) type I collagen (COL) expression. From Chen et al.[39] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
The nanofibrous architecture, the high surface area, the microporosity between nanofibers (several μms), and the selective adsorption of ECM proteins, all likely contribute to the enhanced cellular responses and tissue regeneration in the nanofibrous scaffolds.
3. Nanocomposite Biomaterials
Mineralized tissues such as bone and dentin are in the form of inorganic/organic nanocomposites. In natural bone, the plate-like apatite crystals with dimensions of 50 nm × 25 nm × 2-3 nm (length × width × thickness), are dispersed in the nanofibrous collagen matrices. The nanometer size of both collagen fibers and apatite crystals as well as the organization of the composite contribute significantly to the excellent mechanical properties and functionality of bone.[70, 71] Being similar to the major inorganic component of natural bone, bioceramics such as hydroxyapatite (HAP) is another category of biomaterials besides polymers that have been widely investigated for bone regeneration.[72-74] Nano-sized HAP has shown interesting properties such as hydrophilicity, wettability, surface area and surface roughness, which are different from microsized HAP. These properties would enhance osteoblast adhesion and provide better osteoconductivity and bonding properties to host bone for long-term functionality.[75] However, in developing regenerative biomaterials for mineralized tissues such as bone, one material alone, either organic polymers or inorganic bioceramics, may not meet all the requirements. One promising alternative is to use nanocomposite biomaterials with similar nanostructures and compositions to those of natural bone. In the composite, the inorganic component such as hydroxyapatite (HAP) provides good osteoconductivity and bone bonding ability,[72-74] while the polymer component offers structural continuity and design flexibility to achieve a high porosity and a high total surface area which are necessary for anchorage-dependent cells including bone cells to survive and differentiate.
To mimic the nano features of natural bone, collagen/nano-HAP (nHAP) composite or porous composite matrices were fabricated by precipitation of HAP nanoparticles from an aqueous solution onto collagen.[76, 77] Interfacial new bone formation by osteoblasts was observed after implantation in a rabbit marrow cavity.[78] Biodegradation of the composite was achieved by solution-mediated dissolution and possibly cell-mediated resorption in which the nanometer size of HAP particles was important.[78, 79] The results suggest that the porous collagen/nHAP scaffold may provide a microenvironment similar to in vivo environment favorable for bone regeneration.
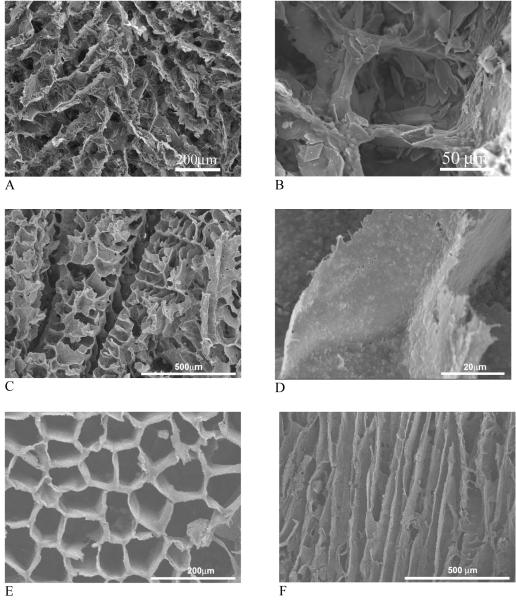
Nanocomposites of synthetic polymers and HAP have been developed to circumvent possible concerns of pathogen transmission and immuno-rejection associated with collagen from animal and cadaver sources and to improve the material processability. By blending and phase separation techniques, polymer/hydroxyapatite composite scaffolds have been fabricated and shown improved mechanical properties and osteoconductivity[36] (Fig. 8A, B). The hydroxyapatite containing scaffolds improved osteoblastic cell seeding uniformity and enhanced expression of osteocalcin and bone sialoprotein over plain polymer scaffolds. Bone tissue formation throughout the scaffold has been demonstrated.[80] Compared to micro-sized HAP (mHAP, 20-50 μm), the incorporation of nano-HAP did not alter the scaffold structure and the nHAP amount as high as 70% was incorporated (Fig. 8C, D). When phase separation was performed with a uniaxial temperature gradient, oriented microtubular pores in the PLLA/nHAP composite scaffolds were obtained whereas such pore morphology could not be obtained from mHAP (Fig. 8E, F). The nHAP/polymer composite scaffolds not only improved the mechanical properties, but also significantly enhanced protein adsorption over microsized HAP/polymer scaffolds.[37] Enhanced protein adsorption improves cell adhesion and suppresses apoptotic cell death.[81]
Fig. 8.
SEM micrographs of polymer/hydroxyapatite composite scaffolds fabricated using phase separation. (A, B) PLLA/mHAP; (C, D) PLLA/nHAP; (E, F) PLGA/nHAP. (A, B) From Zhang and Ma[36] Copyright © 1999 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons; (C-F) From Wei and Ma[37] Copyright © 2004 by Elsevier. Reprinted with permission of Elsevier.
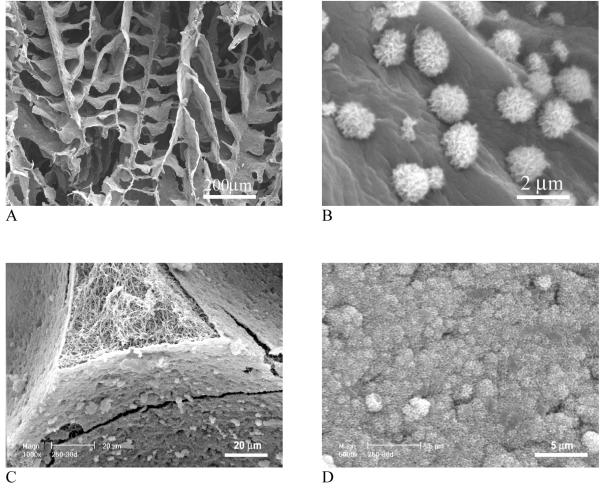
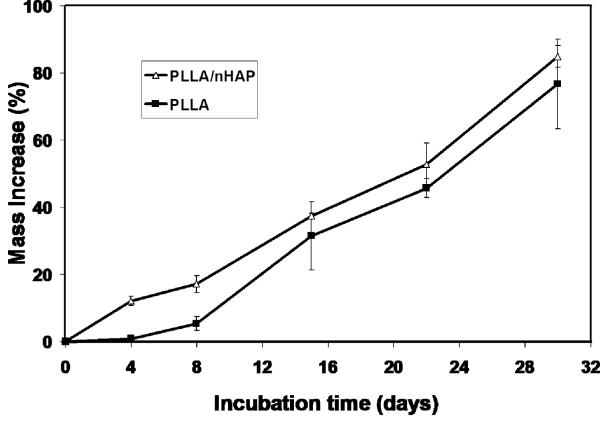
It should be noted that the mineral in natural bone is non-stoichiometric apatite with Ca/P ratio around 1.5 (stoichiometric HAP has a Ca/P ratio of 1.67). The calcium deficient apatite degrades faster than the stoichiometric HAP crystals and should serve as a better scaffold component in terms of bone tissue modeling and remolding. Although one can first synthesize calcium deficient HAP nanoparticles and then mix them with polymers to fabricate nanocomposite scaffolds, a biomimetic approach has been developed to grow bone-like apatite particles on pre-fabricated porous polymer scaffolds in a simulated body fluid (SBF),[44, 82, 83] which efficiently modifies the internal pore wall surfaces without altering the bulk structures of the scaffolds (Fig. 9A, B). The apatite particles have nano-sized features and their growth can be tailored by the sccaffold composition, porous structure, ionic concentration of SBF as well as the pH value.[83] When nanofibrous scaffold was investigated for bone-like apatite deposition, a uniform and dense layer of nano apatite was found to cover the entire internal pore wall surfaces (Fig. 9C, D). However, the same macropores and inter-pore openings were retained as in the starting polymer scaffold. Nano apatite-containing scaffolds showed substantially increased compressive modulus. It was found that pre-incorporation of nHAP in a nanofibrous scaffold promoted the rate of apatite deposition (Fig. 10).[44]
Fig. 9.
SEM micrographs of PLLA/apatite composite scaffolds prepared by a biomimetic approach in a simulated body fluid (SBF). (A, B) PLLA scaffolds prepared by phase separation in dioxane; (C, D) PLLA nanofibrous scaffolds prepared by sugar template leaching and phase separation in THF. Scaffolds were incubated in 1.5x SBF at 37 °C for 30 days. (A, B) From Zhang and Ma [83], Copyright © 1999 by John Wiley & Sons; (C, D) From Wei and Ma[44] Copyright © 2006 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons.
Fig. 10.
Mass increase of nanofibrous PLLA and PLLA/nHAP scaffolds over incubation time in 1.5X SBF. From Wei and Ma[44] Copyright © 2006 by John Wiley & Sons. Reprinted with permission of John Wiley & Sons.
Besides calcium phosphate based nanocomposite materials, carbon nanotube (CNT) based polymer composites have also been explored for tissue regeneration.[14, 84] CNTs have high elastic modulus and strength, low density, chemical inertness, thermal stability as well as the electrical conductivity. The initial motivation of using CNTs was to reinforce polymer materials for improved mechanical properties.[85] Later, it was found that carbon nanotubes could accelerate cell growth.[86-88] Electrically conductive PLLA/CNTs nanocomposites were shown to substantially increase osteoblast proliferation and calcium production upon receiving alternating current stimulation.[88] Despite the osteoconductive properties of CNTs, concerns over their biocompatibility and toxicity remain to be addressed.
4. Drug-delivering Nanostructured Biomaterials
Owing to the rapid advance in recombinant technology and the availability of a large scale of purified recombinant polypeptides and proteins, protein drugs such as growth factors have been widely used to stimulate cellular activity and regulate tissue regeneration.[89-91] However, protein and peptide drugs in general have short plasma half-lives, are unstable in the gastrointestinal tract and also have low bioavailability due to their relatively large molecular weight and high aqueous solubility. These properties have limited their effective clinical applications[92] and a delivery system is required to achieve high therapeutic efficacy of corresponding proteins.[93-95] Nanotechnology is revolutionizing drug delivery. A nano-scale drug delivery system can be devised to tune release kinetics, to regulate biodistribution and to minimize toxic side effect, thereby enhancing the therapeutic efficacy of a given drug. Incorporating nano-scale drug delivery system into a nanostructured biomaterial represents a novel and promising strategy to tissue regeneration.
4.1. Nanospheres
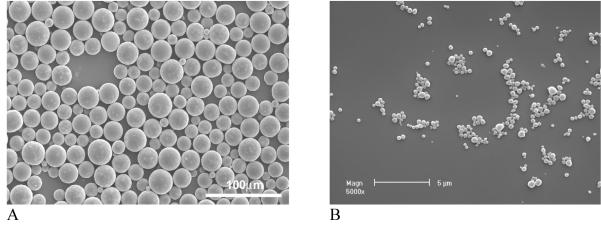
Polymeric particulate carriers (micro- and nano-spheres) have been demonstrated to be an effective way to offer controlled release of contained substance and to protect unstable biologically active molecules from denaturing and degradation after administration.[93, 96] Nanospheres are often made from biodegradable polyesters such as poly(lactic acid) (PLLA) and poly(lactic-co-glycolic acid) (PLGA) due to their excellent biocompatibility and controllable biodegradability through natural pathways.[97, 98] With a double emulsion technique, spherical particles with different sizes (from micro to nano meters) can be produced by adjusting the concentration of surfactant used and the emulsion strength employed in the second emulsification (Fig. 11). The release of proteins from microspheres/nanospheres is controlled in the first stage by diffusion and in the second stage by the degradation of polymer micro- or nano-spheres. By varying the the ratio of LA/GA in the PLGA copolymers and their molecular weight, sustained protein release over days to months can be achieved.[97] Most importantly, the released proteins are able to maintain a high level of biological activity with desired prolonged durations.[97, 99] Recombinant human platelet-derived growth factor BB (rhPDGF-BB) released from PLGA nanospheres was biological active and was able to stimulate the proliferation of human gingival fibroblasts.[100] These results illustrate the feasibility of achieving local delivery of bioactive macromolecules (proteins and polypeptides) to induce cellular responses by a microsphere/nanosphere encapsulation and delivery technique.
Fig. 11.
SEM micrographs of PLGA micro-/nano-spheres with varying sizes. (A) From Wei et al.[97] Copyright © 2004 by Elsevier; (B) From Wei et al.[100] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
4.2. Nanosphere Immobilized Nanofibrous Scaffolds
As discussed earlier, nanostructured biomaterials especially nanofibrous scaffolds are able to mimic natural ECM morphology as well as surface chemistry. When the advantageous nanostructures are integrated with interconnected 3D pore structure, the resulting novel scaffolds show enhanced regeneration performance than scaffolds without such nanostructures. Growth factors, if employed properly, are able to stimulate desired cellular and tissue response. Delivery of growth factors from an advanced 3D scaffold should provide tissue engineers with additional control over regeneration outcome.[13] In this strategy, the 3D scaffold serves both as a temporary substrate for cell function and as a delivery carrier for the controlled release of growth factors.
Proteins can be simply adsorbed onto a scaffold to achieve delivery from the scaffold. However, the burst release is severe and temporal control over release kinetics is very limited in the passive adsorption approach.[101, 102] Growth factors have also been incorporated into porous scaffold using other techniques such as emulsion freeze drying[103] or gas foaming.[104] One disadvantage associated with these two incorporation techniques is the difficulty to achieve controllable macro porosity and open pore structures in the scaffold.
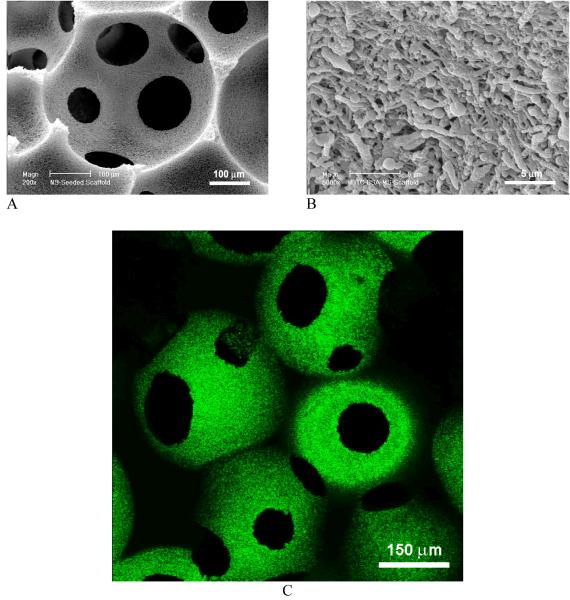
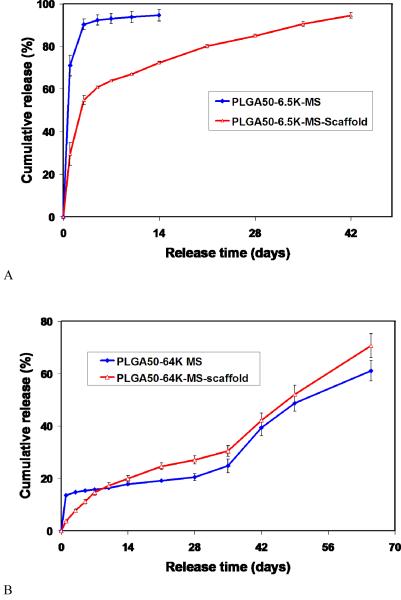
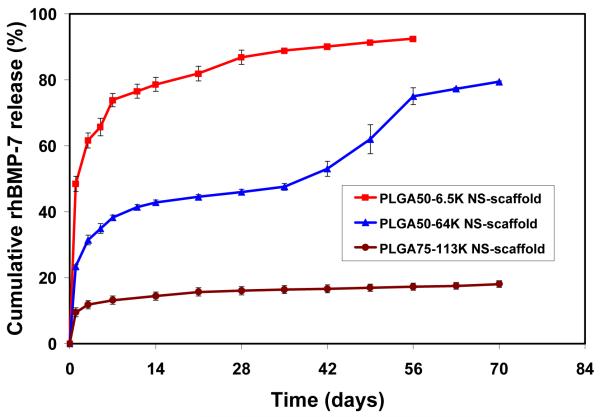
A novel technology has been developed to incorporate nanospheres into prefabricated nanofibrous scaffolds (NS-scaffold).[100, 105] Biological factors were first encapsulated into biodegradable nanospheres, which were then immobilized onto a macroporous and nanofibrous scaffold. Both the size of nanospheres and the macroporosity of the scaffold are important for the immobilization technique. Nanospheres with diameters of 200-500 nm were uniformly distributed throughout the scaffold (Fig. 12). It is advantageous to attach the nanospheres on the nanofibrous surfaces throughout the macropores without altering the macroporous structures of the scaffold. The delivery dose of a protein can be adjusted either by the amount of protein encapsulated in the nanospheres or the amount of nanospheres immobilized onto the scaffold. The initial burst release of PDGF-BB was significantly reduced after immobilizing PDGF containing nanospheres onto a 3D nanofibrous scaffold and different release profiles were achieved through the immobilization of nanospheres with different degradation rates (Fig. 13).[100] The NS-scaffolds can also be used to control the release kinetics of other macromolecules or proteins. By varying the chemical composition (LA/GA ratio) and molecular weight of the PLGA polymer, the release kinetics of the growth factors can be tailored to meet the need of a specific application (Fig. 14).
Fig. 12.
SEM micrographs (A, B) and laser scanning confocal micrograph (C) of PLGA nanosphere-immobilized PLLA nanofibrous scaffolds. FITC-labeled bovine serum albumin was encapsulated in PLGA nanospheres, showing green emission under confocal microscopy (C). From Wei et al.[100] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
Fig. 13.
In vitro release kinetics of PDGF-BB from nanosphere-immobilized PLLA nanofibrous scaffolds. (A) PLGA (50/50, 6.5Kd)-NS-scaffold; and (B) PLGA(50/50, 64Kd)-NS-scaffold. From Wei et al.[100] Copyright © 2006 by Elsevier. Reprinted with permission of Elsevier.
Fig. 14.
In vitro release kinetics of rhBMP-7 from nanosphere-immobilized nanofibrous scaffolds: in 10 mM PBS with a rhBMP-7 loading of 200 ng/scaffold. Three distinct release profiles were achieved from three different PLGA polymers (different LA/GA ratios such as 50/50 vs. 75/25 or different molecular weights). From Wei et al.[105] Copyright © 2007 by Elsevier. Reprinted with permission of Elsevier.
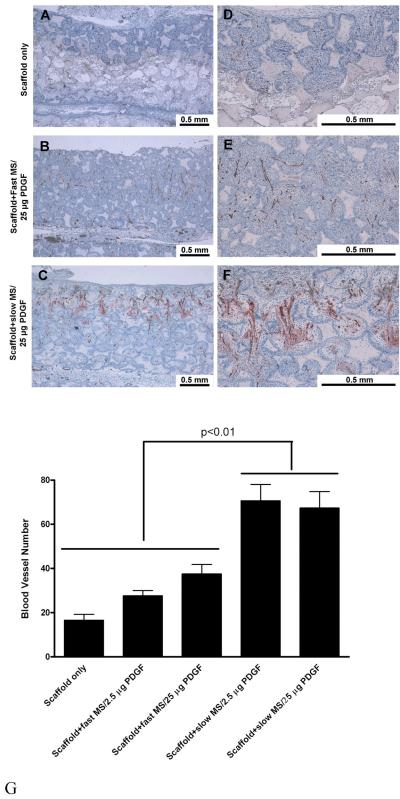
PDGF-delivering NS-scaffolds were evaluated in a soft tissue wound repair model in rats for cell penetration, vasculogenesis and tissue neogenesis.[106] In vivo, PDGF functions in a dose-dependent and release mode-dependent manner. Cells and tissues penetrated completely into all PDGF NS scaffold and there was massive new blood vessel formation (Fig. 15A-F). In the control group (scaffolds with empty NS), the tissue penetrated into only a small portion of the scaffold and only negligible new blood vessels were formed. Significantly more blood vessels were formed in slow-releasing NS-scaffolds than in faster releasing scaffolds, both at low (2.5 μg) and high (25 μg) doses of PDGF (Fig. 15G). Compared to scaffolds with simple coating of PDGF, PDGF-releasing NS-scaffolds showed both significantly more tissue penetration and blood vessel formation. The enhanced tissue neogenesis and neovascularization by sustained PDGF release were found to be associated with changes in PDGF-induced gene expression profiles of chemokine family members, actin, and interleukins.[106]
Fig. 15.
Nanofibrous scaffolds with PDGF nanospheres promote vasculogenesis in vivo. (A-F) Histological observation: left panel is at a lower magnification (A-C: 10x), right panel is at a higher magnification (D-F: 40x). Positive Factor VIII stained blood vessels (brown) are located in the central regions of the pores within penetrated tissues. The blood vessels have also permeated through the interpore openings. (G) Blood vessel number in scaffolds. * indicate p<0.01. From Jin et al.[106].
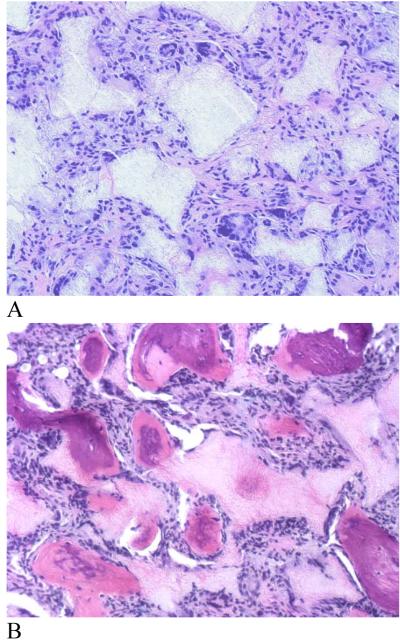
Similarly, BMP-7 delivered from nanosphere-immobilized nanofibrous scaffolds induced significant ectopic bone formation throughout the scaffold (Fig. 16). In contrast, passive adsorption of the same amount of rhBMP-7 onto the scaffold failed to induce bone formation due to either the loss of rhBMP-7 biological function or/and insufficient duration within the scaffold.[105] Once again the result demonstrate that the new NS-scaffold platform utilizes nanosphere encapsulation technology to protect growth factors from denaturation, which may occur readily in a passive adsorption approach. Passive adsorption of growth factors (such as rhVEGF, BMP-4 and bFGF) onto scaffold leads to complete degradation during a very short release time of 3 days.[107]
Fig. 16.
New bone formation in rhBMP-7 incorporated PLLA nano-fibrous scaffolds retrieved 6 weeks after subcutaneous implantation in rats. (A) 5 μg rhBMP-7 adsorbed to scaffold; (B) 5 μg rhBMP-7 incorporated in nanosphere immobilized scaffold. H & E staining with original magnification of 100x. From Wei et al.[105] Copyright © 2007 by Elsevier. Reprinted with permission of Elsevier.
While many current regeneration strategies have focused on delivering single potent growth factors to enhance a specific repair process, it appears that multiple growth factors and cytokines are required to regulate tissue regeneration during different stages.[108, 109] Multiple factor delivery can be achieved by immobilizing multiple types of nanospheres (NS) onto a single macroporous nanofibrous scaffold.[110] Different biodegradable polymer nanospheres are utilized to individually control the release profiles of different biological molecules. The resulting NS-scaffold can release several macromolecules simultaneously, but each with individualized release profile. The released factors may function synergistically to emulate the natural repair process.
Growth factors and cytokines are one type of biological signals that actively participate in regulating cell function and tissue regeneration. Other biological molecules such as DNA and small interfering RNAs (siRNA) are also being explored to enhance tissue regeneration. Based on the understanding of the interactions between these molecules and polymer nanospheres, the NS immobilization approach can also be taken to deliver DNA, siRNA or in combination with growth factors. Thus, the NS-scaffold fabrication technology provides a platform to program a spectrum of biological signals into 3D nanomaterials to regulate cellular activities and to orchestrate more predictable tissue regeneration.
Due to the limited space of a Feature Article we primarily reviewed our own progress in this topic area and certain closely related works. We apologize for not being able to provide an exhaustive review and not being able to cite more publications from other groups.
5. Conclusions
Tissue regeneration entails the successful interplay between cells, biological signals, and biomaterials. It requires the fundamental understandings in both life sciences and materials sciences to develop successful regeneration technologies. With the advent of nanotechnology, enormous advances have been made in the field of biomaterials science and engineering. Various biomaterials including nanofibers, nanocrystals, nanopores, nanospheres, and other nano features have been developed. Nanofibrous biomaterials, mimicking the nano fibrillar structure of the natural extracellular matrix, have been demonstrated to mediate protein interactions and cell function. Nanocomposite biomaterials, mimicking the composition and structure of mineralized tissues, provide excellent mechanical properties besides favorable biological properties. Nanospheres/nanoparticles are designed and incorporated into 3D nanostructured tissue engineering scaffolds to achieve temporally and spatially controlled deliveries of biological molecules, which mimic the signaling cascades in natural development and repair of a living system. Nano-structured biomaterials are providing novel solutions in regenerative medicine and are expanding exponentially with time.
Biography
Dr. Peter X. Ma received his BS in Polymer Chemistry and Chemical Engineering from Tsinghua University (Beijing, China), and Ph.D. in Polymer Science and Engineering from Rutgers University. He then did his postdoctoral research at MIT and Harvard Medical School on Biomaterials and Tissue Engineering. In 1996, he joined the faculty of the University of Michigan, in the Department of Biologic and Materials Sciences (School of Dentistry), the Department of Biomedical Engineering, and the Macromolecular Science and Engineering Center (College of Engineering). He is currently a tenured full professor. He received a DuPont Young Professor Award in 2000, and was elected to Fellow of the American Institute of Medical and Biological Engineering in 2006.

Footnotes
References
- [1].Karp JM, Langer R. Curr. Opin. Biotechnol. 2007;18:454. doi: 10.1016/j.copbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- [2].Dumitriu S. Polymeric Biomaterials. Marcel Dekker, Inc.; New York: 1996. [Google Scholar]
- [3].Maskarinec SA, Tirrell DA. Curr. Opin. Biotechnol. 2005;16:422. doi: 10.1016/j.copbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- [4].Hertz A, Bruce IJ. Nanomed. 2007;2:899. doi: 10.2217/17435889.2.6.899. [DOI] [PubMed] [Google Scholar]
- [5].Malard O, Espitalier F, Bordure P, Daculsi G, Weiss P, Corre P. Expert Rev. Med. Devices. 2007;4:729. doi: 10.1586/17434440.4.5.729. [DOI] [PubMed] [Google Scholar]
- [6].Ma PX. Mater. Today. 2004;7:30. [Google Scholar]
- [7].Ma PX. In: Encyclopedia of Polymer Science and Technology. Kroschwitz JI, editor. John Wiley & Sons; New Jersey: 2004. [Google Scholar]
- [8].Muschler GF, Nakamoto C, Griffith LG. J. Bone Joint Surg. Am. 2004;86A:1541. doi: 10.2106/00004623-200407000-00029. [DOI] [PubMed] [Google Scholar]
- [9].Roy TD, Simon JL, Ricci JL, Rekow ED, Thompson VP, Parsons JR. J. Biomed. Mater. Res. 2003;66A:283. doi: 10.1002/jbm.a.10582. [DOI] [PubMed] [Google Scholar]
- [10].Lu JX, Flautre B, Anselme K, Hardouin P, Gallur A, Descamps M, Thierry B. J. Mater. Sci. Mater. Med. 1999;10:111. doi: 10.1023/a:1008973120918. [DOI] [PubMed] [Google Scholar]
- [11].Ishaug-Riley SL, Crane-Kruger GM, Yaszemski MJ, Mikos AG. Biomaterials. 1998;19:1405. doi: 10.1016/s0142-9612(98)00021-0. [DOI] [PubMed] [Google Scholar]
- [12].Tsuruga E, Takita H, Itoh H, Wakisaka Y, Kuboki Y. J. Biochem. (Tokyo) 1997;121:317. doi: 10.1093/oxfordjournals.jbchem.a021589. [DOI] [PubMed] [Google Scholar]
- [13].Ma PX. Adv. Drug Del. Rev. 2008;60:184. doi: 10.1016/j.addr.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Streicher RM, Schmidt M, Fiorito S. Nanomed. 2007;2:861. doi: 10.2217/17435889.2.6.861. [DOI] [PubMed] [Google Scholar]
- [15].Traykova T, Aparicio C, Ginebra MP, Planell JA. Nanomed. 2006;1:91. doi: 10.2217/17435889.1.1.91. [DOI] [PubMed] [Google Scholar]
- [16].Hasirci V, Vrana E, Zoplutuna P, Ndreu A, Yilgor P, Basmanav FB, Aydin E. J. Biomater. Sci. Polymer. Ed. 2006;17:1241. doi: 10.1163/156856206778667442. [DOI] [PubMed] [Google Scholar]
- [17].Tabata Y. Methods Mol. Biol. 2005;300:81. doi: 10.1385/1-59259-858-7:081. [DOI] [PubMed] [Google Scholar]
- [18].Desai TA. Med. Eng. Phys. 2000;22:595. doi: 10.1016/s1350-4533(00)00087-4. [DOI] [PubMed] [Google Scholar]
- [19].Woodson M, Liu J. Phys. Chem. Chem. Phys. 2007;9:207. doi: 10.1039/b610651j. [DOI] [PubMed] [Google Scholar]
- [20].Betancourt T, Brannon-Peppas L. Int. J. Nanomedicine. 2006;1:483. doi: 10.2147/nano.2006.1.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Elsdale T, Bard J. J. Cell Biol. 1972;54:626. doi: 10.1083/jcb.54.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Strom SC, Michalopoulos G. Methods Enzymol. 1982;82A:544. doi: 10.1016/0076-6879(82)82086-7. [DOI] [PubMed] [Google Scholar]
- [23].Grinnell F, Bennett MH. Methods Enzymol. 1982;82(Pt A):535. doi: 10.1016/0076-6879(82)82085-5. [DOI] [PubMed] [Google Scholar]
- [24].Kuntz RM, Saltzman WM. Biophys. J. 1997;72:1472. doi: 10.1016/S0006-3495(97)78793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Reneker DH, Chun I. Nanotech. 1996;7:216. [Google Scholar]
- [26].Matthews JA, Wnek GE, Simpson DG, Bowlin GL. Biomacromolecules. 2002;3:232. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- [27].Xu CY, Inai R, Kotaki M, Ramakrishna S. Biomaterials. 2004;25:877. doi: 10.1016/s0142-9612(03)00593-3. [DOI] [PubMed] [Google Scholar]
- [28].Nair LS, Bhattacharyya S, Laurencin CT. Expert Opin. Biol. Ther. 2004;4:659. doi: 10.1517/14712598.4.5.659. [DOI] [PubMed] [Google Scholar]
- [29].Yang F, Murugan R, Wang S, Ramakrishna S. Biomaterials. 2005;26:2603. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- [30].Boland ED, Wnek GE, Simpson DG, Pawlowski KJ, Bowlin GL. J. Macromol. Sci. Pure Appl. Chem. 2001;38:1231. [Google Scholar]
- [31].Yoshimoto H, Shin YM, Terai H, Vacanti JP. Biomaterials. 2003;24:2077. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- [32].Zong XH, Ran SF, Kim KS, Fang DF, Hsiao BS, Chu B. Biomacromolecules. 2003;4:416. doi: 10.1021/bm025717o. [DOI] [PubMed] [Google Scholar]
- [33].Luu YK, Kim K, Hsiao BS, Chu B, Hadjiargyrou M. J.Controlled Rel. 2003;89:341. doi: 10.1016/s0168-3659(03)00097-x. [DOI] [PubMed] [Google Scholar]
- [34].Li C, Vepari C, Jin HJ, Kim HJ, Kaplan DL. Biomaterials. 2006;27:3115. doi: 10.1016/j.biomaterials.2006.01.022. [DOI] [PubMed] [Google Scholar]
- [35].Baker RW. Membrane technology and applications. Wiley; New York: 2004. [Google Scholar]
- [36].Zhang R, Ma PX. J. Biomed. Mater. Res. 1999;44:446. doi: 10.1002/(sici)1097-4636(19990315)44:4<446::aid-jbm11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- [37].Wei GB, Ma PX. Biomaterials. 2004;25:4749. doi: 10.1016/j.biomaterials.2003.12.005. [DOI] [PubMed] [Google Scholar]
- [38].Zhang R, Ma PX. J. Biomed. Mater. Res. 2000;52:430. doi: 10.1002/1097-4636(200011)52:2<430::aid-jbm25>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- [39].Chen VJ, Smith LA, Ma PX. Biomaterials. 2006;27:3973. doi: 10.1016/j.biomaterials.2006.02.043. [DOI] [PubMed] [Google Scholar]
- [40].Woo KM, Chen VJ, Ma PX. J. Biomed. Mater. Res. 2003;67:531. doi: 10.1002/jbm.a.10098. [DOI] [PubMed] [Google Scholar]
- [41].Chen VJ, Ma PX. Biomaterials. 2004;25:2065. doi: 10.1016/j.biomaterials.2003.08.058. [DOI] [PubMed] [Google Scholar]
- [42].Ma PX, Zhang R. J. Biomed. Mater. Res. 1999;46:60. doi: 10.1002/(sici)1097-4636(199907)46:1<60::aid-jbm7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- [43].Chen VJ, Ma PX. Biomaterials. 2006;27:3708. doi: 10.1016/j.biomaterials.2006.02.020. [DOI] [PubMed] [Google Scholar]
- [44].Wei GB, Ma PX. J. Biomed. Mater. Res. 2006;78:306. doi: 10.1002/jbm.a.30704. [DOI] [PubMed] [Google Scholar]
- [45].Whitesides GM, Grzybowski B. Science. 2002;295:2418. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- [46].Zhang SG. Biotech. Adv. 2002;20:321. [Google Scholar]
- [47].Ball P. Nature. 1994;367:323. doi: 10.1038/367323a0. [DOI] [PubMed] [Google Scholar]
- [48].Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- [49].Hartgerink JD, Beniash E, Stupp SI. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5133. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Beniash E, Hartgerink JD, Storrie H, Stupp SI. Acta Biomater. 2005;1:387. doi: 10.1016/j.actbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- [51].Holmes TC, de Lacalle S, Su X, Liu GS, Rich A, Zhang SG. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6728. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhang SG, Holmes T, Lockshin C, Rich A. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3334. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhang SG, Holmes TC, Dipersio CM, Hynes RO, Su X, Rich A. Biomaterials. 1995;16:1385. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- [54].Liu G. Adv. Mater. 1997;9:437. [Google Scholar]
- [55].Liu GJ, Qiao LJ, Guo A. Macromolecules. 1996;29:5508. [Google Scholar]
- [56].Liu DJ, De Feyter S, Coltler M, Wiesler UM, Weil T, Herrmann A, Mullen K, De Schryver FC. Macromolecules. 2003;36:8489. [Google Scholar]
- [57].Liu DJ, Zhang H, Grim PCM, De Feyter S, Wiesler UM, Berresheim K, Mullen K, De Schryver FC. Langmuir. 2002;18:2385. [Google Scholar]
- [58].Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- [59].Duan Y, Wang Z, Yan W, Wang S, Zhang S, Jia J. J. Biomater. Sci. Polym. Ed. 2007;18:1153. doi: 10.1163/156856207781554019. [DOI] [PubMed] [Google Scholar]
- [60].Ma Z, He W, Yong T, Ramakrishna S. Tissue Eng. 2005;11:1149. doi: 10.1089/ten.2005.11.1149. [DOI] [PubMed] [Google Scholar]
- [61].Liu X, Won Y, Ma PX. J. Biomed. Mater. Res. 2005;74A:84. doi: 10.1002/jbm.a.30367. [DOI] [PubMed] [Google Scholar]
- [62].Liu X, Won Y, Ma PX. Biomaterials. 2006;27:3980. doi: 10.1016/j.biomaterials.2006.03.008. [DOI] [PubMed] [Google Scholar]
- [63].Liu X, Smith LA, Wei GB, Won Y, Ma PX. J. Biomed. Nanotech. 2005;1:54. [Google Scholar]
- [64].Zhu H, Ji J, Shen J. Biomacromolecules. 2004;5:1933. doi: 10.1021/bm049753u. [DOI] [PubMed] [Google Scholar]
- [65].Zhu H, Ji J, Tan Q, Barbosa MA, Shen J. Biomacromolecules. 2003;4:378. doi: 10.1021/bm025773p. [DOI] [PubMed] [Google Scholar]
- [66].Filmon R, Basle MF, Atmani H, Chappard D. Bone. 2002;30:152. doi: 10.1016/s8756-3282(01)00634-2. [DOI] [PubMed] [Google Scholar]
- [67].Woo KM, Jun J, Chen VJ, Seo J, Baek JH, Ryoo HM, Kim GS, Somerman MJ, Ma PX. Biomaterials. 2007;28:335. doi: 10.1016/j.biomaterials.2006.06.013. [DOI] [PubMed] [Google Scholar]
- [68].Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. Journal of Neuroscience. 2008;28:3814–23. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Li W-J, Jiang YJ, Tuan RS. Tissue Engineering. 2006;12:1775–85. doi: 10.1089/ten.2006.12.1775. [DOI] [PubMed] [Google Scholar]
- [70].Rho JY, Kuhn-Spearing L, Zioupos P. Med. Eng. Phys. 1998;20:92. doi: 10.1016/s1350-4533(98)00007-1. [DOI] [PubMed] [Google Scholar]
- [71].Landis WJ. Bone. 1995;16:533. doi: 10.1016/8756-3282(95)00076-p. [DOI] [PubMed] [Google Scholar]
- [72].Li SH, de Groot K, Layrolle P. Key Eng. Mater. 2002;218:25. [Google Scholar]
- [73].Sun LM, Berndt CC, Gross KA, Kucuk A. J. Biomed. Mater. Res. 2001;58:570. doi: 10.1002/jbm.1056. [DOI] [PubMed] [Google Scholar]
- [74].LeGeros RZ. Clin. Orthop. Rel. Res. 2002;395:81. doi: 10.1097/00003086-200202000-00009. [DOI] [PubMed] [Google Scholar]
- [75].Webster TJ, Ergun C, Doremus RH, Siegel RW, Bizios R. J Biomed Mater Res. 2000;51:475–83. doi: 10.1002/1097-4636(20000905)51:3<475::aid-jbm23>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- [76].Wang RZ, Cui FZ, Lu HB, Wen HB, Ma CL, Li HD. J. Mater. Sci. Lett. 1995;14:490. [Google Scholar]
- [77].Du C, Cui FZ, Feng QL, Zhu XD, Groot K. d. J. Biomed. Mater. Res. 1999;44:407. doi: 10.1002/(sici)1097-4636(19990315)44:4<407::aid-jbm6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- [78].Du C, Cui FZ, Feng QL, Zhu XD, Groot K. d. J. Biomed. Mater. Res. 1998;42:540. doi: 10.1002/(sici)1097-4636(19981215)42:4<540::aid-jbm9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- [79].Cui FZ, Du C, Su XW, Zhu XD, Zhao NM. Cells Mater. 1996;6:31. [Google Scholar]
- [80].Ma PX, Zhang R, Xiao G, Franceschi R. J. Biomed. Mater. Res. 2001;54:284. doi: 10.1002/1097-4636(200102)54:2<284::aid-jbm16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- [81].Webster TJ, Ergun C, Dpremus RH, Siegel RW, Bizios R. J. Biomed. Mater. Res. 2000;51:475. doi: 10.1002/1097-4636(20000905)51:3<475::aid-jbm23>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- [82].Zhang R, Ma PX. J. Biomed. Mater. Res. 1999;45:285. doi: 10.1002/(sici)1097-4636(19990615)45:4<285::aid-jbm2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- [83].Zhang R, Ma PX. Macromol. Biosci. 2004;4:100. doi: 10.1002/mabi.200300017. [DOI] [PubMed] [Google Scholar]
- [84].Harrison BS, Atala A. Biomaterials. 2007;28:344. doi: 10.1016/j.biomaterials.2006.07.044. [DOI] [PubMed] [Google Scholar]
- [85].Wang SF, Shen L, Zhang WD, Tong YJ. Biomacromolecules. 2005;6:3067. doi: 10.1021/bm050378v. [DOI] [PubMed] [Google Scholar]
- [86].Jell G, Verdejo R, Safina L, Shaffer SP, Stevens MM, Bismarck A. J. Mater. Chem. 2008 In press. [Google Scholar]
- [87].MacDonald RA, Laurenzi BF, Viswanathan G, Ajayan PM, Stegemann JP. J. Biomed. Mater. Res. 2005;74A:489. doi: 10.1002/jbm.a.30386. [DOI] [PubMed] [Google Scholar]
- [88].Supronowicz PR, Ajayan PM, Ullmann KR, Arulanandam BP, Metzger DW, Bizios R. J. Biomed. Mater. Res. 2002;59:499. doi: 10.1002/jbm.10015. [DOI] [PubMed] [Google Scholar]
- [89].Babensee JE, McIntire LV, Mikos AG. Pharm. Res. 2000;17:497. doi: 10.1023/a:1007502828372. [DOI] [PubMed] [Google Scholar]
- [90].Reddi AH. Nat. Biotech. 1998;16:247. doi: 10.1038/nbt0398-247. [DOI] [PubMed] [Google Scholar]
- [91].Lee H, Cusick RA, Browne F, Kim TH, Ma PX, Utsunomiya H, Langer R, Vacanti JP. Transplantation. 2002;73:1589. doi: 10.1097/00007890-200205270-00011. [DOI] [PubMed] [Google Scholar]
- [92].Lee VHL. Crc. Crit. Rev. Ther. Drug Carrier Sys. 1988;5:69. [PubMed] [Google Scholar]
- [93].Langer R. Science. 1990;249:1527. doi: 10.1126/science.2218494. [DOI] [PubMed] [Google Scholar]
- [94].Morley P, Whitfield JF, Willick GE. Curr. Pharm. Des. 2001;7:671. doi: 10.2174/1381612013397780. [DOI] [PubMed] [Google Scholar]
- [95].Ferrara N, Alitalo K. Nat. Med. 1999;5:1359. doi: 10.1038/70928. [DOI] [PubMed] [Google Scholar]
- [96].Schwendeman SP. Crit Rev Ther Drug Carrier Syst. 2002;19:73–98. doi: 10.1615/critrevtherdrugcarriersyst.v19.i1.20. [DOI] [PubMed] [Google Scholar]
- [97].Wei GB, Pettway GJ, McCauley LK, Ma PX. Biomaterials. 2004;25:345. doi: 10.1016/s0142-9612(03)00528-3. [DOI] [PubMed] [Google Scholar]
- [98].Uludag H, D’Augusta D, Golden J, Li J, Timony G, Riedel R, Wozney JM. J. Biomed. Mater. Res. 2000;50:227. doi: 10.1002/(sici)1097-4636(200005)50:2<227::aid-jbm18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- [99].Oldham JB, Lu L, Zhu X, Porter BD, Hefferan TE, Larson DR, Currier BL, Mikos AG, Yaszemski MJ. J.Biomech. Eng. Trans. ASME. 2000;122:289. doi: 10.1115/1.429662. [DOI] [PubMed] [Google Scholar]
- [100].Wei G, Jin Q, Giannobile WV, Ma PX. J Control Rel. 2006;112:103. doi: 10.1016/j.jconrel.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Miyamoto S, Takaoka K, Okada T, Yoshikawa H, Hashimoto J, Suzuki S, Ono K. Clin. Orthop. Relat. Res. 1992:274. [PubMed] [Google Scholar]
- [102].Tamura S, Kataoka H, Matsui Y, Shionoya Y, Ohno K, Michi KI, Takahashi K, Yamaguchi A. Bone. 2001;29:169. doi: 10.1016/s8756-3282(01)00498-7. [DOI] [PubMed] [Google Scholar]
- [103].Whang K, Tsai DC, Nam EK, Aitken M, Sprague SM, Patel PK, Healy KE. J. Biomed. Mater. Res. 1998;42:491. doi: 10.1002/(sici)1097-4636(19981215)42:4<491::aid-jbm3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- [104].Sheridan MH, Shea LD, Peters MC, Mooney DJ. J Control Rel. 2000;64:91. doi: 10.1016/s0168-3659(99)00138-8. [DOI] [PubMed] [Google Scholar]
- [105].Wei G, Jin Q, Giannobile WV, Ma PX. Biomaterials. 2007;28:2087. doi: 10.1016/j.biomaterials.2006.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jin Q, Wei G, Lin Z, Sugai JV, Lynch SE, Ma PX, Giannobile WV. PLoS One. 2008;3:1729. doi: 10.1371/journal.pone.0001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ziegler J, Mayr-Wohlfart U, Kessler S, Breitig D, Gunther KP. J. Biomed. Mater. Res. 2002;59:422. doi: 10.1002/jbm.1258. [DOI] [PubMed] [Google Scholar]
- [108].Franceschi RT. J. Dent. Res. 2005;84:1093. doi: 10.1177/154405910508401204. [DOI] [PubMed] [Google Scholar]
- [109].Rasubala L, Yoshikawa H, Nagata K, Iijima T, Ohishi M. Br. J. Oral Maxillofac. Surg. 2003;41:173. doi: 10.1016/s0266-4356(03)00075-5. [DOI] [PubMed] [Google Scholar]
- [110].Wei GB. Biomedical Engineering. The University of Michigan; Ann Arbor: 2006. Growth factor-delivering nano-fibrous scaffolds for bone tissue regeneration. [Google Scholar]