Abstract
Background
Ecologists and evolutionary biologists are becoming increasingly interested in networks as a framework to study plant–animal mutualisms within their ecological context. Although such focus on networks has brought about important insights into the structure of these interactions, relatively little is still known about the mechanisms behind these patterns.
Scope
The aim in this paper is to offer an overview of the mechanisms influencing the structure of plant–animal mutualistic networks. A brief summary is presented of the salient network patterns, the potential mechanisms are discussed and the studies that have evaluated them are reviewed. This review shows that researchers of plant–animal mutualisms have made substantial progress in the understanding of the processes behind the patterns observed in mutualistic networks. At the same time, we are still far from a thorough, integrative mechanistic understanding. We close with specific suggestions for directions of future research, which include developing methods to evaluate the relative importance of mechanisms influencing network patterns and focusing research efforts on selected representative study systems throughout the world.
Key words: Ant–plant interactions, forbidden links, mutualism, neutrality, trait matching, plant–animal interactions, pollination, seed dispersal
INTRODUCTION
Mutualisms between plants and animals pervade nature. Pollination, seed dispersal and ant–plant protection mutualisms are key ecological processes in many terrestrial ecosystems throughout the world. Thus, the study of plant–animal mutualistic interactions is important both for a basic understanding of ecological systems and for their management and conservation (Bronstein et al., 2006; Waser and Ollerton, 2006; Rico-Gray and Oliveira, 2007).
Like any other kind of ecological interaction, plant–animal interactions occur in a community context. Ecologists and evolutionary biologists have realized that it is necessary to study interactions in their community context to draw valid conclusions about ecological and evolutionary processes (Thompson, 1994, 2005; Waser et al., 1996; Strauss and Irwin, 2004). The study of ecological interaction networks – schematics of who interacts with whom in a community (Ings et al., 2009) – is thus a key approach for understanding ecological and evolutionary processes in the ecological context in which they occur.
The study of mutualistic networks has brought about important insights regarding the organization of plant–animal mutualisms. Several apparently general topological features have been described, including the skewed distribution of links per species (many ‘specialists’ and few extreme ‘generalists’; Waser et al., 1996; Jordano et al., 2003; Vázquez and Aizen, 2003), the nested organization of the interaction matrix (Bascompte et al., 2003) and the frequent occurrence of asymmetric interactions (Vázquez and Aizen, 2004; Bascompte et al., 2006). These structural properties have potentially important consequences for ecological and evolutionary processes. For example, the frequent occurrence of asymmetric interactions suggests a low potential for ecological and co-evolutionary coupling of the dynamics of interacting populations of plants and pollinators (Vázquez and Aizen, 2004); furthermore, a nested organization makes networks highly vulnerable to the extinction of species with many links and robust to the extinction of species with few links (Memmott et al., 2004).
Several non-mutually exclusive mechanisms have been proposed to explain the origin of the above network patterns, including neutrality, trait matching among interacting species, phylogenetic constraints and sampling artefacts (Jordano et al., 2003; Burns, 2006; Rezende et al., 2007b; Santamaría and Rodríguez-Gironés, 2007; Stang et al., 2007; Vázquez et al., 2007; Blüthgen et al., 2008). However, relatively little is still known about the relative importance of these mechanisms, and there has been little effort to synthesize this work scattered in the literature. Our aim here is to offer such a synthesis.
We review the evidence on the mechanisms that have been identified as potential determinants of the structure of plant–animal mutualistic networks. We first present a brief overview of the salient patterns described so far. We then discuss the potential mechanisms, trying to go beyond simplistic dicotomies and organizing them into a complex, hierarchical causal model. Finally, a review of the evidence evaluating these mechanisms is presented. As this review will show, researchers of plant–animal mutualisms have made substantial progress in the understanding of the processes behind the patterns observed in mutualistic networks. However, at the same time, we are still far from a thorough mechanistic understanding. It is hoped this review will help pinpoint the unresolved issues and identify avenues for future research.
A BRIEF OVERVIEW OF STRUCTURAL FEATURES OF MUTUALISTIC NETWORKS
Recent years have seen an upsurge of studies of mutualism in a network context. Several properties of mutualistic networks have been described repeatedly enough to qualify as generalizations. In this section we offer a brief summary of these structural features. This summary is not intended to be an in-depth review of network patterns, but rather a short guide for readers with little experience with this literature. We also offer a glossary of network terms used in here (Table 1) to complement this short guide. Readers should consult the references cited below and the recent review by Bascompte and Jordano (2007) for more details on network structural patterns.
Table 1.
A glossary of network terms used in the text. Key references are given for further reading
| Binary/unweighted network | A network in which only the presence/absence of links is recorded (Jordano et al., 2003; Bascompte and Jordano, 2007). |
| Compartmentalization | The existence of clearly defined groups of species (compartments or modules) with many intragroup links and few intergroup links (Dicks et al., 2002; Olesen et al., 2007). |
| Connectance | The proportion of potential links that are actually realized (Jordano, 1987). |
| Degree | The number of species to which a species is linked (Jordano et al., 2003). A measure of ecological specialization. |
| Degree asymmetry | The imbalance in the degree of two interacting species (Vázquez and Aizen, 2004). |
| Degree distribution | The frequency histogram of the degree of the species in a network (Jordano et al., 2003). |
| Dependence | An estimate of the extent to which a species depends on another species (Jordano, 1987; Bascompte et al., 2006). See also interaction strength. |
| Interaction neutrality | The occurrence of interactions resulting from the random encounter of individuals, so that all individuals have the same probability of interacting, regardless of their identity (Vázquez et al., 2007; Krishna et al., 2008). |
| Interaction strength | An estimate of the ecological impact of one species on another (Vázquez et al., 2005; Bascompte et al., 2006). Note that ‘dependence’ of a species A on another species B is usually defined as the interaction strength of B on A (Bascompte et al. 2006). |
| Link | An interespecific interaction, usually used for binary networks (Jordano, 1987). |
| Nestedness | The tendency of specialized species to interact with a subset of the interaction partners of more generalized species (Bascompte et al., 2003); the degree of symmetry in the distribution of unexpected absences and presences around the boundary line defining perfect nestedness (Almeida-Neto et al., 2007). |
| Quantitative/weighted network | A network in which links are given a weight, usually a measure of interaction strength (Memmott, 1999; Bascompte et al., 2006). |
| Sampling effects | The distortion of ‘true’ network patterns by sampling/observation errors (Vázquez and Aizen, 2006; Nielsen and Bascompte, 2007). |
| Strength/dependence asymmetry | The imbalance in the interaction strength/dependence of two interacting species (Bascompte et al., 2006). |
| Trait matching | The occurrence of interactions resulting from the matching of the phenotypic traits of interacting species (Stang et al., 2006, 2007, 2009; Santamaría and Rodríguez-Gironés, 2007). There are two types of matching: trait complementarity (e.g. flower colour or nectar sugar concentration correspond with the pollinator's preferences) and exploitation barriers or thresholds (e.g. long corollas exclude flower visitors with short proboscises). |
Only a few of the potential interspecific interactions actually occur
In other words, using network jargon, connectance – the proportion of potential interspecific interactions that are actually realized – is low in mutualistic networks (Jordano, 1987; Olesen and Jordano, 2002), as has been observed in predator–prey networks (‘food webs’; Montoya and Solé, 2003).
There is an imbalance in the number of species of plants and animals in the network
That is, most plant–animal mutualistic networks deviate from a 1 : 1 ratio in the species richness of plants and animals. This deviation is strong for plant–pollinator and ant–nectar networks, with almost four times more animal than plant species, but more modest for plant–seed disperser and ant–myrmecophyte networks (with animal : plant ratios of 1·2 and 1·6, respectively; Blüthgen et al., 2007; Guimarães et al., 2007a).
Most species have few links, few have many links
In network parlance, there is a right-skewed distribution of degree (the number of other species to which a given species is connected; Waser et al., 1996; Jordano et al., 2003; Vázquez and Aizen, 2003). As observed in other ecological and non-ecological networks (Barabási and Albert, 1999; Dunne et al., 2002), some mutualistic networks exhibit a power-law degree distribution (a decaying straight line in a log–log plot of number of species per degree category vs. degree) while a majority exhibit a ‘truncated power-law’ (a straight line in a log–log plot with a sharp cut-off at high degree values; Jordano et al., 2003). The presence of a cut-off means that the frequency of species with an extremely high number of links is lower than under a pure power-law distribution.
Most links are weak, few are strong
As observed in food webs, interaction strength in mutualistic networks is also right-skewed (Bascompte et al., 2006). Interaction strength – sometimes also called ‘dependence’ (Jordano, 1987; Bascompte et al., 2006) – is usually approximated using interaction frequency (Vázquez et al., 2005; Sahli and Conner, 2006), which has allowed moving from binary to quantitative networks (Bascompte et al., 2006; Blüthgen et al., 2006a, 2007; Vázquez et al., 2007).
Most interactions are asymmetric
Plant–animal mutualistic interactions are usually asymmetric, in terms of both degree and strength (Bascompte et al., 2003, 2006; Vázquez and Aizen, 2004). Degree asymmetry means that species with low degree (‘specialists’) tend to interact with highly connected species (‘generalists’; Vázquez and Aizen, 2004). Strength asymmetry means that species with strong effects usually experience weak reciprocal effects from their interaction partners (Bascompte et al., 2006).
Mutualistic networks tend to be nested
Most mutualistic networks exhibit a significant degree of nestedness – the tendency of little connected species to interact with a subset of interaction partners of highly connected species (Bascompte et al., 2003). Nestedness implies both a high occurrence of degree asymmetry and the existence of a network ‘core’ – a relatively small group of highly connected species (Bascompte et al., 2003).
Mutualistic networks tend to be compartmentalized
Many mutualistic networks exhibit some degree of compartmentalization or modularity – the existence of clearly defined groups of species (compartments or modules) with many intragroup links and few intergroup links (Dicks et al., 2002; Guimarães et al., 2007a; Olesen et al., 2007). Although strict nestedness is not compatible with strict compartmentalization, Lewinsohn et al. (2006) proposed that plant–animal interaction networks usually exhibit both types of patterns simultaneously, with a compartmentalized structure superimposed with a nested structure within compartments.
MECHANISMS: THE PROCESSES BEHIND THE PATTERNS
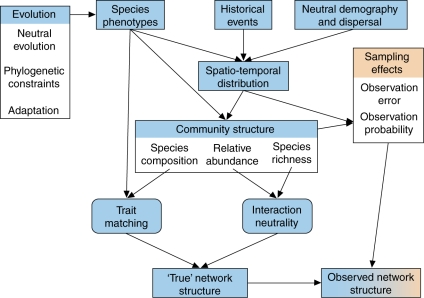
The observed network structure can be envisaged as a function of multiple factors including species phenotypes, neutral demographic and dispersal processes, and the resulting spatio-temporal distribution of species, community structure (species richness, composition and relative abundances) and sampling effects (Vázquez et al., 2009). However, these factors are likely to have a hierarchical, complex causal structure and, in some cases, influence the observed network structure in multiple ways (Fig. 1).
Fig. 1.
Causal model of potential determinants of network structure.
First of all we must distinguish between the ‘true’ network structure (what we want to describe) and the observed structure (what we actually record in the field). The former is a product of ecological, evolutionary and historical processes (blue boxes in Fig. 1), while the latter is also influenced by sampling artefacts (orange boxes in Fig. 1; Ollerton and Cranmer, 2002; Herrera, 2005; Vázquez and Aizen, 2006; Nielsen and Bascompte, 2007; Blüthgen et al., 2008).
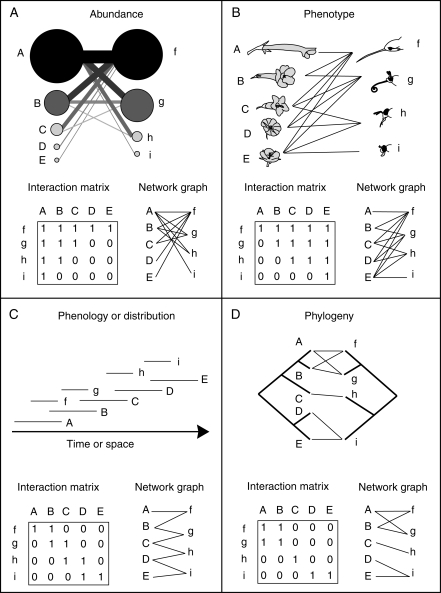
The ‘true’ network structure results directly from the cumulative interaction events of individuals, dictated by the combination of two distinct types of processes: interaction neutrality and trait matching (Fig. 1). Interaction neutrality refers to the occurrence of interactions resulting from the random encounter among individuals, so that all individuals have the same probability of interacting with other individuals, regardless of their taxonomic identity (Vázquez, 2005; Vázquez et al., 2007; Santamaría and Rodríguez-Gironés, 2007; Krishna et al., 2008). Because under interaction neutrality all individuals have the same interaction probability, abundant species will interact more frequently and with more species than rare species (Fig. 2A). Specifically, the probability of interaction between two species is equal to the product of their relative abundances. Thus, a neutral interaction process depends on relative species abundance and the number of species in the community. Interactions will be more evenly distributed in the network under an even distribution of abundance than under a skewed distribution of abundance.
Fig. 2.
Illustration of how network structure can result from (A) species abundance, (B) trait matching, (C) species spatio-temporal distribution and (D) phylogenetic relationships.
In contrast, trait matching refers to a set of interaction rules resulting from the correspondence between phenotypic traits of interacting individuals, either in a complementary fashion (e.g. flower colour or nectar sugar concentration correspond to the pollinator's preferences) or as a barrier (e.g. long corollas exclude flower visitors with short proboscises; Jordano, 1987; Jordano et al., 2003; Santamaría and Rodríguez-Gironés, 2007; Stang et al., 2006, 2007). Trait matching will depend chiefly on the identities of species in the community (species composition) and their phenotypes (Fig. 2B). We refer here to traits in their broadest sense, including not only morphology but also behavioural traits such as feeding preferences. Note that our use of ‘matching’ differs from that in Stang et al. (2009): whereas our definition includes both trait complementarity and exploitation barriers, Stang et al. refer specifically to ‘size matching’, a type of trait complementarity.
Community structure (species composition, richness and relative abundance), which influences both neutral and phenotypically driven interactions, is a direct result of the spatio-temporal distribution of individuals, populations and species (Fig. 1). Spatio-temporal distribution will impose constraints to interactions – species that do not overlap spatially or temporally cannot interact, even if their phenotypes match and even if they are abundant at a particular time in the flowering season (Fig. 2C). These constraints can occur at multiple scales, from local to regional and from short-term to long-term. Spatio-temporal distribution in turn depends on neutral demographic and dispersal processes, historical events and species phenotypes (which determine the species' niches; Ricklefs, 1987, 2004; Bell, 2000, 2001; Pulliam, 2000; Hubbell, 2001; Chase and Leibold, 2003). Community structure is also affected directly by species phenotypes, which will influence the outcome of species interactions.
Species phenotypes result from adaptation, neutral evolution and phylogenetic constraints (Fig. 1). Thus, phylogenetic constraints may influence mutualistic interactions, imprinting a phylogenetic signal on network structure (Ives and Godfray, 2006; Rezende et al., 2007a, b; Fig. 2D). The presence of a phylogenetic signal would suggest that network patterns are constrained by past evolutionary history and not exclusively explained by current ecological processes.
There are a number of ways in which sampling artefacts can influence the observed network. As in any observation, the image depends both on the target of the observation and on observation uncertainty. Sampling interactions in the field involves many methodological assumptions and is subject to many potential observation errors. Thus, the observed network structure is determined both by the ‘true’ network structure and sampling effects (observation uncertainty). Sampling effects are a function of observer error (anything that can distort the real pattern that is due to the observer or to the methods she uses) and of the relative abundance of the species in the network, which influences the observation probability of interactions (Fig. 1) – those involving abundant species are more likely to be observed than interactions involving rare species. The influence of relative abundance on sampling effects is a key confounding factor in the study of interaction networks, because relative abundance influences both the true occurrence of interactions and the occurrence of sampling effects.
Viewing putative determinants of network structure only in one direction (Fig. 1) may miss some important feedbacks, most notably those between network structure and some of the factors that affect it directly or indirectly. For example, co-evolution between interacting species could influence phenotypes, and the presence of particular species (i.e. species composition) could affect the abundance of species that depend on them. Other potential feedbacks include relative abundance → spatio-temporal distribution, spatio-temporal distribution → species traits, and trait matching → species traits. Considering these feedbacks is, however, beyond the scope of this review, as our aim here is to evaluate the forces that lead to the intriguing structural patterns exhibited by plant–animal mutualistic networks.
EVIDENCE: WHAT DO WE REALLY KNOW ABOUT MECHANISMS?
There is a growing literature evaluating the influence of one or more of the potential determinants of network structure depicted in Fig. 1. In this section, we attempt a review of this literature, a summary of which is presented in the Appendix. Our review is probably incomplete, but we are confident that we have been able to cover the majority of studies evaluating the determinants of the structure of mutualistic networks.
Species richness
Several studies have evaluated the influence of species richness of mutualists in a network (network size) on several structural properties. Jordano (1987) showed that the number of links in binary plant–pollinator and plant–seed disperser networks was positively correlated and connectance negatively correlated with the number of species in the network, as had been previously observed in food webs (Montoya and Solé, 2003). Jordano's finding was later confirmed by several other studies using improved data sets covering a broader set of communities than were available to Jordano in 1987 (Olesen and Jordano, 2002; Devoto et al., 2005; Olesen et al., 2006; Thébault and Fontaine, 2008). This result means that as networks grow larger, the mean number of links per species decreases.
Other properties of binary networks such as degree distribution, nestedness and degree asymmetry have also been shown to be correlated with network size. Nestedness and degree asymmetry increase with increasing network size in plant–pollinator, plant–seed disperser and plant–ant networks (Bascompte et al., 2003; Vázquez and Aizen, 2004; Vázquez and Stevens, 2004; Guimarães et al., 2006, 2007b), while truncation of the degree distribution becomes more pronounced as the total number of species in the network and in the network core decrease (Guimarães et al., 2005; Medan et al., 2007).
What is the interpretation of the above results? As shown in Fig. 1, this scaling of network properties with network size may be a result of multiple, non-exclusive processes. Firstly, richness can act on network properties directly, by imposing constraints on the structural properties the network can exhibit. For example, a plant species cannot interact with 100 pollinator species if there are only ten pollinator species in the network; this constraint would impose a limit on maximum degree, resulting in a greater truncation of the degree distribution and lower nestedness (Guimarães et al., 2005; Medan et al., 2007). Secondly, richness can also influence the observed network structure through sampling effects. For example, the decline in connectance with increasing network size may be explained simply by a decreasing relative sampling intensity in larger networks (i.e. lower number of observations per putative link). We discuss sampling effects at length below.
Network structure has also been suggested to be influenced by variations in species richness at a continental scale, such as the well-known latitudinal gradient in species richness (Willig et al., 2003). For example, Olesen and Jordano (2002) and Ollerton and Cranmer (2002) have found a latitudinal effect on several attributes of binary networks, so that connectance and degree tend to be lower in tropical than temperate regions. However, Ollerton and Cranmer (2002) point out that this effect of latitude is most probably a sampling artefact, because the few available data sets for tropical regions tend to have lower sampling efforts than their temperate counterparts. Furthermore, this latitudinal effect of sampling effort is exacerbated by the sampling effect of richness discussed above, which is likely to increase with increasing latitude because of the latitudinal gradient in species richness.
Abundance
Available evidence indicates that abundance, or some surrogate of it (e.g. total interaction frequency), influences aggregate network properties such as connectance, degree distribution, nestedness, degree and strength asymmetry, and interaction evenness (Dupont et al., 2003; Ollerton et al., 2003; Vázquez and Aizen, 2004; Vázquez, 2005; Burns, 2006; Santamaría and Rodríguez-Gironés, 2007; Stang et al., 2006, 2007; Vázquez et al., 2007, 2009; Krishna et al., 2008; Morales and Vázquez, 2008). Furthermore, combining information on local abundance with spatial and temporal distribution (i.e. a spatio-temporally explicit definition of abundance) allows even greater predictive ability of aggregate network patterns (Vázquez et al., 2009). The overall conclusion of these studies is that information on the relative abundance of species in a network goes a long way in predicting aggregate properties of binary and quantitative networks.
The influence of abundace on aggregate network structure has been interpreted as evidence of the influence of neutrality. However, abundance alone (or in combination with spatio-temporal distribution) is not sufficient to predict the detailed structure of the network (i.e. the occurrence and frequency of each pairwise interaction in the network; Vázquez et al., 2009). Thus, the key prediction of the neutrality hypothesis is met only partially, which suggests that although interaction neutrality plays a role in determining network structure, other processes must also be at play.
It is important to bear in mind that abundance can also affect observed network structure through sampling effects (Fig. 1), particularly the detection probability of pairwise interactions (Vázquez and Aizen, 2006; Blüthgen et al., 2008). Thus, as with richness, the effects of abundance on the true network structure must be disentangled from those operating on the observed network structure through sampling effects. For instance, interactions involving rare species are less likely to be observed in the field than those involving abundant species, so that the correlation between total abundance or frequency of interaction and degree frequently observed in mutualistic networks (Dupont et al., 2003; Vázquez and Aizen, 2003) may be simply a sampling artefact. Again, sampling effects are discussed at length below.
Another important issue when evaluating the influence of abundance on network patterns is how abundance is measured. Unlike predator–prey interactions, in which the prey individual is usually the unit of consumption, in plant–animal mutualisms it is not just the abundance of plant individuals that matters, but the abundance/biomass of the functionally important tissues directly involved in the interactions (e.g. flowers, pollen and nectar). Most studies have so far considered individual abundance, but the little available evidence suggests that taking a more appropriate measure of abundance (such as flower abundance) may influence the results significantly (Stang et al., 2006, 2007; Vázquez et al., 2009). Similarly, measuring abundance for animal mutualists is problematic. For example, most studies of plant–pollinator networks have used visitation frequency as an estimate of flower visitor abundance. However, visitation frequency may not represent the true abundance of animals and is not independent of the network we want to predict. Fortunately, encouraging progress comparing and refining sampling methods is being made (Westphal et al., 2008), which will hopefully allow more accurate estimates of species abundance in the future.
Spatio-temporal distribution of individuals and species
A few recent studies have considered temporal variation in network structure (Basilio et al., 2006; Alarcón et al., 2008; Olesen et al., 2008; Petanidou et al., 2008; Vázquez et al., 2009; N. P. Chacoff and D. P. Vázquez, unpubl. res.). Several conclusions can be drawn from these studies. First, aggregate network properties such as connectance and nestedness seem to exhibit little temporal variation, both within and between years. Secondly, there is high temporal variation in the occurrence of pairwise interactions. For example, Petanidou et al. (2008) found that only 5 % of interactions were consistently observed in all 4 years of study, Alarcón et al. (2008) found 31 % of links present in 3 years, and Chacoff and Vázquez (unpub. res.) found 15 % of interactions in 3 years of study. Thirdly, as mentioned above, information on abundance and temporal distribution allows the prediction of several aggregate properties of binary and quantitative networks; however, such information is not sufficient to predict the occurrence of pairwise interactions (Vázquez et al., 2009).
Likewise, recent studies have found that spatial distribution of individuals and species influences network structure. A recent simulation study has shown that the spatial aggregation of individuals may strongly affect several commonly used network statistics by influencing the degree of ‘mixing’ of plant and animal individuals (Morales and Vázquez, 2008). Jordano et al. (2006) point out that 29 % of the unobserved interactions in the plant–hummingbird network constructed with data described by Snow and Snow (1972) can be attributed to lack of spatial overlap between pairs of species. Finally, Vázquez et al. (2009) found that spatial information, in combination with abundance and temporal distribution, allows partial prediction of the occurrence of pairwise interactions.
Another way in which spatial distribution can influence network patterns is through the size of geographic ranges. Ollerton et al. (2007) found that geographic ranges of anemonefish and their anemone mutualists partly determine the nested structure of their interaction network. (Notice that although this example about the anemone–anemonefish mutualism is not really a plant–animal interaction, we decided to include it anyway because it involves a mutualism between sessile and mobile organisms and is thus functionally comparable with mutualisms between plants and animals.) Similarly, Jordano and Bascompte (unpubl. manuscript, cited in Bascompte and Jordano, 2007) found that geographic ranges of interacting species partly predict degree, as has been found for phytophagous insects and parasites (Strong et al., 1984; Combes, 2001; Poulin, 2006).
Thus, both spatial and temporal distribution seem to impose constraints on plant–animal mutualistic interactions, influencing network patterns. As before, however, spatio-temporal distribution can also influence the observed network structure through sampling effects, as interactions involving species with short phenological spans or narrow geographic distributions may be harder to observe than those involving species with long phenologies and wide geographic ranges. Again, sampling effects are dealt with at greater length below.
Trait matching
Several studies have shown that trait matching also influences network patterns. Stang et al. (2006) have shown that nectar holder depth and width impose a size threshold on nectar-feeding flower visitors, so that the number of visitor species visiting a plant species decreases with increasing nectar holder depth. Stang et al. (2007) have shown that this interaction rule also influences nestedness and asymmetry in degree. Finally, Stang et al. (2009) incorporated size complementarity besides the threshold used by Stang et al. (2006, 2007), showing that both trait complementarity and exploitation barriers are important in structuring mutualistic networks. Chamberlain and Holland (2009) found that body size predicts ant degree in plant–ant interaction networks, which suggests that these interactions may also be influenced by trait complementarity. Santamaría and Rodríguez-Gironés (2007) have shown that simulation models that incorporate two kinds of rules of trait matching (interaction barriers and trait complementarity) partly predict the correlation between connectance or nestedness and network size in 40 plant–pollinator networks. Finally, Rezende et al. (2007a) found that simulating trait complementarity involving multiple traits resulted in nested networks, and analysis of one plant–seed disperser network found a weak but statistically detectable effect of trait complementarity on network structure. Note that all these studies evaluating the influence of trait matching on network structure considered only morphological traits. Although, to our knowledge, no studies have evaluated other types of traits, such as foraging behaviour, they are likely to be important, as has been shown in food webs (Beckerman et al., 2006; Petchey et al., 2008).
The importance of trait matching as a determinant of network structure appears to vary among interaction types. Blüthgen et al. (2007) have shown that different types of plant–animal mutualisms show consistent differences in specialization that may correspond to variation in the extent to which trait matching determines network structure. Defining network specialization as the deviation from the null expectation under neutrality (Blüthgen et al., 2006a), Blüthgen et al. (2007) found that pollination networks are significantly more specialized than seed–disperser networks. While many flowers restrict the visitor spectrum by morphological barriers and concealed rewards (Stang et al., 2006, 2007), fleshy fruits are highly exposed. Although other limitations to frugivore assemblages may apply (e.g. fruit size, defences), this difference in accessibility of consumable rewards may contribute to the observed difference in specialization. Similarly, in ant–plant mutualisms, the accessibility of the ants' benefits differs and may explain variation in specialization. Two types of ant–plant mutualisms can be distinguished: myrmecophytic plants, which provide shelter for resident ant colonies and are inaccessible to a number of ant species, and plants with extrafloral nectaries, which attract ants to the foliage and provide open, easily accessible rewards (Rico-Gray and Oliveira, 2007). Correspondingly, myrmecophyte–ant networks are much more specialized (Blüthgen et al., 2007) and compartmentalized (Fonseca and Ganade, 1996; Guimarães et al., 2007a) than networks comprising ants and extrafloral nectaries. A similar observation was made by Blüthgen et al. (2006b) for a plant–hemipteran–ant network, in which specialization was higher for plant–hemipteran than for hemipteran–ant interactions, also presumably because of the greater accessibility of hemipteran rewards for the ants than the plant resources for the hemipterans.
A problem with evaluating the influence of trait matching in determining the structure of plant–animal mutualistic networks lies in the multidimensional nature of phenotypes. An organism is composed of many traits, which in turn form trait complexes. In addition there are many possible ways of ‘matching’ those multitrait phenotypes. Thus, coming up with sensible rules of interaction is not an easy task. The successful attempts we know of to date (most notably Stang et al., 2006, 2007, 2009) have restricted the analysis to plant–animal assemblages sharing a single type of reward (e.g. nectar). However, deriving rules for broader assemblages encompassing all plants and animal mutualists in a community is surely much harder. For example, what plant and pollinator traits determine that a particular pollinator will visit a particular flower? Flower and inflorescence shape, size, colour, scent and rewards, and pollinator body size, vision, olfaction, mobility and nutritional needs are just some obvious examples. Coming up with sensible rules for all these traits requires a deep knowledge of the natural history of all plants and animals in the network.
An alternative approach to study the contribution of trait matching to network structure is to eliminate the influence of other proximate determinants (i.e. interaction neutrality and sampling effects). This is precisely what the method of Blüthgen et al. (2006a) does, using quantitative indices to estimate species-level and network-level specialization after eliminating the contributions of neutrality and sampling effects. So, if what we want to know is whether networks are influenced by traits of interacting species, this method is certainly useful.
Phylogenetic relatedness
Rezende et al. (2007b) evaluated the presence of a phylogenetic signal on 36 plant–pollinator and 37 plant–seed disperser networks. They found that the phylogenies of plants resulted in a significant phylogenetic signal for roughly half of the analysed data sets, whereas the phylogenetic signal of the animal phylogenies was detectable in one-third of the data sets. Thus, at least in some cases, current ecological processes shaping the structure of plant–animal mutualistic networks appear to be constrained by phylogenetic history. However, although the phylogenetic signal was detectable in some cases, the influence of phylogeny on network attributes such as degree or species strength was rather low, suggesting that the influence of phylogenetic effects on network structure is weak compared with current ecological processes.
Historical effects
No studies have evaluated the influence of historical events on mutualistic networks. However, we know that at least in some systems, such as the Mediterranean region of southern Europe, historical events explain much variation in fruit dispersal syndromes (Herrera, 1992, 1995). Historical events may also partly determine local community structure (Ricklefs, 1987, 2004). Thus, it is at least plausible that the influences of traits, richness and abundance on network structure are ultimately determined by historical events (Fig. 1).
Sampling effects
As we have repeatedly discussed above, sampling effects can influence the observed network structure in a number of ways. Firstly, the observed scaling of network properties with species richness can be the result of a sampling artefact. For example, the decline in connectance with increasing network size may be explained simply by a decreasing relative sampling intensity in larger networks (i.e. a lower number of observations per putative link; Blüthgen et al., 2008).
Secondly, abundance can also affect the observed network structure through sampling effects (Fig. 1), particularly the detection probability of pairwise interactions (Herrera, 2005; Vázquez and Aizen, 2006; Blüthgen et al., 2008). A heterogeneous distribution of detection probabilities of interactions (such as the log-normal) suffices to generate most of the aggregate structural properties usually measured in studies of mutualistic networks (Blüthgen et al., 2008). Thus, as with richness, it is important to disentangle the effects of abundance on the true network structure from those operating on the observed network structure through sampling effects. For instance, interactions involving rare species are less likely to be observed in the field than those involving abundant species, so that the correlation between total abundance or frequency of interaction and degree frequently observed in mutualistic networks (Dupont et al., 2003; Vázquez and Aizen, 2003) may be simply a sampling artefact (although it can also result from real ecological processes; see Fontaine et al., 2008).
Thirdly, because phenotypes can influence species' detectability (conspicuous species are more detectable than inconspicuous species), species identities and thus community composition are also a source of sampling effects.
Fourthly, spatio-temporal distribution can also influence the observed network structure through sampling effects; interactions involving species with short phenological spans or narrow geographic distributions may be harder to observe than those involving species with long phenologies and wide geographic ranges. For example, it is likely that some of the 95 % of interactions not observed in all 4 years of study by Petanidou et al. (2008) did actually occur but were simply unrecorded.
What can we do about sampling effects? We can adopt two main strategies: apply indices that correct for sampling effects and increase sampling effort to minimize sampling biases. Regarding the first approach, it is possible to apply rarefaction to correct estimates of degree (i.e. richness of interaction partners) to account for sampling effort (Vázquez and Simberloff, 2002; Herrera, 2005). As an alternative, Blüthgen et al. (2006a) proposed indices of specialization in ecological networks that quantify the residual deviation of interaction strengths from neutrality. This allows focusing on non-neutral, trait-based mechanisms, thus comparing fundamental specialization among species and among networks. Accordingly, Blüthgen et al. (2007) found that pollination, seed dispersal and ant–plant networks differed consistently in their degree of specialization, but within each type of mutualism there was no change across networks with different sizes. Moreover, rare species were not inevitably more ‘specialized’: although rarely visited flowers were indeed more specialized than common ones, common pollinators tended to be more specialized than rare ones. However, it should be noted that the method of Blüthgen et al. (2006a) removes both sampling effects and true effects of abundance and richness through neutrality, which may not be desired if what we want is to describe the ‘true’ network structure regardless of whether it originated by trait-based or neutral interactions.
Several studies have considered the second approach – increasing sampling effort to minimize sampling biases. Vázquez and Aizen (2006) conducted a simulation to evaluate whether increasing sampling effort could improve estimates of degree for rare species. Interestingly, the answer was no: the slope of the correlation between degree and total interaction frequency remained unchanged, so that rare species always tended to have lower degree than abundant species. Thus, in this simulation, increasing sampling effort did not eliminate the sampling problem, but simply kept its influence constant. However, the simulation assumed no saturation of community composition, an arguably unrealistic assumption given the limits imposed by regional species richness. Thus, sampling effects would be minimized if sample effort were large enough to reach an asymptote in the accumulation curves of interaction partners for each species in the network. However, such a sampling design may require a monumental sampling effort, making it unfeasible as a practical solution to sampling problems.
Nielsen and Bascompte (2007) conducted another study evaluating the effect of increasing sampling effort on number of species and number of links in the network and nestedness. They found that although increasing sampling effort led to ever-increasing numbers of links and species, nestedness stabilized relatively quickly, suggesting that this network property is robust to sampling effects.
In summary, sampling effects are ubiquitous and should be considered seriously as one of the processes generating the observed network structure. There are proposed solutions to this crucial problem, but all solutions are imperfect and should be applied with caution. As stressed by Kay and Schemske (2004) and Herrera (2005), new methods of analysis cannot compensate for the current scarcity of reliable field data on plant–animal mutualistic interactions, and sophisticated analytical tools can hardly redeem biased or otherwise messy data.
PUTTING THE PIECES TOGETHER
As our review of the evidence shows, no single mechanism accounts for all the structural variation observed in mutualistic networks. Rather, it is likely that each of the above processes contributes to some extent to network structure. The challenge now is to put these pieces together and understand their relative importance.
A few studies have attempted a simultaneous evaluation of multiple mechanisms. For example, Stang et al. (2006, 2007, 2009) and Krishna et al. (2008) have evaluated simultaneously the influence of abundance and trait matching on network structure, finding that considering both factors simultaneously allows greater predictive ability of network structure than any of them considered separately. Similarly, Rezende et al. (2007a) found that both trait matching and phylogenetic relationships of plants and seed-dispersers influenced nestedness simultaneously. Finally, Vázquez et al. (2009) considered abundance, spatial–temporal overlap and phylogenetic relatedness as predictors of network structure; they found that abundance and temporal overlap together conferred the best predictive ability, with a minor contribution of spatial overlap and an almost nil effect of phylogenetic relatedness.
A difficulty with evaluating more than one potential determinant is methodological: how do we compare their relative contribution? Vázquez et al. (2009) proposed a likelihood method, which allows comparison of the observed interaction matrix with the interaction probability matrices expected under each potential determinant and all their combinations. In this way, it is possible to calculate a likelihood (or an information-based criterion such as Akaike's information criterion) for each probability model. Conceptually similar approaches have been proposed by Stang et al. (2009) to compare observed and expected matrices of trait classes of plants and pollinators, and by Allesina et al. (2008) to compare multiple models of food web structure.
As is hopefully clear from our review, sampling effects are likely to be a pervasive influence on network structure, because many of the ecological mechanisms hypothesized to determine true network structure can also influence the observed structure through sampling effects. Thus, developing methods for accounting for or eliminating sampling effects seems of as much importance as developing methods to evaluate simultanously the influence of ecological and evolutionary processes. Although, as we have shown in this review, there has been some promising progress towards this goal, we are still far from a thorough understanding of the contribution of sampling effects.
Finally, we believe further progress in the understanding of plant–animal mutualistic networks requires improved data sets that allow the evaluation of multiple mechanisms simultanously. Specifically, we need spatio-temporally explicit data sets with detailed natural history information that may allow the derivation of sensible rules of trait complementarity. We believe this goal will be facilitated if research efforts are focused on a sample of representative study systems with contrasting ecological conditions throughout the world.
ACKNOWLEDGEMENTS
D.P.V. thanks the Annals of Botany Company and the Ecological Society of America for financial support to attend the Symposium on the Ecology and Evolution of Plant–Pollinator Interactions on which this special issue is based. N.P.C. and D.P.V. are career researchers, and L.C. is a post-doctoral fellow with CONICET. Research was funded through grants to D.P.V. from CONICET (PIP 6564), FONCYT–ANPCYT (PICT 20805) and the BBVA Foundation (BIOCON03-162). Collaboration between D.P.V. and N.B. was supported by the Sonderforschungsbereich ‘Mechanisms and Evolution of Arthropod Behaviour’ (SFB-554) of the German Research Foundation (DFG).
APPENDIX
Studies that have evaluated potential determinants of network structure
| Interaction type | Explanatory variable | Predicted network attribute | No. of networks | Result | Reference |
|---|---|---|---|---|---|
| Plant–pollinator | Abundance | Nestedness | 1 | Abundance partly predicts nestedness | Ollerton et al. (2003) |
| Plant–pollinator | Abundance | Nestedness | 1 | Abundance partly predicts degree | Dupont et al. (2003) |
| Plant–seed disperser | Abundance | Pairwise interactions | 1 | Abundance explains 83 % of variation of interactions | Burns (2006) |
| Plant–ant, plant–pollinator | Abundance | Strength asymmetry | 1 | Abundance partly predicts strength asymmetry | Vázquez et al. (2007) |
| Plant–pollinator | Abundance and trait matching (complementarity and barriers) | Connectance–richness and nested–richness correlations | 40 | Abundance and complementarity (but not barriers) separately partly predict these correlations | Santamaría and Rodríguez-Gironés (2007) |
| Plant–pollinator | Abundance and trait matching (complementarity and barriers) | Size-specific interaction patterns | 1 | Abundance, complementarity and barriers predict interactions among size classes of plants and flower visitors | Stang et al. (2009) |
| Plant–pollinator | Abundance and trait matching (exploitation barriers) | Interaction asymmetry, nestedness | 1 | Abundance and exploitation barriers predict number of interaction partners and degree asymmetry | Stang et al. (2007) |
| Theoretical | Abundance and trait matching (complementarity) | Nestedness | 2 | Abundance explains most variation in nestedness, but trait complementarity also contributes | Krishna et al. (2008) |
| Plant–pollinator | Abundance and trait matching (exploitation barriers) | Plant degree | 1 | Abundance and exploitation barriers explain 71 % of variation in plant degree | Stang et al. (2006) |
| Plant–pollinator | Abundance and spatio-temporal distribution | Connectance, nestedness, interaction evenness, strength asymmetry, overall network structure | 1 | Abundance and spatio-temporal distribution predict aggregate properties, but only partially overall structure | Vázquez et al. (2009) |
| Plant–seed disperser | Abundance, geographic range, phenological spread and phylogenetic structure | Degree | 2 | Abundance, geographic range, phenological spread and phylogenetic structure partly predict degree | Jordano and Bascompte unpubl. (cited in Bascompte and Jordano, 2007) |
| Plant–ant | Body size | Degree | 8 | Body size partly predicts degree (R2 = [0·05, 0·20]) | Chamberlain and Holland (2009) |
| Simulation | Evenness of species observation records | Connectance, nestedness, degree distribution, strength asymmetry, interaction evenness, generality, standardized diversity (H2′) | 0 | Evenness in species observation records influences network statistics | Blüthgen et al. (2008) |
| Anemone–anemonefish | Geographic range | Nestedness | 1 | Geographic range partly predicts nestedness | Ollerton et al. (2007) |
| Plant–pollinator, plant–seed disperser | Interaction frequency | Shape of degree distribution | 12 (p–p), 5 (p–sd) | Interaction frequency predicts degree distribution | Vázquez (2005) |
| Plant–ant | Interaction intimacy | Nestedness and compartmentalization | 19 | Interaction intimacy explains both nestedness and compartmentalization | Guimarães et al. (2007) |
| Plant–hemiptera–ant | Interaction type | Network specialization (H2′) | 1 | H2′ higher for plant–hemiptera than ant–hemiptera and ant–plant associations | Blüthgen et al. (2006a) |
| Plant–ant, plant–pollinator, plant–seed disperser | Interaction type, species richness, total frequency, network asymmetry | Network specialization (H2'), species-level specialization (d') | 51 | Interaction type influences specialization after correcting for the effect of total frequency | Blüthgen et al. (2007) |
| Plant–pollinator | Latitude, sampling effort | Degree | Sampling effort partly predicts degree | Ollerton and Cramer (2002) | |
| Simulation | Trait matching and phylogenetic relatedness | Nestedness | 1 | Trait complementarity and phylogenetic relationships can result in observed network patterns | Rezende et al. (2007a) |
| Plant–seed disperser | Phylogenetic structure | Degree, species strength and ecological–phylogenetic distance correlation | 36 (p–p), 37 (p–sd) | There is a detectable phylogenetic signal in some of the networks analysed | Rezende et al. (2007b) |
| Plant–pollinator | Sampling effort | Degree–frequency correlation | 1 | Sampling effort does not drive this correlation | Vázquez and Aizen (2006) |
| Plant–pollinator | Sampling effort, species richness and number of links | Nestedness | 4 | Nestedness is more influenced by sampling than by species richness and number of links | Nielsen and Bascompte (2007) |
| Plant–pollinator | Sampling effort, species richness, richness ratio, precipitation | Connectance, number of links | 8 | Sampling effort, species richness and richness ratio influence connectance and number of links | Devoto et al. (2005) |
| Simulation | Spatial aggregation and scale of animal movement | Connectance, nestedness, strength asymmetry, interaction evenness, CV of rare interactions | 0 | Spatial aggregation and animal movement influence network properties | Morales and Vázquez (2008) |
| Plant–seed disperser | Spatial distribution and trait matching | Occurrence of pairwise interactions | 1 | Spatio-temporal segregation and trait matching partly explain absence of interactions | Jordano et al. (2006) |
| Plant–pollinator | Species richness | Connectance | 24 | Connectance decreases with increasing richness | Thébault and Fontaine (2008) |
| Plant–pollinator | Species richness | Connectance and other metrics | Species richness predicts network attributes | Olesen et al. (2006) | |
| Plant–pollinator, plant–seed disperser | Species richness | Connectance, number of links | 33 (p–p), 19 (p–sd) | Connectance decreases and number of links increases with increasing richness | Jordano (1987) |
| Plant–pollinator | Species richness | Degree asymmetry | 18 | Asymmetric specialization increasing with increasing richness | Vázquez and Aizen (2004) |
| Theoretical, plant–pollinator | Species richness | Degree distribution, nestedness | 5 | Truncation of degree distribution and nestedness depend on network size | Medan et al. (2007) |
| Plant–pollinator | Species richness | Modularity, nestedness | 51 | Networks >150 plants always modular, <50 never; most networks nested | Olesen et al. (2007) |
| Plant–ant | Species richness | Nestedness | 4 | Nestedness increases with increasing richness | Guimarães et al. (2006) |
| Plant–pollinator, plant–seed disperser | Species richness | Nestedness | 25 (p–p), 27 (p–sd) | Nestedness increases with increasing richness | Bascompte et al. (2003) |
| Plant–pollinator | Species richness | Proportion of species with only one link (extreme specialists) | 23 | Proportion of species with one link increases with network size | Vázquez and Stevens (2004) |
| Simulation | Species richness of network core | Shape of degree distribution | 0 | Truncation increases with increasing network core | Guimarães et al. (2005) |
| Plant–pollinator | Species richness, latitude | Connectance | 29 | Connectance decreases with increasing richness | Olesen and Jordano (2002) |
| Plant–pollinator, plant–seed disperser | Species richness, richness ratio | Shape of degree distribution | 29 (p–p), 24 (p–sd) | Species richness and richness ratio may determine truncation of degree distribution | Guimarães et al. (2007) |
| Plant–pollinator | Temporal variation (between years) | Between-year similarity, matrix size, connectance, degree centralization, clustering, nestedness, average distance, network diameter | 1 | Aggregate network properties temporally invariant, but identity of interactions highly variable | Petanidou et al. (2008) |
| Plant–pollinator | Temporal variation (between years) | Between-year similarity, matrix size, connectance, degree distribution, nestedness | 1 | Aggregate network properties temporally invariant, but identity of interactions highly variable | Chacoff and Vázquez (unpubl.) |
| Plant–pollinator | Temporal variation (between years) | Number of unique links, comparison of entire matrix composition, nestedness, centrality scores | 1 | Invariant nestedness, high variation in identity of generalized core and composition of reciprocally specialized groups | Alarcón et al. (2008) |
| Plant–pollinator | Temporal variation (within and between years) | Similarity, matrix size, connectance, linkage level, shape of degree distribution | 1 | Aggregate properties temporally invariant, but identity of interactions highly variable | Olesen et al. (2008) |
| Plant–pollinator | Temporal variation (within year) | Matrix size, connectance, assemblage similarity, shape of degree distribution | 1 | Month-to-month fluctuation in partners' identity, matrix size and connectance | Basilio et al. (2006) |
LITERATURE CITED
- Alarcón R, Waser NM, Ollerton J. Year-to-year variation in the topology of a plant–pollinator interaction network. Oikos. 2008;117:1796–1807. [Google Scholar]
- Allesina S, Alonso D, Pascual M. A general model for food web structure. Science. 2008;320:658–661. doi: 10.1126/science.1156269. [DOI] [PubMed] [Google Scholar]
- Almeida-Neto M, Guimarães PR, Jr, Lewinsohn TM. On nestedness analyses: rethinking matrix temperature and anti-nestedness. Oikos. 2007;116:716–722. [Google Scholar]
- Barabási AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Bascompte J, Jordano P. Plant–animal mutualistic networks: the architecture of biodiversity. Annual Review of Ecology, Evolution and Systematics. 2007;38:567–593. [Google Scholar]
- Bascompte J, Jordano P, Melián CJ, Olesen JM. The nested assembly of plant–animal mutualistic networks. Proceedings of the National Academy of Sciences of the USA. 2003;100:9383–9387. doi: 10.1073/pnas.1633576100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascompte J, Jordano P, Olesen JM. Asymmetric coevolutionary networks facilitate biodiversity maintenance. Science. 2006;312:431–433. doi: 10.1126/science.1123412. [DOI] [PubMed] [Google Scholar]
- Basilio AM, Medan D, Torretta JP, Bartoloni NJ. A year-long plant–pollinator network. Austral Ecology. 2006;31:975–983. [Google Scholar]
- Beckerman AP, Petchey OL, Warren PH. Foraging biology predicts food web complexity. Proceedings of the National Academy of Sciences of the USA. 2006;103:13745–13749. doi: 10.1073/pnas.0603039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. The distribution of abundance in neutral communities. American Naturalist. 2000;155:606–617. doi: 10.1086/303345. [DOI] [PubMed] [Google Scholar]
- Bell G. Neutral macroecology. Science. 2001;293:2413–2418. doi: 10.1126/science.293.5539.2413. [DOI] [PubMed] [Google Scholar]
- Blüthgen N, Menzel F, Blüthgen N. Measuring specialization in species interaction networks. BMC Ecology. 2006;a 6:9. doi: 10.1186/1472-6785-6-9. doi:10.1186/1472-6785-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüthgen N, Mezger D, Linsenmair KE. Ant–hemipteran trophobioses in a Bornean rainforest: diversity, specificity and monopolisation. Insectes Sociaux. 2006;b 53:194–203. [Google Scholar]
- Blüthgen N, Menzel F, Hovestadt T, Fiala B, Blüthgen N. Specialization, constraints, and conflicting interests in mutualistic networks. Current Biology. 2007;17:341–346. doi: 10.1016/j.cub.2006.12.039. [DOI] [PubMed] [Google Scholar]
- Blüthgen N, Fründ J, Vázquez DP, Menzel F. What do interaction network metrics tell us about specialization and biological traits? Ecology. 2008;89:3387–3399. doi: 10.1890/07-2121.1. [DOI] [PubMed] [Google Scholar]
- Bronstein JL, Alarcón R, Geber M. The evolution of plant–insect mutualisms. New Phytologist. 2006;172:412–428. doi: 10.1111/j.1469-8137.2006.01864.x. [DOI] [PubMed] [Google Scholar]
- Burns KC. A simple null model predicts fruit–frugivore interactions in a temperate rainforest. Oikos. 2006;115:427–432. [Google Scholar]
- Chamberlain SA, Holland JN. Body size predicts degree in ant–plant mutualistic networks. Functional Ecology. 2009;23:196–202. [Google Scholar]
- Chase JM, Leibold MA. Ecological niches: linking classical and contemporary approaches. Chicago: University of Chicago Press; 2003. [Google Scholar]
- Combes C. Parasitism: the ecology and evolution of intimate interactions. Chicago: University of Chicago Press; 2001. [Google Scholar]
- Devoto M, Medan D, Montaldo NH. Patterns of interaction between plants and pollinators along an environmental gradient. Oikos. 2005;109:461–472. [Google Scholar]
- Dicks LV, Corbet SA, Pywell RF. Compartmentalization in plant–insect flower visitor webs. Journal of Animal Ecology. 2002;71:32–43. [Google Scholar]
- Dunne JA, Williams RJ, Martinez ND. Food-web structure and network theory: the role of connectance and size. Proceedings of the National Academy of Sciences of the USA. 2002;99:12917–12922. doi: 10.1073/pnas.192407699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont YL, Hansen DM, Olesen JM. Structure of a plant–flower–visitor network in the high-altitude sub-alpine desert of Tenerife, Canary Islands. Ecography. 2003;26:301–310. [Google Scholar]
- Fonseca CR, Ganade G. Asymmetries, compartments and null interactions in an Amazonian ant–plant community. Journal of Animal Ecology. 1996;65:339–347. [Google Scholar]
- Fontaine C, Collin CL, Dajoz I. Generalist foraging of pollinators: diet expansion at high density. Journal of Ecology. 2008;96:1002–1010. [Google Scholar]
- Guimarães PR, Aguilar MAMd, Bascompte J, Jordano P, Furtado dos Reis S. Random initial conditions in small Barabasi–Albert networks and deviations from the scale-free behavior. Physical Review E. 2005;71 doi: 10.1103/PhysRevE.71.037101. 037101. doi: 10.1103/PhysRevE.71.037101. [DOI] [PubMed] [Google Scholar]
- Guimarães PR, Rico-Gray V, Furtado dos Reis S, Thompson JN. Asymmetries in specialization in ant–plant mutualistic networks. Proceedings of the Royal Society of London B, Bological Sciences. 2006;273:2041–2047. doi: 10.1098/rspb.2006.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães P, Rico-Gray V, Oliveira PS, Izzo T, dos Reis S, Thompson J. Interaction intimacy affects structure and coevolutionary dynamics in mutualistic networks. Current Biology. 2007;a 17:1797–1803. doi: 10.1016/j.cub.2007.09.059. [DOI] [PubMed] [Google Scholar]
- Guimarães PR, Machado G, de Aguiar MA, et al. Build-up mechanisms determining the topology of mutualistic networks. Journal of Theoretical Biology. 2007;b 249:181–189. doi: 10.1016/j.jtbi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Herrera CM. Historical effects and sorting processes as explanations for contemporary ecological patterns: character syndromes in Mediterranean woody plants. American Naturalist. 1992;140:421–446. [Google Scholar]
- Herrera CM. Plant–vertebrate seed dispersal systems in the Mediterranean: ecological, evolutionary, and historical determinants. Annual Review of Ecology and Systematics. 1995;26:705–727. [Google Scholar]
- Herrera CM. Plant generalization on pollinators: species property or local phenomenon? American Journal of Botany. 2005;92:13–20. doi: 10.3732/ajb.92.1.13. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. The unified neutral theory of biodiversity and biogeography. Princeton: Princeton University Press; 2001. [DOI] [PubMed] [Google Scholar]
- Ings TC, Montoya JM, Bascompte J, et al. Ecological networks – beyond food webs. Journal of Animal Ecology. 2009;78:253–269. doi: 10.1111/j.1365-2656.2008.01460.x. [DOI] [PubMed] [Google Scholar]
- Ives AR, Godfray HCJ. Phylogenetic analysis of trophic associations. American Naturalist. 2006;168:E1–E14. doi: 10.1086/505157. [DOI] [PubMed] [Google Scholar]
- Jordano P. Patterns of mutualistic interactions in pollination and seed dispersal: connectance, dependence asymmetries, and coevolution. American Naturalist. 1987;129:657–677. [Google Scholar]
- Jordano P, Bascompte J, Olesen JM. Invariant properties in coevolutionary networks of plant–animal interactions. Ecology Letters. 2003;6:69–81. [Google Scholar]
- Jordano P, Bascompte J, Olesen JM. The ecological consequences of complex topology and nested structure in pollination webs. In: Waser NM, Ollerton J, editors. Plant–pollinator interactions: from specialization to generalization. Chicago: University of Chicago Press; 2006. pp. 173–199. [Google Scholar]
- Kay KM, Schemske DW. Geographic patterns in plant–pollinator mutualistic networks: comment. Ecology. 2004;85:875–878. [Google Scholar]
- Krishna A, Guimarães PR, Jr, Jordano P, Bascompte J. A neutral–niche theory of nestedness in mutualistic networks. Oikos. 2008;117:1609–1618. [Google Scholar]
- Lewinsohn TM, Prado PI, Jordano P, Bascompte J, Olesen J. Structure in plant–animal interaction assemblages. Oikos. 2006;113:174–184. [Google Scholar]
- Medan D, Perazzo RPJ, Devoto M, et al. Analysis and assembling of network structure in mutualistic systems. Journal of Theoretical Biology. 2007;246:510–521. doi: 10.1016/j.jtbi.2006.12.033. [DOI] [PubMed] [Google Scholar]
- Memmott J. The structure of a plant–pollinator food web. Ecology Letters. 1999;2:276–280. doi: 10.1046/j.1461-0248.1999.00087.x. [DOI] [PubMed] [Google Scholar]
- Memmott J, Waser NM, Price MV. Tolerance of pollination networks to species extinctions. Proceedings of the Royal Society B, Biological Sciences. 2004;271:2605–2611. doi: 10.1098/rspb.2004.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JM, Solé RV. Topological properties of food webs: from real data to community assembly models. Oikos. 2003;102:614–622. [Google Scholar]
- Morales JM, Vázquez DP. The effect of space in plant–animal mutualistic networks: insights from a simulation study. Oikos. 2008;117:1362–1370. [Google Scholar]
- Nielsen A, Bascompte J. Ecological networks, nestedness and sampling effort. Journal of Ecology. 2007;95:1134–1141. [Google Scholar]
- Olesen JM, Jordano P. Geographic patterns in plant–pollinator mutualistic networks. Ecology. 2002;83:2416–2424. [Google Scholar]
- Olesen J, Bascompte J, Dupont Y, Jordano P. The smallest of all worlds: pollination networks. Journal of Theoretical Biology. 2006;240:270–276. doi: 10.1016/j.jtbi.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Olesen JM, Bascompte J, Dupont YL, Jordano P. The modularity of pollination networks. Proceedings of the National Academy of Sciences of the USA. 2007;104:19891–19896. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen JM, Bascompte J, Elberling H, Jordano P. Temporal dynamics in a pollination network. Ecology. 2008;89:1573–1582. doi: 10.1890/07-0451.1. [DOI] [PubMed] [Google Scholar]
- Ollerton J, Cranmer L. Latitudinal trends in plant–pollinator interactions: are tropical plants more specialised? Oikos. 2002;98:340–350. [Google Scholar]
- Ollerton J, Johnson SD, Cranmer L, Kellie S. The pollination ecology of an assemblage of grassland asclepiads in South Africa. Annals of Botany. 2003;92:807–834. doi: 10.1093/aob/mcg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerton J, Stott A, Allnutt E, Shove S, Taylor C, Lamborn E. Pollination niche overlap between a parasitic plant and its host. Oecologia. 2007;151:473–485. doi: 10.1007/s00442-006-0605-y. [DOI] [PubMed] [Google Scholar]
- Petanidou T, Kallimanis AS, Tzanopoulos J, Sgardelis SP, Pantis JD. Long-term observation of a pollination network: fluctuation in species and interactions, relative invariance of network structure and implications for estimates of specialization. Ecology Letters. 2008;11:564–575. doi: 10.1111/j.1461-0248.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- Petchey OL, Beckerman AP, Riede OJ, Warren PH. Size, foraging, and food web structure. Proceedings of the National Academy of Sciences of the USA. 2008;105:4191–4196. doi: 10.1073/pnas.0710672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R. Evolutionary ecology of parasites. Princeton: Princeton University Press; 2006. [Google Scholar]
- Pulliam HR. On the relationship between niche and distribution. Ecology Letters. 2000;3:349–361. [Google Scholar]
- Rezende E, Jordano P, Bascompte J. Effects of phenotypic complementarity and phylogeny on the nested structure of mutualistic networks. Oikos. 2007;a 116:1919–1929. [Google Scholar]
- Rezende EL, Lavabre JE, Guimarães PR, Jordano P, Bascompte J. Non-random coextinctions in phylogenetically structured mutualistic networks. Nature. 2007;b 448:925–928. doi: 10.1038/nature05956. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE. Community diversity: relative roles of local and regional processes. Science. 1987;235:167–171. doi: 10.1126/science.235.4785.167. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE. A comprehensive framework for global patterns in biodiversity. Ecology Letters. 2004;7:1–15. [Google Scholar]
- Rico-Gray V, Oliveira PS. The ecology and evolution of ant–plant interactions. Chicago: University of Chicago Press; 2007. [Google Scholar]
- Sahli H, Conner J. Characterizing ecological generalization in plant-pollination systems. Oecologia. 2006;148:365–372. doi: 10.1007/s00442-006-0396-1. [DOI] [PubMed] [Google Scholar]
- Santamaría L, Rodríguez-Gironés MA. Linkage rules for plant–pollinator networks: trait complementarity or exploitation barriers? PLoS Biology. 2007;5:e31. doi: 10.1371/journal.pbio.0050031. doi:10.1371/journal.pbio.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BK, Snow DW. Feeding niches of hummingbirds in a Trinidad valley. Journal of Animal Ecology. 1972;41:471–485. [Google Scholar]
- Stang M, Klinkhamer PGL, van der Meijden E. Size constraints and flower abundance determine the number of interactions in a plant–flower visitor web. Oikos. 2006;112:111–121. [Google Scholar]
- Stang M, Klinkhamer PGL, van der Meijden E. Asymmetric specialization and extinction risk in plant–flower visitor webs: a matter of morphology or abundance? Oecologia. 2007;151:442–453. doi: 10.1007/s00442-006-0585-y. [DOI] [PubMed] [Google Scholar]
- Stang M, Klinkhamer PGL, Waser NM, Stang I, van der Meijden E. Size-specific interaction patterns and size matching in a plant–pollinator interaction web. Annals of Botany. 2009;103 doi: 10.1093/aob/mcp027. doi:10.1093/aob/mcp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss SY, Irwin RE. Ecological and evolutionary consequences of multispecies plant–animal interactions. Annual Review of Ecology, Evolution, and Systematics. 2004;35:435–466. [Google Scholar]
- Strong DR, Lawton JH, Southwood TRE. Insects on plants. Oxford: Blackwell Scientific Publishers; 1984. [Google Scholar]
- Thébault E, Fontaine C. Does asymmetric specialization differ between mutualistic and trophic networks? Oikos. 2008;117:555–563. [Google Scholar]
- Thompson JN. The coevolutionary process. Chicago: University of Chicago Press; 1994. [Google Scholar]
- Thompson JN. The geographic mosaic of coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- Vázquez DP. Degree distribution in plant–animal mutualistic networks: forbidden links or random interactions? Oikos. 2005;108:421–426. [Google Scholar]
- Vázquez DP, Aizen MA. Null model analyses of specialization in plant–pollinator interactions. Ecology. 2003;84:2493–2501. [Google Scholar]
- Vázquez DP, Aizen MA. Asymmetric specialization: a pervasive feature of plant–pollinator interactions. Ecology. 2004;85:1251–1257. [Google Scholar]
- Vázquez DP, Aizen MA. Community-wide patterns of specialization in plant–pollinator interactions revealed by null-models. In: Waser NM, Ollerton J, editors. Plant–pollinator interactions: from specialization to generalization. Chicago: University of Chicago Press; 2006. pp. 200–219. [Google Scholar]
- Vázquez DP, Simberloff D. Ecological specialization and susceptibility to disturbance: conjectures and refutations. American Naturalist. 2002;159:606–623. doi: 10.1086/339991. [DOI] [PubMed] [Google Scholar]
- Vázquez DP, Stevens RD. The latitudinal gradient in niche breadth: concepts and evidence. American Naturalist. 2004;164:E13–E31. doi: 10.1086/421445. [DOI] [PubMed] [Google Scholar]
- Vázquez DP, Morris WF, Jordano P. Interaction frequency as a surrogate for the total effect of animal mutualists on plants. Ecology Letters. 2005;8:1088–1094. [Google Scholar]
- Vázquez DP, Melián CJ, Williams NM, Blüthgen N, Krasnov BR, Poulin R. Species abundance and asymmetric interaction strength in ecological networks. Oikos. 2007;116:1120–1127. [Google Scholar]
- Vázquez DP, Chacoff NP, Cagnolo L. Evaluating multiple determinants of the structure of plant–animal mutualistic networks. Ecology. 2009 doi: 10.1890/08-1837.1. (in press). [DOI] [PubMed] [Google Scholar]
- Waser NM, Chittka L, Price MV, Williams NM, Ollerton J. Generalization in pollination systems, and why it matters. Ecology. 1996;77:1043–1060. [Google Scholar]
- Waser NM, Ollerton J, editors. Plant–pollinator interactions: from specialization to generalization. Chicago: University of Chicago Press; 2006. [Google Scholar]
- Westphal C, Bommarco R, Carre G, et al. Measuring bee diversity in different European habitats and biogeographical regions. Ecological Monographs. 2008;78:653–671. [Google Scholar]
- Willig MR, Kaufman DM, Stevens RD. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annual Review of Ecology, Evolution, and Systematics. 2003;34:273–309. [Google Scholar]