Abstract
To detect the positions of disease loci, lod scores are calculated at multiple chromosomal positions given trait and marker data on members of pedigrees. Exact lod score calculations are often impossible when the size of the pedigree and the number of markers are both large. In this case, a Markov Chain Monte Carlo (MCMC) approach provides an approximation. However, to provide accurate results, mixing performance is always a key issue in these MCMC methods. In this paper, we propose two methods to improve MCMC sampling and hence obtain more accurate lod score estimates in shorter computation time. The first improvement generalizes the block-Gibbs meiosis (M) sampler to multiple meiosis (MM) sampler in which multiple meioses are updated jointly, across all loci. The second one divides the computations on a large pedigree into several parts by conditioning on the haplotypes of some ‘key’ individuals. We perform exact calculations for the descendant parts where more data are often available, and combine this information with sampling of the hidden variables in the ancestral parts. Our approaches are expected to be most useful for data on a large pedigree with a lot of missing data.
Key Words: Meiosis indicator, Block-Gibbs MCMC, Haplotype update, Multiple meiosis update
Introduction
In this paper we develop improved methods for accurate estimation of lod scores for genetic linkage analyses [1], using data jointly at multiple genetic markers on members of extended pedigrees with substantial missing information. To map the genes underlying complex diseases, the usual strategy has been to first localize genes to regions on a scale of centiMorgans (cM) using data on pedigrees. Fine-scale mapping using associations between genotype and phenotype can then identify candidate genes. A recent example is the study of age-related macular degeneration (AMD) [2].
The development of genotyping technology and publicly available information on human genetic variation such as that provided by the HapMap project [3] have made possible the direct testing of associations between genotype and phenotype on a genome-wide scale. Such genome-wide association studies (GWA) raise the possibility of identifying disease-related genes from large samples of unrelated individuals, which are much more easily obtained than are pedigree data. However, it remains to be determined how far GWA studies can replace pedigree linkage analyses. As yet, GWA studies have provided conflicting results in comprehensive nationwide studies of Parkinson disease [4,5,6,7,8,9] and of obesity [10,11,12,13]. Problems of population structure and of genetic heterogeneity, at both the locus and allelic level, impact GWA severely but have little impact on pedigree analyses. Further, the power of linkage analysis is substantially increased by joint analysis of marker data at multiple polymorphic markers observed on members of extended pedigrees ascertained for multiple affected individuals [14]. Thus it remains important to develop computationally efficient methods for obtaining accurate lod scores using such data.
To obtain linkage lods scores, we assume availability of data on the genotypes at multiple linked genetic marker loci (YM) and trait characteristics (YT) of some subset of the members of extended pedigrees. Genotyping error is not considered in this paper. The genetic marker model, including the genetic map of the marker loci and the population frequencies of marker alleles, is assumed known. We also consider a single-locus model for the trait data YT. The parameter of interest is then the location γ of the trait locus on the chromosome of the markers, with γ = ∞ denoting that the trait locus is not on this chromosome. The lod score is the log-likelihood statistic
| (1) |
since, in the absence of linkage, trait data YT and marker data YM are independent. On extended pedigrees with a substantial proportion of missing observations, a major challenge remains the computation of this lod score [15]. Note that the subscript γ is dropped when a probability, such as P(YT), does not depend on the location of trait locus.
When exact computation is infeasible or impractical, Markov chain Monte Carlo (MCMC) methods provide a way to estimate the lod score [16, 17]. Where there are multiple genetic marker loci, both exact and Monte Carlo methods use some form of meiosis indicators [18] or inheritance vectors [19] to achieve the computation. The inheritance of DNA at any locus can be specified by binary meiosis indicators Sij, with Sij = 0 or 1 as in meiosis i at locus j the maternal or paternal DNA of the parent is transmitted to the offspring. Here i = 1, …, m indexes the meioses in the pedigree. We assume that the linked marker loci j = 1, …, n are ordered along the chromosome. Let SM denote the meiosis indicators relating to all the markers. Then the probability required for the lod score (1) may be written
The form of equation (2) shows the challenge for exact computation when m and n are both large; SM consists of mn binary indicators. Equation (2) also shows how Monte Carlo estimation may be achieved through sampling SM conditionally on YM and averaging the resulting values of Pγ(YT ∣ SM) for each γ of interest.
There are many ways to sample the components of SM conditionally on YM using MCMC, but among the simplest and most effective are block Gibbs samplers. The locus (or L) sampler [20] jointly resamples {Sij; i = 1, …, m} successively over loci j. The meiosis (or M) sampler jointly resamples {Sij; j = 1,…, n} successively over meioses i. These two block-Gibbs samplers may be combined to form the LM-sampler [21] which has been the mainstay of our MCMC lod score estimation approaches [22]. The LM-sampler, together with the estimation approach indicated by equation (2), forms the basis of the MORGAN [23] program lm_markers. In comparisons with other software, the lm_markers programs has been found to be quite competitive for lod score estimation on extended pedigrees [24]. We therefore take lm_markers (MORGAN V2.8.1) as the base-point for comparison of the improved MCMC methods presented in this paper. Our new programs are also implemented in the MORGAN package.
In this paper, we describe two ways in which MCMC sampling may be improved in order to obtain more accurate lod score estimates more efficiently. Our first improvement is a generalization of the block-Gibbs meiosis (M) sampler in which multiple meioses are updated jointly, across all loci. The simplest proposal is to update some randomly chosen subset of k meioses, which can be achieved in time of order k2k using the factored Hidden Markov Model (HMM) method [25]. Alternative proposals are to update specific subsets, such as the two meioses of an individual, the maternal and/or paternal meioses in a sibship, or the meioses of a 3-generation pedigree segment. Where the number of such meioses is too large for simple computation a restricted sampling of such updates has been implemented, similar to that of Thomas et al. [26]. These proposals have been implemented in the program lm_multiple released in version 2.8.2 of MORGAN package [23].
The second method augments the latent state space with the haplotypes of a few ‘key’ individuals. Sobel and Lange [27] describe two sets of latent variables, descent graphs and descent states. Descent graphs are equivalent to the meiosis indicators Sij which define the descent of DNA at every locus j. Descent states specify in addition the haplotypes of founders: the alleles carried on each founder chromosome. The founder allelic types and the meiosis indicators together determine the genotypes of all individuals. Thus use of descent states makes computations simpler, but the much larger and more constrained space impairs the mixing performance of the MCMC. Our approach specifies the haplotypic states not of the founders but of key pedigree members who divide the pedigree. Segments of the pedigree are then independent conditional on these haplotypes, permitting either exact computation and independent realizations or MCMC methods to be performed on each segment and the results combined. Both the multiple-meiosis sampling and the augmentation with haplotypes of key individuals are implemented in the program lm_haplotype within the framework of the MORGAN version 2.8 package [23]. The program lm_haplotype is not yet publicly released.
In this paper we first provide the details of these generalizations of our MCMC methods. We then compare the performance of the three MORGAN [23] programs lm_markers, lm_multiple and lm_haplotype, using simulated data at 10 linked markers on a 52-member pedigree. In our simulations and in the paper, we use sex-averaged genetic maps, but this is for ease of presentation only. As with all MORGAN programs [23], these three programs can equally use gender-specific maps. On an extended pedigree, with a substantial portion of missing data, and when marker loci are tightly linked, the lm_markers program can perform poorly. We show how lm_multiple improves estimation of a multipoint lod score, and lm_haplotype provides further improvement. For comparison with exact results using VITESSE [28], we use just 4 markers on the full 52-member pedigree. We also compare the results for all 10 markers on a 14-member subset of the pedigree, using MERLIN [29]. We show that, using our new programs, MCMC is both an accurate and a computationally efficient approach to computation of multipoint lod scores.
Methods
Meiosis Indicators, Recombination, and Genetic Maps
We first review the probability model and conditional independence structure of the meiosis indicators Sij. This structure underlies all multilocus computations and sampling methods. At any single locus j, the meiosis indicators Sij, i = 1, …, m are independent. However, for each meiosis i, the Sij, j = 1, …, n are dependent. In meiosis i, recombination occurs between two loci j and l if Sij ≠ Sil. That is, the genes segregating to the offspring are from different grandparents. At any two loci j and l, the pairwise distribution of (Sij, Sil) is determined by the recombination rate θjl between the two loci. That is,
For loci that are close together on a chromosome, θjl is close to 0. For independently segregating loci, such as loci on different chromosomes, θjl = 1/2. Although, in reality and in our software, recombination rates may differ between male and female meioses, for simplicity we use sex-averaged rates in this paper.
To define the joint distribution of Sij over j = 1, …, n, an additional assumption is required. In this paper, we assume absence of genetic interference so that {Sij}j = 1, …, n are independent Markov chains (i = 1, …, m). In this case, the Haldane map function [30] provides the conversion between the genetic distance between j and l and the recombination rate θjl. Let S.j = (S1j, …, Smj) denote the meiosis indicators and Y.j denote the observed marker genotype data at locus j over the whole pedigree. At locus j, conditional on meiosis indicators S.j, the observed data Y.j are independent of observed data and meiosis indicators at other loci.
This hidden Markov structure permits use of the factored HMM method [25] to obtain the forward cumulative probabilities αj(S.j) = P(S.j ∣ Y.1,…, Y.j) for j = 1, …, n in time of order mn2m. Then S.n may be resampled from αn(S.n). For j = n − 1, …, 1, S.j may be successively resampled from P(S.j ∣ S.j+ 1, Y.1,…, Y.j), which may be written as
since S.j+ 1 is independent of Y.1, …, Y.j conditional on S.j due to hidden Markov structure.
Multiple Meiosis Sampler
Except on small pedigrees, the number of meioses m is too large for the above exact forward computation and backwards sampling to be feasible. Thus, only a small subset of the total set of meioses can be sampled jointly. MCMC procedures resample a subset of the indicators Sij conditional on the data and current values of the remaining indicators. An MCMC iteration (or scan) consists of a sequence of such resampling steps repeated until all the indicators have been resampled. In the sampling implemented in the programs lm_markers and lm_multiple only inheritance vectors at marker loci are resampled. Thus each MCMC scan provides the next realization of SM. Once a sequence of realizations of SM is obtained, equation (2) then provides a Monte Carlo estimate of the lod score as a function of the location of the trait locus.
At each MCMC iteration, our programs first randomly determine whether the scan is to be by locus (L-sampler) or by meiosis (M-sampler). Our new multiple meiosis (MM) sampler is a generalization of the M-sampler of Thompson and Heath [21] in which meioses are updated singly. For a subset I of meioses, we write S.j = (SIj, S–Ij), where SIj (S–Ij) denotes meiosis indicators in (not in) subset I at locus j. Instead of one meiosis i, MM-sampler jointly resamples {SIj; j = 1, …, n} successively over subsets I of meioses conditionally on {S–Ij; j = 1, …, n} and the marker data YM. The factored HMM structure holds for this subset of meioses, so that the method of Fishelson and Geiger [25] can be applied for the calculations.
We have defined several possible subsets I: their benefits for MCMC mixing performance are considered further in the Discussion. Specifically, we consider subsets I such as
1 random update: a random subset of the meioses, or
2 individual update: both maternal and paternal meioses for an individual, or
3 complete sib update: all the meioses from parents to children in a nuclear family, or
4 complete 3-generation update: all the meioses from grandparents to parents and from parents to children in a 3-generation family, which is defined as a nuclear family together with any grandparents present in the pedigree.
At each MCMC iteration of sampling inheritance indicators, a sampling type of a random update, individual update, sib update or 3-generation update is chosen according to probabilities pr, pi, ps and p3, where pr + pi + ps + p3 = 1. If a random update is chosen, then a random integer will first determine the size of this subset of meioses. For example, if the allowed maximum number of meioses in a subset is 8, then a number will be selected from the integers from 1 to 8 each with probability 1/8. This number of meioses will be selected randomly from the overall set of meioses and the indicators are updated according to its posterior distribution. Then a second integer from 1 to 8 will be randomly selected and a second subset of meioses randomly selected from the remaining meioses (excluding the ones previously selected), and so on until all the meiosis indicators are updated. If an individual update is chosen, the two meioses of each individual are updated jointly for each individual of the pedigree in a random order. Similarly, if a sib or 3-generation update is chosen, then the meioses included in these subsets are jointly updated, with the subsets being updated in random order. The subsets in a sib update are disjoint while the subsets in a 3-generation update can overlap since the grandparents in a 3-generation family can be parents or children in other 3-generation families.
Restricted Multiple-Meiosis Updates
The computational time is exponential in the number k of meioses in subset I. When k is too large, it could be computationally expensive or even impossible to do a complete sib or 3-generation update. In this case, a restricted sib or 3-generation update is possible.
For the restricted sib update, consider I to be the set of meioses from parents to children in a nuclear family. Write I = (Im, If), where Im and If are the maternal and paternal meioses in subset I. Given a current realization of meiosis indicators sI. = (sI1, …, sIn), for each locus j = 1, …, n, we define Xj to be an indicator function of flipping paternal (or maternal) meiosis indicators or not in the next updating proposal. Specifically, we have
Similarly, when the complete 3-generation update is impractical, a restricted 3-generation update can be applied. Consider I to be the set of meioses in a three generational family of grandparents, parents and children. Let sI = (sIm, sIf, sIc) be the current realization of meiosis indicators in subset I, where sIm and sIf are the maternal and paternal meiosis indicators from grandparents to parents, and sIc are meiosis indicators from parents to children. For j = 1, …, n, we define Xj to be the following
That is, there are two restricted choices to update meiosis indicators in I: (1) the same as current realization (2) swapping maternal and paternal meiosis indicators for meioses from grandparents to parents and flipping all the meiosis indicators for meioses from parents to children.
We show in the Appendix that {Xj}j = 1, …, n is a Markov chain for the case of restricted sib update. An analogous result holds for the case of the restricted 3-generation update. Hence, given data Y.j and fixed S–Ij for j = 1, …, n, we retain the HMM structure with the {Xj}j = 1, …, n replacing the much larger space of SIj. Since each Xj takes only 2 or 4 values, it is possible to jointly sample X = (X1, …, Xn) for all the marker loci in time of order n instead of the previous nk2k. The total number of choices over all n loci is then 4n or 2n at each resampling step, providing a wide space of potential updated meiosis indicators. Updates similar to our restricted updates were proposed by Thomas et al. [26], for a single marker locus. However, they did not demonstrate how all the loci can be updated jointly.
Haplotype Sampler: Combination of Exact and Approximate Calculations
Conditional on the complete haplotypes at marker and trait loci of individuals who divide the pedigree into subsets of individuals, data on the different pedigree subsets are independent. The conditional independence of pedigree segments given the haplotypes of individuals who divide the pedigree is the basis of the Elston-Stewart algorithm [31] for likelihood computations on pedigrees. However, here we have multiple marker loci, so our method of computation on each pedigree segment is based on the Lander-Green approach [19]. We augment the space of hidden variables with the haplotypes of such ‘key’ individuals. If a subset separated by a ‘key’ individual is small, exact computation is feasible. In genetic studies, data are more often available for extant individuals at the bottom of the pedigree, constraining the haplotypes of immediate ancestors. It is therefore practical and efficient to do exact calculation on small subpedigrees of current individuals, taking as a ‘key’ individual the ancestor who connects the subpedigree to the remainder of the pedigree.
The haplotypes used to augment the space of hidden variables include the trait locus in addition to the marker loci. Without loss of generality, assume the trait locus T is between the dth and d+1st marker loci. Then, for each meiosis i, {Sij}j = 1, …, d,T,d+1,…,n is a Markov chain, and the trait data depend only on the trait model and the inheritance at the trait locus. That is, the HMM structure and all the computational and sampling methods of the previous sections follow exactly as before, with the trait locus now being included.
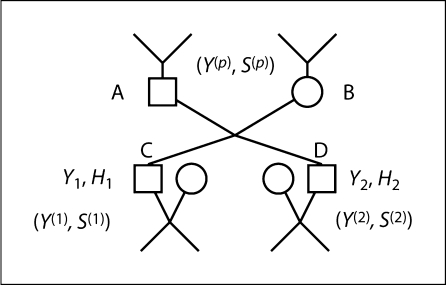
For example, in the pedigree segment shown in figure 1, let Y1 = (Y1T, Y1M) and Y2 = (Y2T, Y2M) be the observed marker and trait data of ‘key’ individuals C and D, and H1 and H2 be the pairs of haplotypes of C and D who are offspring of the couple A and B. The total pedigree is then divided into three parts. The first descendant pedigree consists of C, C's descendants and all relatives of C's spouses. The second descendant pedigree is defined analogously for D. Finally, the ancestral pedigree consists of the remainder of the pedigree, including C and D themselves. Let Y(p), S(p) be data and meiosis indicators involved in the ancestral pedigree, Y(1); S(1) (Y(2), S(2)) be data and meiosis indicators in the 1st (2nd) descendant pedigree. Note that, since the key individuals themselves are in both descendant and ancestral pedigree segments, Y1 is included in both Y(1) and Y(p), and Y2 is included in both Y(2) and Y(p). Then the probability distribution of the observed data Y can be written as
| (3) |
Fig. 1.
An example of part of a complete pedigree, which includes ancestors and collateral relatives of individuals A and B, and descendants and siblings of individuals C and D. The two children C and D of the couple A and B are chosen as ‘key’ individuals, dividing the pedigree into three subpedigrees. For further details, see text.
Notice that in equation (3) the first term Pγ(Y(1) ∣ Y1, H1) and second one Pγ(Y(2) ∣ Y2, H2) are the same type, which only involves data in descendant pedigrees. The term Pγ(Y(p), S(p)) considers ancestral pedigree only. The last term P(H1, H2 ∣ Y(p), S(p)) is a connection between descendant and ancestral pedigrees. We analyze these terms and explain in detail how to calculate them in the following four paragraphs.
First, consider the term Pγ(Y(1) ∣ Y1, H1), or equivalently Pγ(Y(2) ∣ Y2, H2). This part is independent of S(p), the meiosis indicators in the ancestral pedigree. Now
The probability for the first descendant part Pγ(Y(1)) can either be calculated exactly or, if it is too large, estimated by MCMC. For a fixed genetic map, Pγ(Y(1)) is calculated only once summing over all possible values of H1 and H2. The prior probability, P(H1), of the ordered haplotype pair H1, is assumed to be the product of the allele frequencies at each marker/trait locus. The multilocus penetrance probability P(Y1 ∣ H1) is a product over the marker and trait loci, 1 or 0 for genotypic data, or a more general penetrance for discrete or quantitative trait data. When P(Y1 ∣ H1) = 0, Pγ(Y(1) ∣ Y1, H1) is defined to be 0. The haplotype pair h1 can be sampled according to the posterior distribution Pγ(H1 ∣ Y(1)). When the number of possible haplotypes is not large, the full distribution Pγ(H1 ∣ Y(1)) can be calculated exactly. However, when the exact calculation is infeasible, we have
| (4) |
Equation (4) indicates that we can first sample s(1) conditional on Y(1). We then sample h1, locus by locus, conditional on Y(1) and a realization of S(1).
Second, consider the term Pγ(Y(p), S(p)) in equation (3). We use MCMC to sample s(p) conditional on data Y(p) using the new MM-sampler to improve mixing. Now
Thus we may sample s(p) = (sM(p), sT(p)) by first sampling sM(p) from Pγ(SM(p) ∣ YM(p)) and then sT(p) from Pγ(ST(p) ∣ sM(p)). The trait-locus penetrance probability on the ancestral pedigree, P(YT(p) ∣ sT(p)), can be easily calculated for a given sT(p). The last term P(YM(p)) is free of parameter γ and the values of S(p), H1 and H2. Thus to obtain a lod score, it is not necessary to calculate this term.
Finally, consider the term P(H1, H2 ∣ Y(p), S(p)) in equation (3). Similarly to equation (4), this probability can be easily calculated locus by locus
To summarize, we estimate the likelihood of equation (3) by first sampling h1∼ Pγ(H1 ∣ Y(1)) and h2 ∼ Pγ(H2 ∣ Y(2)); then sampling sM(p) ∼ P(SM(p) ∣ YM(p)), and sT(p) ∼ Pγ(ST(p) ∣ sM(p)); finally calculating
The average of this quantity over MCMC realizations provides an estimate of Pγ(Y ∣ YM(p)), which is proportional to the likelihood
Of course, this method is not limited to the special case of two ‘key’ individuals who are siblings. One can choose any small number of individuals anywhere in the pedigree as ‘key’ individuals. However, different choices of ‘key’ individuals can make a difference in the efficiency of estimation. As a guideline, one should choose individuals whose descendants have enough information to at least partially restrict the space of haplotypes of each key individual. Additionally, the procedure is much more effective if exact computation is used on the descendant pedigree segments. Thus, the number of each individual's descendants should be not large so that the computation of the exact haplotype distribution is infeasible.
The methods of this section have been implemented in the program lm_haplotype. In this program, a subset of ‘key’ individuals must be predefined by users. Then the program lm_haplotype will automatically divide the whole pedigree into parts and calculate each part exactly or approximately according to the size of each part and the maximum number of meiosis allowed for exact calculations. When Monte Carlo approximation is necessary, the MM-sampling of lm_multiple, rather than the M-sampler of lm_markers, is applied. The final results are then combined as described above and estimates of lod scores are returned.
Results
Set Up for Simulation and Analysis
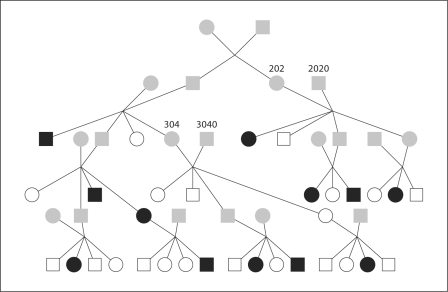
We illustrate our methods on the single pedigree, ped52, shown in figure 2. The pedigree has 52 individuals in 5 generations: 12 individuals are founders, 32 are observed, 12 are affected, and 20 are unaffected. There are 80 meioses to be sampled. Data were simulated at 10 linked marker loci, labeled from 1 to 10, at chromosomal positions (0, 10, 20, 28, 29, 31, 40, 50, 60, 61) cM. Each marker locus has 4 alleles, with allele frequencies (0.4, 0.3, 0.2, 0.1). The trait locus is simulated at position 30 cM. Note that the trait locus is in a region of tightly linked markers: markers 4, 5 and 6 and the trait locus present a particular challenge for MCMC methods. The trait locus has 2 alleles with frequencies (0.5, 0.5). An affectation status with penetrances (0.95, 0.6, 0.05) is simulated. In the analysis, trait-locus genotypes are unobserved, but affectation status is available for each of the 32 observed individuals. The assumption of ‘known’ phenotypes of unaffected individuals results in a stronger linkage signal compared to many real data analyses, where often only the more clearly defined ‘affected’ phenotype is specified. However, this fact has no (in lm_multiple) or little (in lm_haplotype) effect on the mixing performance of MCMC.
Fig. 2.
Ped52 pedigree used for simulation of data. The individuals in grey are not observed – both marker genotype and affectation status are missing. The white individuals are unaffected and the black ones are affected.
All the programs were run on a Dell Precision 360 workstation with Pentium 4 (3 GHz) processor, 2 GB memory and Red Hat Enterprise Linux 4 WS system. In all three MCMC programs, we use 3K (3,000) sequential imputation realizations to obtain the initial realization of meiosis indicators SM. The value chosen is the realization that gives the highest value of P(YT ∣ SM). In lm_markers and lm_multiple, the probabilities for L and M sampling are 0.5 and 0.5 respectively. Sampling is by scan in both lm_markers and lm_multiple. When lm_multiple selects an M-sample scan, the probabilities for individual, sib, 3-generation and random updates are 0.3, 0.3, 0.2 and 0.2 respectively. In the random update, the number k of meioses jointly updated is uniformly distributed from 1 to 8. In the sib and 3-generation updates, the maximum allowed number of meioses for complete updating is 8. That is, when the number of involved meioses in these updates is greater than 8, restricted updates are applied. For lm_haplotype, whenever MCMC sampling is necessary on either ancestral or descendant parts of the pedigree, lm_multiple is used with the same parameter values as above.
To choose the ‘optimal’ key individuals for lm_haplotype, 50 short lm_multiple runs (each of length 5K MCMC scans) are performed on some subpedigrees of ped52. For example, individuals 202, 2,020 and their descendants would be one such subpedigree (fig. 2). Another example subpedigree would be individuals 304, 3040 and their descendants. The idea is that special attention should be paid to the subpedigrees that give significant contribution to the overall lod scores but have large variation among independent runs. Using this method, we find that in this example, the subpedigree that might be the source of large variation is the one consisting of individuals 202, 2020 and their descendants. There are 14 individuals and 20 meioses in this subpedigree. For lm_haplotype, we then choose individual 202 as the key individual and sample her haplotype, conditional the data on this subpedigree. The computation time for this preliminary analysis is not included in table 2.
Table 2.
Comparison of programs: computation time in seconds
| Pedigree | Exact | # of MCMC scans | MCMC programs |
||
|---|---|---|---|---|---|
| mrk | mul | hap | |||
| ped52-4 | VITESSE (19,000) | 10K | 49 | 137 | 145 |
| 30K | 128 | 404 | 420 | ||
| 1M | 4,112 | 12,467 | 12,530 | ||
| ped14-10 | MERLIN (2) | 10K | 17 | 34 | 38 |
| 30K | 46 | 98 | 106 | ||
| ped52-10 | – | 10K | 86 | 317 | 332 |
| 30K | 223 | 931 | 945 | ||
Each MCMC computation time is an average over the corresponding set of 50 runs of table 1. The computation time of VITESSE and MERLIN are shown in parentheses in the exact column.
Ped52 with 4 Markers
Our aim is to obtain accurate MCMC lod score estimates for large pedigrees with multiple markers. However, on ped52 we first consider markers 3, 4, 6, 7 only (ped52-4), in order to make possible a comparison with exact computations.
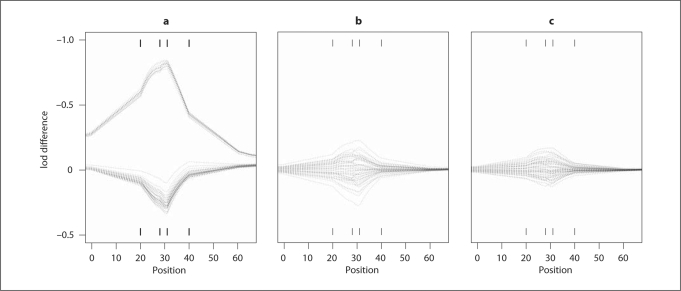
The exact lod scores are calculated using VITESSE [28] and is shown in figure 5 (dashed line). The differences between exact lod scores and estimated ones from 50 runs using the three MCMC programs lm_markers, lm_multiple and lm_haplotype (30K MCMC scans for each run) are shown in figure 3. For lm_markers (fig. 3a), there are two clear separate clusters of estimates, indicating poor MCMC mixing. Therefore, the estimated lod scores are not accurate and reliable in this case. To determine whether a longer run can overcome this mixing problem, we choose 10 runs from the upper cluster and 10 runs from the lower one and extend the number of MCMC scans from 30K to 1M (1,000,000) for each run. All the 20 runs are still in their original cluster after such a long run, although the between-run variation of lod scores within each cluster does decrease compared to the lod score estimates based on 30K MCMC scans (results not shown). This indicates that the lm_markers sampling procedure has negligible chance of moving between subsets of SM that provide lod scores in different clusters in this example.
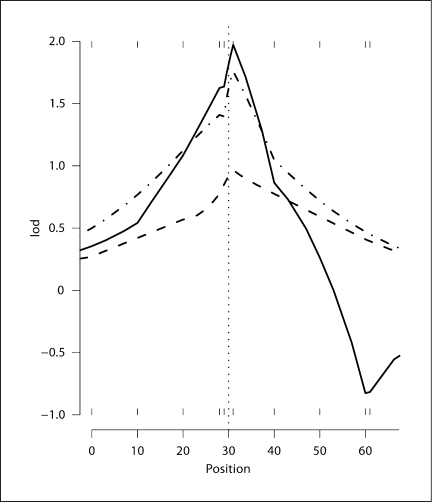
Fig. 5.
The solid line is the estimated lod score curve for ped52-10 using lm_haplotype with 1M scans. The dot-dashed line is the estimated lod score curve for ped52-5 at positions 20, 28, 29, 31, 40 cM and dashed line is the exact lod score curve for ped52-4 at positions 20, 28, 31, 40 cM. The vertical dotted line at 30 cM represents the position of simulated trait locus.
Fig. 3.
Differences between exact lod scores and MCMC estimates for ped52 with 4 markers at positions 20, 28, 31 and 40 cM. The exact lod scores are calculated using VITESSE. From left to right, the three MCMC programs used in estimation are lm_markers (a), lm_multiple (b), lm_haplotype (c). For each program, the number of runs is 50. For each run, the number of scans is 30K.
Figure 3b shows that lm_multiple successfully overcomes the mixing difficulties and obtains lod scores varying around true lod-score curve. Figure 3c shows that lm_haplotype not only overcomes the mixing difficulties of lm_markers, but also has smaller variation compared to lm_multiple. In the worst scenarios of bad choices of key individuals, lm_haplotype performs at least as well as lm_multiple (results not shown).
To see how the number of MCMC scans affects the final estimates, we also did 50 short runs (10K scans per run), 50 long runs (30K scans per run) and 50 extremely long runs (1M scans per run). For a given position, we consider two measures of accuracy and precision of the estimated lod score: the discrepancy which is the mean (over runs) of the absolute difference from the truth, and the range which is the difference between maximum and minimum over runs. For each program, these 2 statistics are calculated at three positions: marker-3, marker-6, and marker-7. The results are shown in table 1.
Table 1.
Comparison of programs: lod score accuracy and variation
| Pedigree | Position (lod) | # of MCMC scans | Discrepancy | Range |
||||
|---|---|---|---|---|---|---|---|---|
| mrk | mul | hap | mrk | mul | hap | |||
| ped52-4 | Marker-3 (0.568) | 10K | 0.217 | 0.073 | 0.050 | 0.827 | 0.342 | 0.248 |
| 30K | 0.249 | 0.044 | 0.032 | 0.754 | 0.245 | 0.163 | ||
| 1M |
0.156 |
0.008 |
0.008 |
0.675 |
0.042 |
0.034 |
||
| Marker-6 (0.965) | 10K | 0.405 | 0.143 | 0.095 | 1.231 | 0.594 | 0.487 | |
| 30K | 0.439 | 0.082 | 0.057 | 1.195 | 0.504 | 0.299 | ||
| 1M |
0.335 |
0.014 |
0.012 |
1.091 |
0.081 |
0.071 |
||
| Marker-7 (0.776) | 10K | 0.140 | 0.040 | 0.028 | 0.554 | 0.176 | 0.131 | |
| 30K | 0.162 | 0.023 | 0.017 | 0.510 | 0.148 | 0.095 | ||
| 1M | 0.093 | 0.004 | 0.004 | 0.472 | 0.023 | 0.020 | ||
| ped14-10 | Marker-3 (0.224) | 10K | 0.012 | 0.006 | 0.004 | 0.056 | 0.032 | 0.024 |
| 30K |
0.007 |
0.004 |
0.003 |
0.038 |
0.021 |
0.014 |
||
| Marker-6 (0.646) | 10K | 0.027 | 0.012 | 0.007 | 0.135 | 0.065 | 0.041 | |
| 30K |
0.020 |
0.008 |
0.006 |
0.122 |
0.042 |
0.031 |
||
| Marker-7 (0.636) | 10K | 0.018 | 0.010 | 0.006 | 0.108 | 0.055 | 0.039 | |
| 30K | 0.016 | 0.006 | 0.004 | 0.120 | 0.034 | 0.026 | ||
The number 10K means 10,000 and 1M means 1 million. The numbers included in parentheses in the position column are exact lod scores calculated by VITESSE or MERLIN at these positions. mrk, mul and hap represent lm-markers, lm-multiple and lm-hap-plotype respectively.
From table 1 we see that for ped52 with 4 markers, both lm_haplotype and lm_multiple have much smaller discrepancy than lm_markers. For example, for the long runs at marker-6, lm_haplotype, lm_multiple and lm_markers have discrepancies 0.057, 0.082, 0.439 respectively, which are 6, 8 and 45% of the true lod score 0.965 at this position. With extremely long runs, the discrepancies for these three programs decreased to 0.012, 0.014, 0.335 respectively. However, the discrepancy for lm_markers (0.335) is still too large to be reliable. This shows that lm_markers can not overcome the mixing difficulties even with extremely long runs (1M scans). Moreover, lm_haplotype and lm_multiple also have smaller range than lm_markers in most cases.
For a given number of MCMC scans, the computation time for lm_haplotype is about the same as for lm_multiple and about three times the time for lm_markers (table 2). That is, a short run (10K scans) of lm_multiple or lm_haplotype uses about the same computation time as a long run (30K scans) of lm_markers. By comparison, note that exact computation using VITESSE [28] takes much longer time to compute the lod scores at the same points as the MCMC programs estimate them (table 2). On this pedigree, four 4-allele markers is the limit of computation feasibility for VITESSE, showing the importance of having accurate and reliable MCMC methods.
For the three MCMC programs, a fair comparison with respect to time is thus of the long run results using lm_markers with the short run results using lm_multiple and lm_haplotype. Table 1 shows that even short runs (10K scans) of lm_haplotypes and lm_multiple perform much better than long runs (30K scans) of lm_markers, with both smaller discrepancy and smaller range over runs. Notice that, in this example, more MCMC scans of lm_markers do not necessarily increase accuracy or decrease variability because of poor mixing of the Markov chain. However, increasing the number of MCMC scans for lm_haplotype or for lm_multiple does decrease both the discrepancy and the range.
Ped14 with 10 Markers
To use all 10 markers and to compare the performance of these with exact calculation, we can use only a small part (ped14–10) of the pedigree. We consider the right part of ped52 consisting of founders 202, 2,020 and their descendants: 14 individuals in total (see fig. 2). MERLIN [29] can compute exact lod scores on this subpedigree. For each of the three MCMC programs, the discrepancy and range at marker-3, at marker-6, and at marker-7 are shown in table 1. Comparing the long run results from lm_markers with short run results from lm_multiple and lm_haplotype, we find that the latter two programs always have smaller discrepancy and less variation (table 1). In fact, in this example, lm_markers requires about 50% more computation time for 30K MCMC scans than do the other two programs for 10K scans (table 2).
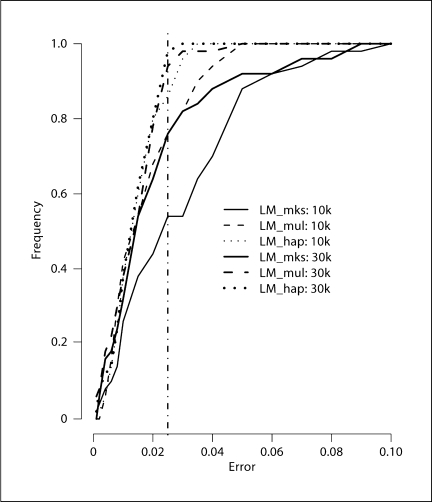
A further comparison of the accuracy of MCMC estimates as compared to exact results from MERLIN is shown in figure 4. For all three MCMC programs, and for both short and long runs, this figure shows the empirical CDF (over 50 runs) of absolute error in lod score at the true trait locus (position 30 cM, at the midpoint between marker-5 and marker-6). For example, consider the number of runs (out of 50) with this error less than or equal to 0.025, which is 4.8% of the true lod score of 0.518. For short and long runs, respectively, this number is about 27 and 38 for lm_markers, 38 and 47 for lm_multiple, and 43 and 49 for lm_haplotypes. In this example, lm_markers does not have the obvious mixing problem that it had on the full pedigree; there are no distinct clusters of lod score estimates among the 50 short or long runs. However, lm_multiple and lm_haplotype still perform better, providing more accurate estimates with less variability in shorter time.
Fig. 4.
Empirical CDF of errors of lod scores at true trait locus using lm_markers, lm_multiple and lm_haplotype.
Ped52 with 10 Markers
We estimate the lod score for the complete ped52 pedigree with all 10 markers (ped52-10) using these three MCMC programs. As before, we obtain 50 independent long runs (30K MCMC scans) for each program. Now, no comparison with exact results is possible, so the discrepancy measure is not obtainable. The lod score ranges at marker-6, for lm_markers, lm_multiple and lm_haplotype, are 0.466, 0.059, and 0.032, respectively. As before we get the smallest range for lm_haplotype and the largest range for lm_markers. Comparing with the corresponding results for ped52-4 (table 1), we see that there is much less between-run variation in the lod score using all 10 markers.
To check how the between-run variation in the lod score is affected by markers included in the analysis, we estimate the lod score for the complete ped52 pedigree with 5 markers of 3, 4, 5, 6, 7 (ped52-5). Exact results are not possible here. Using the results from 50 independent long runs (30K MCMC scans) for each program (results not shown), the lod score ranges at marker-6, for lm_markers, lm_multiple and lm_haplotype, are 0.307, 0.031, and 0.021, respecitvely. The between run variations are not only less than the ones for ped52 using 4 markers, but also less than the ones for ped52 using all 10 markers. This shows that marker-5 plays an important role in estimating lod scores at marker-6.
Finally, the lod-score curve is estimated for ped52-10 using our best program lm_haplotype with 1M scans (fig. 5). For comparison, figure 5 shows also the exact lod scores for ped52-4 and the estimated lod scores for ped52-5. The estimated lod score curve for ped52-5 is the average over the 50 long runs using lm_haplotype. It takes about 2 hours to get the MCMC-based ped52-10 lod score curve. Based on the consistent performance of lm_haplotype for ped52-4 and ped14-10, we are confident that this lod-score curve is reliable. The maximum lod score 1.972 is obtained at marker-6 for ped52-10, which is more than the maximum lod score of 1.771 from ped52-5 and about twice of the maximum lod score from ped52-4. This shows that the increase in maximum lod score is mainly, but not entirely, due to the marker data at marker 5. For extended pedigrees with many missing data, we thus see that joint use of data at multiple markers increases the power to detect linkage.
Discussion
We have developed new MCMC methods for accurate estimation of multilocus likelihoods using pedigree data. MCMC methods make linkage analysis feasible for large pedigree data when exact computational methods cannot be applied (ped52-10). Even in the case that exact computation is feasible, MCMC methods can give reasonable results using much less computational time (ped52-4).
Compared to the lm_markers method, our new MCMC methods improve the accuracy of lod score estimates and decrease the variation of lod score estimates between runs. The procedure lm_multiple outperforms lm_markers by jointly considering multiple meioses, and lm_haplotypes outperforms lm_multiple by additionally considering haplotypes of some individuals.
By updating jointly over loci, a single meiosis sampler (M-sampler) avoids problems of poor mixing due to tight linkage [21], but inheritances in different meioses are jointly constrained by data. Further, although the L-sampler and hence also the LM-sampler are irreducible, the M-sampler alone can never be guaranteed irreducible. Clearly, the random, individual, complete sib and 3-generation updates improve mixing; they may sometimes even ensure irreducibility of the sampler. Often the joint updating of meioses within nuclear families may be sufficient to improve mixing, whereas the random update could (in principle) choose meioses whose inheritances are independent given the data. In this extreme case, the MM-update process is equivalent to M-sampler updates of this block of meioses but requires longer computation time. However, as shown by the example of Sobel and Lange [27], two meioses well separated in the pedigree may be jointly constrained by the data on descendants, causing reducibility of single-meiosis updates. Thus it seems advisable to always give positive probability to the updating of randomly chosen subsets, and not restrict the process only to local sib or even 3-generation updates.
In lm_multiple, the probabilities for random, individual, sib and 3-generation updates are pre-defined. Higher probability of complete sib and 3-generation updates may provide better estimates of lod scores because the meiosis indicators are jointly updated according to full conditional distribution. However, it remains to be investigated whether the gains outweigh the additional computational burden. Similarly, for lm_haplotype, exact calculations can be done for small parts of the pedigree. However, exact calculation is computationally intensive and the choice of how much exact computation to do is always a compromise between accuracy and time.
In lm_haplotype, the choice of key individuals has effects on both the performance of MCMC and the computation time required. In principle, any individual can be a key individual. However, we would like to do as much exact calculation as possible since this computation can be done once only, and the more exact computation is done the smaller the Monte Carlo variation in the lod score estimate. Thus we prefer to choose a key individual that has a deeper descendant pedigree provided exact computation can still be done on this sub-pedigree. In the current analysis, we use short runs in partial pedigrees to find the underlying problematic part of the pedigree. Although this worked well here, more theoretical exploration is needed to support generalization. An automated procedure to choose optimal key individuals is also desirable.
All the three simulation studies are based on data from simple structured pedigree ped52. For complicated pedigree with loops, lm_markers and lm_multiple can be used without any modification, although loops may result in poor mixing in some cases. To use lm_haplotype, a little bit more modification in likelihood equation in equation (3) is needed, when some key individuals and their descendants actually form a loop. More investigation is needed on the effect of loops on MCMC mixing performance.
The examples of this paper have used microsatellite type markers, but increasingly data are available for large numbers of dense SNP markers. For a given number of MCMC scans, both lm_markers and lm_multiple have computation time linear in the number of marker loci n. The program lm_haplotype is also linear in n in the case that the haplotypes of key individuals are sampled conditional on realizations of meiosis indicators in sub-pedigrees. Wijsman et al. [24] have shown that lm_markers can obtain accurate lod score estimates using dense simulated SNP marker data. In an analysis of the real SNP data of GAW15 [32], lm_multiple shows better performance than lm_markers. Therefore, we are optimistic on the general performance of lm_multiple using dense SNP data. More work is needed to evaluate the performance of lm_haplotype on SNP data.
Although it is always very challenging to deal with real, large pedigrees with multiple tightly linked marker loci and a lot of missing data, our new methods are promising in improving the performance over previous MCMC methods.
Acknowledgements
This work is supported by NIH grant GM-46255. The authors are very grateful to a referee whose many detailed and constructive comments have much improved the paper.
Appendix 1
1. Proof of Markov Property in Restricted Sib Update
For j = 1, …, n, let s*Ij = (s*Imj, s*Ifj) be one of the four possible choices based on current realization sIj (flip or not for paternal or maternal meiosis indicators). Then for kj ∈ {0, 1, 2, 3},
where g(k) is a function of k. This indicates that {Xj}j = 1, …, n is a Markov chain. Thus, using the HMM algorithm of Baum and Petrie [33] and Rabiner [34], we are able to sample P(X) jointly over n marker loci using block Gibbs sampling method in time of order O(n).
References
- 1.Ott J. Analysis of Human Genetic Linkage. ed 3. Baltimore, MD: The Johns Hopkins University Press; 1999. [Google Scholar]
- 2.Abecasis GR, Yashar BM, Zhao Y, Ghiasvand NM, Zareparsi S, Branham KEH, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–494. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Hapmap Consortium A haplotype map of the human genome. Nature. 2005;237:1299–1319. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PVK, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers RH. Considerations for genomewide association studies in Parkinson disease. Am J Hum Genet. 2006;78:1081–1082. doi: 10.1086/504730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarimon J, Scholz S, Fung HC, Hardy J, Eerola J, Hellstrm O, Chen CM, Wu YR, Tienari PJ, Singleton A. Conicting results regarding the semaphorin gene (SEMA5A) and the risk for Parkinson disease. Am J Hum Genet. 2006;78:1082–1084. doi: 10.1086/504727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrer MJ, Haugarvoll K, Ross OA, Stone JT, Milkovic NM, Cobb SA, Whittle AJ, Lincoln SJ, Hulihan MM, Heckman MG, White LR, Aasly JO, Gibson JM, Gosal D, Lynch T, Wszolek ZK, Uitti RJ, Toft M. Genomewide association, Parkinson disease, and PARK10. Am J Hum Genet. 2006;78:1084–1088. doi: 10.1086/504728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goris A, Williams-Gray CH, Foltynie T, Compston DAS, Barker RA, Sawcer SJ. No evidence for association with Parkinson disease for 13 single-nucleotide polymorphisms identified by whole-genome association screening. Am J Hum Genet. 2006;78:1088–1090. doi: 10.1086/504726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Rowland C, Schrodi S, Laird W, Tacey K, Ross D, Leong D, Catanese J, Sninsky J, Grupe A. A case-control association study of the 12 single-nucleotide polymorphisms implicated in Parkinson disease by a recent genome scan. Am J Hum Genet. 2006;78:1090–1092. doi: 10.1086/504725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 11.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkau B, Froguel P. Comment on ‘a common genetic variant is associated with adult and childhood obesity’. Science. 2007;315:187. doi: 10.1126/science.1129402. [DOI] [PubMed] [Google Scholar]
- 12.Loos RJF, Barroso I, O'Rahilly S, Wareham NJ. Comment on ‘a common genetic variant is associated with adult and childhood obesity’. Science. 2007;315:187. doi: 10.1126/science.1130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosskopf D, Bornhorst A, Rimmbach C, Schwahn C, Kayser A, Kruger A, Tessmann G, Geissler I, Kroemer HK, Volzke H. Comment on ‘a common genetic variant is associated with adult and childhood obesity’. Science. 2007;315:187. doi: 10.1126/science.1130571. [DOI] [PubMed] [Google Scholar]
- 14.Wijsman EM, Amos CI. Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: Summary of gaw10 contributions. Genet Epidemiol. 1997;14:719–735. doi: 10.1002/(SICI)1098-2272(1997)14:6<719::AID-GEPI28>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 15.Silberstein M, Tzemach A, Dovgolesky N, Fishelson M, Schuster A, Geiger D. Online system for faster multipoint linkage analysis via parallel executation on thousands of personal computers. Am J Hum Genet. 2006;78:922–935. doi: 10.1086/504158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange K, Sobel E. A random walk method for computing genetic location scores. Am J Hum Genet. 1991;49:1320–1334. [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson EA. Monte Carlo likelihood in genetic mapping. Stat Sci. 1994;9:355–366. doi: 10.1098/rstb.1994.0073. [DOI] [PubMed] [Google Scholar]
- 18.Donnelly KP. The probability that related individuals share some section of genome identical by descent. Theor Popul Biol. 1983;23:34–63. doi: 10.1016/0040-5809(83)90004-7. [DOI] [PubMed] [Google Scholar]
- 19.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Nat Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson EA, Heath SC. Estimation of conditional multilocus gene identity among relatives. In: Seillier-Moiseiwitsch F, editor. Statistics in Molecular Biology and Genetics: Selected Proceedings of a 1997 Joint AMS-IMS-SIAM Summer Conference on Statistics in Molecular Biology, IMS Lecture Note – Monograph Series Volume 33. Hayward, CA: Institute of Mathematical Statistics; 1999. pp. 95–113. [Google Scholar]
- 22.Thompson EA. MCMC in the analysis of genetic data on pedigrees. In: Liang F, Wang JS, Kendall W, editors. Markov Chain Monte Carlo: Innovations and Applications. Singapore: World Scientific Co Pte Ltd; 2005. pp. 183–216. [Google Scholar]
- 23.MORGAN: a package for Markov chain Monte Carlo in Genetic Analysis (Version 2.8). http://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan. shtml, 2005.
- 24.Wijsman EM, Rothstein JH, Thompson EA. Multipoint linkage analysis with many multiallelic or dense diallelic markers: Markov chain Monte Carlo provides practical approaches for genome scans on general pedigrees. Am J Hum Genet. 2006;79:846–858. doi: 10.1086/508472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischelson M, Geiger D. Optimizing exact linkage computations. J Comput Biol. 2004;11:263–275. doi: 10.1089/1066527041410409. [DOI] [PubMed] [Google Scholar]
- 26.Thomas A, Gutin A, Abkevich V. Multilocus linkage analysis by blocked Gibbs sampling. Stat Comput. 2000;10:259–269. [Google Scholar]
- 27.Sobel E, Lange K. Descent graphs in pedigree analysis: Applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell JR, Weeks DE. The algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nature Genet. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- 29.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene ow trees. Nature Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 30.Haldane JBS. The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet. 1919;8:229–309. [Google Scholar]
- 31.Elston RC, Stewart J. A general model for the analysis of pedigree data. Hum Hered. 1971;21:523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- 32.Sung YJ, Di Y, Fu AQ, Rothstein JH, Sieh W, Tong L, Thompson EA, Wijsman EM: Comparison of multipoint linkage analyses for quantitative traits: Parametric lod scores, variance components lod scores and bayes factors in the ceph data. Biomed Central Genet 2007; in press. [DOI] [PMC free article] [PubMed]
- 33.Baum LE, Petrie T. Statistical inference for probabilistic functions of finite state markov chains. Ann Math Stat. 1966;37:1554–1563. [Google Scholar]
- 34.Rabiner LR. A tutorial on hidden markov models and selected applications in speech recognition. Proc IEEE. 1989;77:257–286. [Google Scholar]