Abstract
Background and Aims
Expression of the mitochondrial gene orf138 causes Ogura cytoplasmic male sterility (CMS) in Raphanus sativus, but little is known about the mechanism by which CMS takes place. A preliminary microarray experiment revealed that several nuclear genes concerned with flavonoid biosynthesis were inhibited in the male-sterile phenotype. In particular, a gene for one of the key enzymes for flavonoid biosynthesis, chalcone synthase (CHS), was strongly inhibited. A few reports have suggested that the inhibition of CHS causes nuclear-dependent male sterile expression; however, there do not appear to be any reports elucidating the effect of CHS on CMS expression. In this study, the expression patterns of the early genes in the flavonoid biosynthesis pathway, including CHS, were investigated in normal and male-sterile lines.
Methods
In order to determine the aberrant stage for CMS expression, the characteristics of male-sterile anthers are observed using light and transmission electron microscopy for several stages of flower buds. The expression of CHS and the other flavonoid biosynthetic genes in the anthers were compared between normal and male-sterile types using real time RT-PCR.
Key Results
Among the flavonoid biosynthetic genes analysed, the expression of CHS was strongly inhibited in the later stages of anther development in sterility cytoplasm; accumulation of putative naringenin derivatives was also inhibited.
Conclusions
These results show that flavonoids play an important role in the development of functional pollen, not only in nuclear-dependent male sterility, but also in CMS.
Key words: Chalcone Synthase, flavonoids, Ogura cytoplasmic male sterility, CMS, pollen, Raphanus sativus
INTRODUCTION
Cytoplasmic male sterility (CMS) is a maternally inherited defect that only affects pollen production. Generally, CMS is induced by specific nuclear/mitochondrial interactions, which mainly arise from mitochondrial mutations and/or nuclear–mitochondrial incompatibilities (Delourme and Budar, 1999; Hanson and Bentolila, 2004; Chase, 2007; Pelletier and Budar, 2007). The expression of CMS genes encoded by the mitochondrial genome is under the influence of the corresponding nuclear genes (Rf), which are able to suppress the mitochondrial defect and restore fertility to plants. Hence, CMS is also called nuclear-cytoplasmic sterility.
In radish (Raphanus sativus), a stable CMS-system specified by a novel mitochondrial gene, orf138 (Bonhomme et al., 1992) or orf125, a partial deletion of orf138 (Iwabuchi et al., 1999), exists and is called Ogura cytoplasm. Ogura cytoplasm was first found in male-sterile R. sativus in an escaped radish population in Japan (Ogura, 1968), and it is now the main CMS cytoplasm used for F1 hybrid production in Brassicaceae across the world (Delourme and Budar, 1999). The sequence of orf138 has an unknown origin (Bonhomme et al., 1992). However, orf138 has recently been demonstrated to have originated from R. raphanistrum, as shown by studies of sequence variation in orf138 and nearby loci in wild and cultivated radishes (Terachi et al., 2001; Yamagishi and Terachi, 2001).
So far, CMS expression has been studied intensively with a focus on the interaction between nuclear-coded Rf genes and the novel mitochondrial genes (Hanson and Bentolila, 2004). In radish, one dominant Rf gene for the Ogura CMS was cloned and it was clarified that the gene, orf687, encodes a member of the pentatricopeptide repeat (PPR) protein family (Koizuka et al., 2003). It has been observed that another Rf gene other than orf687 is distributed widely in Japanese wild radishes (Yasumoto et al., 2008). However, it is still unknown how the mitochondrial gene participates in the CMS process without affecting plant development. It is difficult to consider that orf138 directly controls the pollen disruption event in radish because ORF138 protein is expressed not only in the reproductive organs but also in vegetative tissues (Krishnasamy and Makaroff, 1994; Bellaoui et al., 1999). It has been reported that the CMS-specific protein ORF138 could have a toxic effect on mitochondria (Duroc et al., 2005), but there is no evidence as to why the mitochondrial disfunction should specifically disrupt pollen development.
It is assumed that the novel mitochondrial gene/protein acts on nuclear gene expression related to pollen development via an unknown cytoplasm–nuclear interaction. In order to dissect the process of Ogura cytoplasmic male sterility, a microarray analysis was performed using an Arabidopsis oligo microarray to screen specifically expressed or suppressed nuclear genes in a CMS line of radish containing orf138 (S. Yang et al., unpubl. res.). This analysis identified several flavonoid biosynthetic genes with reduced expression in the CMS line, including chalcone synthase (CHS). Here we describe a detailed analysis of the expression of this radish CHS in CMS lines of Raphanus sativus in order to explore its potential role in CMS. It is shown that expression of CHS was drastically suppressed prior to the morphological induction of male sterility. This is the first report that shows the possible role of flavonoids in pollen development in CMS radish.
MATERIALS AND METHODS
Plant material
Raphanus sativus ‘MS-Gensuke’ (‘MS-G’) and ‘Uchiki-Gensuke’ (‘UC-G’) were used in this study. Male sterile ‘MS-G’ was bred by introducing the Ogura male sterile cytoplasm into ‘UC-G’ (M. Yamabe, Agricultural Research Station of Ishikawa Prefecture, Japan, pers. comm.), which is a local variety of Ishikawa prefecture, Japan. The two varieties are alloplasmic lines, i.e. the difference is that ‘MS-G’ has the cytoplasm that induces male sterility in radish while the background of the nuclear genome is same between these two lines. ‘UC-G’, with normal cytoplasm, produces abundant pollen and was used as a control for the gene expression analysis of Ogura cytoplasm. Flower buds of these two lines were harvested on the same day from plants grown in the greenhouse.
In addition to these two lines, other radish lines having orf138 were used to investigate the relationship between pollen fertility restoration and gene expression. Buds of another male-sterility variety ‘60-Wase’ (‘60-W’) with orf138 were collected and studied. In addition, three F1 crosses whose pollen fertility was normal because of an Rf gene derived from the pollen parent were examined: ‘60-W’ × ‘Comet’, ‘MS-G’ ×' Tomioka' and ‘MS-G’ × ‘Iwasaki’.
Determination of the aberrant anther stage
To determine the aberrant stage of anther development in ‘MS-G’, flower buds including anthers representing a series of developmental stages were excised from ‘MS-G’ and ‘UC-G’. The buds (2–6 mm in length) were fixed in FAA (ethanol : water : formalin : acetic acid = 12 : 6 : 1 : 1, v/v) solution and dehydrated in an ethanol gradient to absolute ethanol. The buds were then embedded in Technovit 7100 resin (Heraeus Kulzer, Wehrheim, Germany) and sectioned transversely at 5 µm thickness with a microtome (Leica, RM2155, Wetzlar, Germany). Thin sections were stained for 1 min with a toluidine blue solution (0·05 %) and observed under a light microscope.
For transmission electron microscopy, flower buds were fixed in 2·5 % (v/v) glutaraldehyde in 0·025 m phosphate buffer (pH 6·8) and were de-aerated under vacuum for 3 h. Following a wash with phosphate buffer, samples were postfixed in 1 % (v/v) osmium tetroxide for 3 h at room temperature followed by dehydration in a graded acetone series. Dehydrated samples were then embedded in epoxy resin and propylene oxide. Transverse ultra-thin sections were collected on coated grids and stained with saturated uranyl acetate and Reynold's lead citrate. Sections were observed with a JEM-1200EX electron microscope at 80 kV (Japan Electron Datum Co.).
For visualization of absorbance of UV light by anthers of ‘MS-G’ and ‘UC-G’, anthers 2–4 mm in length were put on a cross linker (345 nm) and photographed using a digital camera.
Reverse transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from buds with a length of <2 mm, anthers and pistils of young buds (<3 mm in length), and leaves and roots from young seedlings (3-week-old plants) of ‘MS-G’ and ‘UC-G’ using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). More than ten plants of each cultivar were used for RNA extraction from bud material and several plants were used for leaves and roots. Contaminating DNA was completely digested on a column (Qiagen). Concentration and purity were determined by spectrophotometry and analysis of 18S and 16S ribosome bands by agarose gel electrophoresis. The first strands of the cDNA mixture were synthesized from 2·0 µg of total RNA and 2·5 µm Oligo d(T) primer using the SuperScriptTM First-Strand Synthesis System (Invitrogen, USA). The total volume of the reaction mixture was 20 µL, which was prepared according to the manufacturer's protocol. PCR was conducted with radish gene-specific primers. A partial sequence of radish chalcone synthase (CHS) was obtained with primers designed from Arabidopsis CHS AT4G00040·1, which was identified by our preliminarily microarray experiment (Yang et al., unpubl. res.). Radish-specific CHS primers were then designed to this partial sequence. Direct sequencing was performed using a CEQ2000 System (Beckman Coulter, USA) following the protocols of the supplier after the PCR products were purified using a QIAquick PCR purification kit (Qiagen). The primer sequences for radish CHS were 5′-AGTGAGTTCCGTTTGCCCGG (forward) and 5′-TTCTTGCAGAACTCCTCTATGTTG (reverse) and the amplified product was 402 bp in length. The radish Actin gene was used as a control and the primer sequences were 5′-ATCTTCATGCTGCTTGGTGC (forward) and 5′-GAGGCTGATGATATTCAACC (reverse). In the PCR mixture, Ex taq (TAKARA) was used. Thirty-five cycles of PCR (94 °C for 1 min, 50–55 °C for 2 min, 72 °C for 2 min) were carried out. PCR products were separated by electrophoresis in 2 % agarose gels, stained with ethidium bromide and visualized under UV light.
Real-time RT-PCR analysis
Real-time RT-PCR was conducted for CHS and four other genes selected from the flavonoid biosynthetic pathway that function before and after CHS (Harborne, 1993). Each clone-specific primer was initially designed based on the cDNA sequence of the corresponding gene in R. sativus or A. thaliana and then new primer sets were designed according to the sequence of the cloned cDNA fragments from R. sativus. The primers used for real-time PCR were: CHS, 5′-TAGCAACGAGGTGCAGAGG (forward) and 5′-CGTAAGGGCGAGCTTTGTT (reverse); cinnamate 4-hydroxylase (C4H), 5′-TGTCAAGATGTGAAAGACAGGAG (forward) and 5′-CATTTCAATCCTTCGCTCC (reverse); 4-coumarate coA ligase (4CL), 5′-AACTCGATCATGCTGTGTGG (forward) and 5′-TCTCCGTCTCCTGCGACTT (reverse); chalcone isomerase (CHI), 5′-GTCATCTCACCGGCTTCCT (forward) and 5′-TCCGTTAGTTCCTCTGTCGTT (reverse); and flavonol 3-hydroxylase (F3H) 5′-CGTGAACGTATTTCTCGTACCC (forward) and 5′-TCTTCTCGCTGTACTCCTC (reverse). The primers for Actin were 5′-ACAACACCATGCTCAATAGG (forward) and 5′-ATCATGGTGTCATGGTTGGG (reverse). PCR reactions were performed in a 96-well plate with a GeneAmp 5700 sequence detection system, using SYBR Green master mix (Toyobo, Osaka, Japan) to monitor cDNA amplification, according to the manufacturer's protocol. The cDNA standard samples were diluted to 20 000, 10 000, 5000, 2000 and 1000 copies μL−1. Four replications of quantitative assays were performed on 1 µL of each cDNA sample. The following standard thermal profile was used: 1 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Isoform specificity was ensured by analysis of the dissociation curves of the PCR products. Data was analysed using the GeneAmp 5700 SDS software. The standard curve method was used to evaluate quantitative variations between replicates. For all experimental samples, target quantity was determined from the standard curve and divided by the target quantity of the calibrator, and Actin was used as an internal control to normalize all data.
RESULTS
Detection of the abnormal stage of ‘MS-G’ anthers
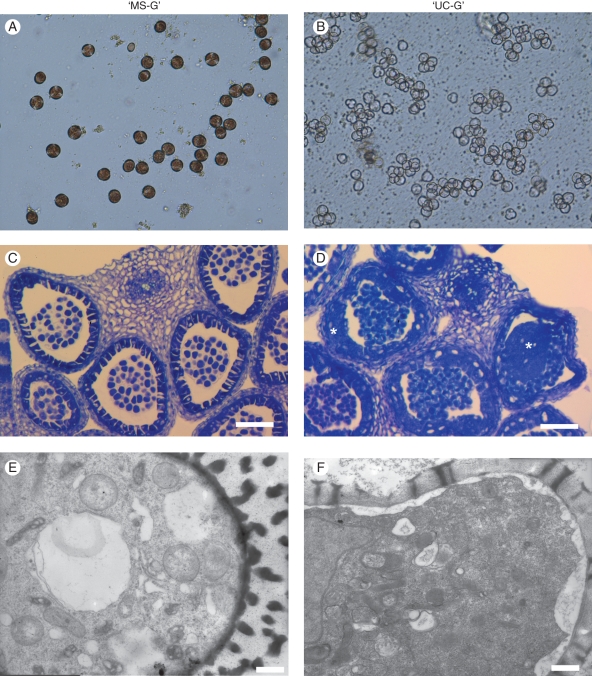
Anthers of ‘MS-G’ and ‘UC-G’, representing a wide range of developmental stages, were examined. Up to the size of 2-mm buds, microsporogenesis and tapetal development were similar in ‘UC-G’ and ‘MS-G’, representing the meiotic division and tetrad stages. Abnormality in the anthers of ‘MS-G’ was first observed at the subsequent stages (Fig. 1C, D). In ‘MS-G’, abnormal division and vacuolization of tapetal cells were obvious from this stage, whereas these were not observed in ‘UC-G’. ‘MS-G’ anthers contained a proliferating tapetum, which partially occluded the anther locules (Fig. 1D) and eventually filled most of the locule, concomitant with abortion of the microspores. Moreover, as was apparent from the unstained microspores, ‘MS-G’ did not colorize as compared to ‘UC-G’, which had already started coloration at this stage (Fig. 1A, B). From this stage (>2 mm), ‘UC-G’ anthers strongly absorbed UV light, whereas ‘MS-G’ anthers did not (Fig. 2). The microspores of 3-mm buds in ‘MS-G’ were substantially different from those of ‘UC-G’ (Fig. 1E, F): the exine of ‘MS-G’ microspores was irregularly developed or partly empty, and the cytoplasm of the microspore showed plasmolysis. It was therefore concluded that the abnormal pollen development of ‘MS-G’ begins approximately at the 2-mm bud stage, in which the anthers were at the tetrad stage or later.
Fig. 1.
Aberrant stage of anthers and microspores of radish from (A, C, E) a sterile line, ‘MS-G’, and (B, D, F) a fertile line, ‘UC-G’. (A, B) Light micrographs of ‘UC-G’ and ‘MS-G’ at the tetrad stage in 2-mm buds. Microspores of ‘MS-G’ do not colorize compared to ‘UC-G’. (C, D) Light micrographs of toluidine blue-stained sections. The images are cross-sections of anthers in 2-mm buds. The shape of microspores is similar between ‘UC-G’ and ‘MS-G’; however, the tapetum is abnormal in ‘MS-G’ due to the presence of large vacuoles and has proliferated to fill the anther locule (*). (E, F) Transmission electron micrographs of microspores in 3-mm buds. Free microspores in ‘UC-G’ have developed sporopollenin material in their exines. In ‘MS-G’, the exines are abnormal due to irregular development and are partly empty. The cytosol of ‘MS-G’ shows plasmolysis. Scale bars: (C,D) = 50 µm; (E, F) = 500 nm.
Fig. 2.
Absorbance of UV light (345 nm) by anthers in 2–4-mm buds of radish lines ‘UC-G’ (fertile) and ‘MS-G’ (sterile). Note that anthers of ‘UC-G’ strongly absorb UV light, compared with the emitted fluorescence of ‘MS-G’.
Expression analysis of flavonoid biosynthetic genes in buds of ‘MS-G’ and ‘UC-G’
The expression of five flavonoid biosynthetic genes including CHS was investigated. Two main genes were chosen, cinnamate 4-hydroxylase (C4H) and 4-coumarate coA ligase (4CL), which function before CHS in flavonoid biosynthesis, and two other genes, chalcone isomerase (CHI) and flavonol 3-hydroxylase (F3H), that function after CHS (Harborne, 1993). These genes were partially cloned from R. sativus ‘UC-G’ according to database sequences from R. sativus or Arabidopsis in order to determine the exact transcript sequences in radish.
As shown in Table 1, the expression levels of C4H, 4CL, CHI and F3H were highly similar between early buds (<2 mm) of ‘MS-G’ and ‘UC-G’. The expression levels of these genes were relatively low compared to that of CHS in ‘UC-G’ (Table 1). The expression of CHS was inhibited significantly in ‘MS-G’ buds, in which the relative amount of CHS mRNA was 35-fold lower than that of ‘UC-G’. In accordance with these results, CHS is a key enzyme that is strongly inhibited in ‘MS-G’ buds in a specific manner.
Table 1.
Real-time RT-PCR analysis of expression of flavonoid biosynthetic genes in buds of ‘MS-G’ (sterile) and ‘UC-G’ (fertile) radish lines
| Relative transcript* |
|||||
|---|---|---|---|---|---|
| Cultivar | C4H | 4CL | CHS | CHI | F3H |
| ‘MS-G’ | 0·56 ± 0·06 | 0·42 ± 0·03 | 0·10 ± 0·05 | 0·46 ± 0·01 | 1·17 ± 0·06 |
| ‘UC-G’ | 0·51 ± 0·12 | 0·36 ± 0·05 | 4·29 ± 1·04 | 0·60 ± 0·05 | 1·34 ± 0·17 |
* The relative transcript level was calculated as target quantity divided by the quantity of a constitutive control gene, Actin.
Abbreviations: C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate coA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; and F3H, flavonol 3-hydroxylase. Each clone-specific primer set was designed from the corresponding gene in radish. Total RNA was isolated from buds (<2 mm) of ‘MS-G’ and ‘UC-G’ lines. Values are mean ± s.e. calculated from four technical replicates of a total of two independent experiments.
Expression analysis of chalcone synthase (CHS) between ‘MS-G’ and ‘UC-G’
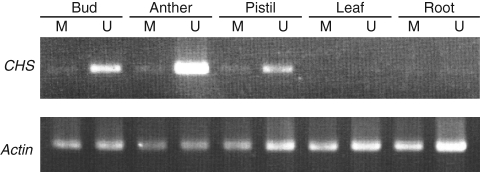
From our preliminarily microarray analysis, it was found that a large portion of genes expressed in ‘UC-G’ are expressed similarly in the male-sterile type ‘MS-G’. The array was conducted with flower buds smaller than 2 mm in length in order to trace the initiation of gene expression alterations before morphological changes in ‘MS-G’. CHS was selected as a substantially inhibited gene in ‘MS-G’, which showed 24-fold and 6·2-fold inhibition in two array experiments. The sequence of the full length cDNA of radish CHS (1407 bp) was determined and it was confirmed that the gene is a member of the chalcone synthase family. As shown in Fig. 3, the expression of CHS was detected strongly in ‘UC-G’ anthers but was almost completely inhibited in ‘MS-G’ anthers. Expression was also detected in the pistil of ‘UC-G’ but not in the pistil of ‘MS-G’. The expression levels of CHS were below the limit of detection in leaves and roots, confirming the inflorescence-specificity of the isolated clones (Fig. 3). The nucleotide sequence of the radish CHS obtained here was registered in the DNA Data Bank of Japan (DDBJ) with the accession number of AB436782.
Fig. 3.
Expression of CHS in reproductive and vegetative organs of radish lines ‘MS-G’ (M) and ‘UC-G’ (U). The first strands of the cDNA were generated from total RNA of buds (<2 mm), anther and pistil (2–3-mm buds), and young leaves and roots.
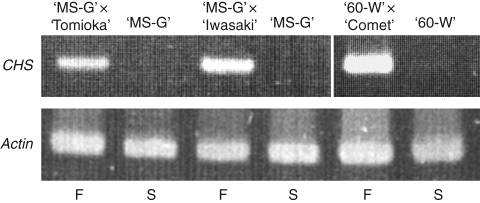
CHS expression in anthers in stages ranging from 2- to 6-mm buds was also confirmed (Table 2). The observations showed that microspores in the anther of ‘MS-G’ were completely degenerated at the stage of a 6-mm bud. Compared to the high expression levels detected in anthers corresponding to 2- to 4-mm buds in ‘UC-G’, no detectable levels of expression were observed in anthers of all 2- to 6-mm buds in ‘MS-G’. In the anthers of 3- to 4-mm buds, a 430-fold lower expression was seen in ‘MS-G’ compared to ‘UC-G’. The results show that CHS is specifically expressed in later stages of anther development and uniquely suppressed in ‘MS-G’. CHS expression was also inhibited in the buds of another CMS radish, ‘60-Wase’; but expression was restored in conjunction with restoration of fertility in CMS radishes by the introduction of the Rf gene (Fig. 4).
Table 2.
Transcript levels of CHS at various stages of radish anther development, assessed by real-time RT-PCR in ‘MS-G’ (sterile) and ‘UC-G’ (fertile) lines
| Bud length |
|||
|---|---|---|---|
| Cultivar | 2–3 mm | 3–4 mm | 5–6 mm |
| ‘MS-G’ | 0·07 ± 0·02 | 0·02 ± 0·02 | 0·04 ± 0·01 |
| ‘UC-G’ | 5·0 ± 1·8 | 8·6 ± 2·6 | 0·6 ± 0·6 |
Values are mean (± s.e.) relative transcript levels from three independent anther samples of total RNA, calculated as target quantity divided by the quantity of a constitutive control gene, Actin.
Fig. 4.
Transcript level of CHS in the buds of male-sterile (S) and fertility-restored (F) radish plants detected by real-time RT-PCR. Total RNA was isolated from buds (<2 mm) of each plant. ‘MS-G’ and ‘60-W’ are male-sterile with orf138; ‘MS-G’ × ‘Tomioka’, ‘MS-G’ × ‘Iwasaki’ and ‘60-W’ × ‘Comet’ are F1 plants that produce fertile pollen because of the Rf gene derived from the pollen parents (‘Tomioka’, ‘Iwasaki’ and ‘Comet’).
DISCUSSION
Until now, there have been no detailed observations of the anatomical features of Ogura cytoplasmic male sterility (CMS). From our anatomical observations, the degeneration of microspores in ‘MS-G’ anthers was first detected in flower buds of ≤2 mm length (Fig. 1); specifically, abnormal formation of the tapetum was the first sign of male sterility. The anatomical process of male sterility may result from the loss of intricate biochemical communication between the tapetum and microspores, which possibly stimulates the tapetal cells to further divide abnormally. The results confirmed the observations in radish made by Ogura (1968), notably excessive vacuolization of the tapetal cells. In many CMS types, the late steps of anther development or pollen maturation are impaired (Linke and Borner, 2005). In the CMS system induced by ‘Tokumasu’ cytoplasm in Brassica napus, degeneration of the tapetum begins after the tetrad stage, leading to empty exines and proliferating tapetum in the anthers (Theis and Robbelen, 1990). The ‘Ogura’ CMS system (Gourret et al., 1992) and ctr CMS lines (Grant and Beversdorf, 1986) of B. napus also demonstrated partial correspondence with the current observation made in this study. In addition, in the PET1-type of CMS in sunflower and petunia the tapetum function is impaired, which is seen as vacuolation or early degradation of tapetal cells (Smart et al., 1994; Balk and Leaver, 2001).
Generally, tapetal cells play an essential role in pollen development, producing protein, lipids and flavonols that are secreted into the pollen sac and form part of the exine (Goldberg et al., 1993). Moreover, several genes involved in secondary metabolism, including CHS, are specifically or predominantly expressed in the tapetum (Shen and Hsu, 1992). The dysfunction of the tapetum in Ogura CMS radish is concomitant with the inhibition of anther-specific CHS expression; as a result, the level of flavonoids might decrease to below the level required to generate the pollen exine. CHS is one of the main enzymes in the flavonoid biosynthesis pathway, and an alteration in CHS activity would be expected to affect the accumulation of all classes of these compounds. As shown in Fig. 2, ‘MS-G’ anthers did not absorb UV light and fluorescence emission was observed, whereas ‘UC-G’ anthers strongly absorbed UV light. These results indicate that CMS anthers do not contain any UV-absorbing compounds, such as flavonoid derivatives. Preliminary observations using thin-layer chromatography and high-performance liquid chromatography analysis suggest that ‘MS-G’ anthers completely lack naringenin derivatives (S. Yang and H. Yamagishi, unpubl. res.), and this is considered as the reason why the microspores of ‘MS-G’ could not colorize (Fig. 1B). Therefore, the present results suggest that the inhibition of CHS expression and the resulting deficiency of flavonoids are related to pollen abortion in CMS anthers.
This study focused on the CHS gene, which was differentially expressed between the Ogura-type male-sterile and normal-type anthers of radish, prior to the morphological pollen abortion initiated in Ogura-type anthers. It was confirmed that CHS, amongst several flavonoid biosynthetic genes, is specifically inhibited in ‘MS-G’ buds (anthers; Table 1, Fig. 3). CHS was also inhibited in the pistil of ‘MS-G’ (Fig. 3), but no morphological or functional abnormalities could be found. In addition, it was confirmed that another CMS radish with orf138 also had reduced CHS expression (Fig. 4). These findings strongly suggest that the pollen abortion mechanism in Ogura CMS is related to inhibition of the biosynthesis of flavonoid compounds. It was observed that CHS expression was increased and decreased along with the development of anthers in normal-type ‘UC-G’ (Table 2). This suggests that CHS is essential in a particular stage of development in the anther. It is of considerable interest that CHS has been demonstrated to be essential for pollen development in several species. Disruptions to CHS activity in the anthers of petunia (Taylor and Jorgensen, 1992; van der Meer et al., 1992), maize (Coe et al., 1981; Mo et al., 1992) and tobacco (Fischer et al., 1997; Atanassov et al., 1998) resulted in the production of sterile pollen; however, these cases are all nuclear types of male sterility. In addition to male sterility, CHS is reported to be an important gene in the reproductive process, for example as a sex-determination gene that is expressed in male flower buds of Silene latifolia (Ageez et al., 2005) and in male cones of Pinus radiate (Walden et al., 1999). CHS-dependent flavonols are required for functional pollen and are involved in the pollination process in a number of species (Taylor and Jorgensen, 1992; van der Meer et al., 1992; van Eldik et al., 1997). Perturbation of phenylpropanoid biosynthesis also produces anthers with decreased pollen viability (Elkind et al., 1990). To our knowledge, this is the first report demonstrating that CHS is involved in a CMS plant.
In general, interactions between the nucleus and mitochondria play a role in the exhibition of cytoplasmic malesterility. Until now, it was unclear how expression of the mitochondrial gene orf138 induces male sterility. It is of interest that the current results confirmed that CHS expression was restored when pollen production was restored in CMS radishes by the introduction of the Rf gene, which inhibits the translation of orf138 (Fig. 4). This result is consistent with our hypothesis that CHS expression is under the control of orf138 and that CHS expression regulates CMS expression. This study suggests that flavonoids play an important role in the development of functional pollen, not only in nuclear-dependent male sterility but also in cytoplasmic male sterility.
ACKNOWLEDGEMENTS
We thank Dr Shuichi Date (Kyoto Prefectural University, Japan) for technical assistance with the TEM. This research was supported by the Japan Society for the Promotion of Science (no. P04459).
LITERATURE CITED
- Ageez A, Kazama Y, Sugiyama R, Kawano S. Male-fertility genes expressed in male flower buds of Silene latifolia include homologs of anther-specific genes. Genes & Genetic Systems. 2005;80:403–413. doi: 10.1266/ggs.80.403. [DOI] [PubMed] [Google Scholar]
- Atanassov I, Russinova E, Antonov L, Atanassov A. Expression of an anther-specific chalcone synthase-like gene is correlated with uninucleate microspore development in Nicotiana sylvestris. Plant Molecular Biology. 1998;38:1169–1178. doi: 10.1023/a:1006074508779. [DOI] [PubMed] [Google Scholar]
- Balk J, Leaver CJ. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell. 2001;13:1803–1818. doi: 10.1105/TPC.010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M, Grelon M, Pelletier G, Budar F. The restorer Rf0 gene acts post-translationally on the stability of the ORF138 Ogura CMS-associated protein in reproductive tissues of rapeseed cybrids. Plant Molecular Biology. 1999;40:893–902. doi: 10.1023/a:1006223908044. [DOI] [PubMed] [Google Scholar]
- Bonhomme S, Burdar F, Lancelin D, Samll I, Defrance MF, Pelletier G. Sequence and analysis of Nco2·5 Ogura-specific fragment correlated with male sterility in Brassica cybrids. Molecular and General Genetics. 1992;235:240–248. doi: 10.1007/BF00279379. [DOI] [PubMed] [Google Scholar]
- Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends in Genetics. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Coe EH, McCormick SM, Modena SA. White pollen in maize. Journal of Heredity. 1981;72:318–320. [Google Scholar]
- Delourme R, Budar F. Male sterility. In: Gómez-Campo C., editor. Biology of Brassica coenospecies. Amsterdam: Elsevier Science; 1999. pp. 185–216. [Google Scholar]
- Duroc Y, Gaillard G, Hiard S, Defrance MC, Pelletier G, Budar F. Biochemical and functional characterization of ORF138, a mitochondrial protein responsible for Ogura cytoplasmic male sterility in Brassiceae. Biochimie. 2005;87:1089–1100. doi: 10.1016/j.biochi.2005.05.009. [DOI] [PubMed] [Google Scholar]
- van Eldik GJ, Reijnen WH, Ruiter RK, van Herpen MMA, Schrauwen AM, Wullems GJ. Regulation of flavonol biosynthesis during anther and pistil development, and during pollen tube growth in Solanum tuberosum. Plant Journal. 1997;11:105–113. doi: 10.1046/j.1365-313x.1997.11010105.x. [DOI] [PubMed] [Google Scholar]
- Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ. Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proceedings of the National Academy of Sciences of the USA. 1990;87:9057–9061. doi: 10.1073/pnas.87.22.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Budde I, Hain R. Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant Journal. 1997;11:489–498. [Google Scholar]
- Goldberg R, Beals T, Sanders P. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourret JP, Delourme R, Renard M. Expression of Ogura cytoplasmic male sterility in hybrids of Brassica napus. Theoretical and Applied Genetics. 1992;83:549–556. doi: 10.1007/BF00226898. [DOI] [PubMed] [Google Scholar]
- Grant I, Beversdorf WE. A comparative light and electron microscopic study of microspore and tapetal development in male fertile and cytoplasmic male sterile oilseed rape (Brassica napus) Canadian Journal of Botany. 1986;64:1055–1068. [Google Scholar]
- Hanson MR, Bentolila S. Interaction of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16:S154–S169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, editor. The flavonoids: advances in research since 1986. Reading, UK: University of Reading Press; 1993. [Google Scholar]
- Iwabuchi M, Koizuka N, Fujimoto H, Sakai T, Imamura J. Identification and expression of the kosena radish (Raphanus sativus cv. Kosena) homologue of the ogura radish CMS-associated gene, orf138. Plant Molecular Biology. 1999;39:183–188. doi: 10.1023/a:1006198611371. [DOI] [PubMed] [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, et al. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant Journal. 2003;34:407–415. doi: 10.1046/j.1365-313x.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Makaroff CA. Organ-specific reduction in the abundance of a mitochondrial protein accompanies fertility restoration in cytoplasmic male-sterile radish. Plant Molecular Biology. 1994;26:935–946. doi: 10.1007/BF00028860. [DOI] [PubMed] [Google Scholar]
- Linke B, Borner T. Mitochondrial effects on flower and pollen development. Mitochondrion. 2005;5:389–402. doi: 10.1016/j.mito.2005.10.001. [DOI] [PubMed] [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JNM, Stuitje AR. Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell. 1992;4:253–262. doi: 10.1105/tpc.4.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proceedings of the National Academy of Sciences of the USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura H. Studies on the new make sterility in Japanese radish, with special reference to the utilization of this sterility towards the practical raising of hybrid seeds. Memorials of the Faculty of Agriculture Kagoshima University. 1968;6:39–78. [Google Scholar]
- Pelletier G, Budar F. The molecular biology of cytoplasmically inherited male sterility and prospects for its engineering. Current Opinion in Biotechnology. 2007;18:121–125. doi: 10.1016/j.copbio.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Shen JB, Hsu FC. Brassica anther-specific genes: characterization and in situ localization of expression. Molecular and General Genetics. 1992;234:379–389. doi: 10.1007/BF00538697. [DOI] [PubMed] [Google Scholar]
- Smart CJ, Moneger F, Leaver CJ. Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell. 1994;6:811–825. doi: 10.1105/tpc.6.6.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Jorgensen R. Conditional male fertility in chalcone synthase-deficient petunia. Journal of Heredity. 1992;83:11–17. [Google Scholar]
- Terachi T, Yamaguchi K, Yamagishi H. Sequence analysis on the mitochondrial orfB locus in normal and Ogura male-sterile cytoplasms from wild and cultivated radishes. Current Genetics. 2001;40:276–281. doi: 10.1007/s00294-001-0256-9. [DOI] [PubMed] [Google Scholar]
- Theis R, Robbelen G. Anther and microspore development in different male sterile lines of oilseed rape (Brassica napus L.) Angewandte Botanik. 1990;64:419–434. [Google Scholar]
- Walden AR, Walter C, Gardner RC. Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiology. 1999;121:1103–1116. doi: 10.1104/pp.121.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Terachi T. Intra- and inter-specific variations in the mitochondrial gene orf138 of Ogura-type male-sterile cytoplasm from Raphanus sativus and Raphanus raphanistrum. Theoretical and Applied Genetics. 2001;103:725–732. [Google Scholar]
- Yasumoto K, Matsumoto Y, Terachi T, Yamagishi H. Restricted distribution of orf687 as a pollen fertility restorer gene for Ogura male sterility in Japanese wild radish. Breeding Science. 2008;58:177–182. [Google Scholar]