Abstract
Background and Aims
Anaerobic or low oxygen conditions occur when maize plants are submerged or subjected to flooding of the soil. Maize survival under low oxygen conditions is largely dependent on metabolic, physiological and morphological adaptation strategies; the regulation mechanisms of which remain unknown. MicroRNAs (miRNAs) play critical roles in the response to adverse biotic or abiotic stresses at the post-transcriptional level. The aim of this study was to understand submergence-responsive miRNAs and their potential roles in submerged maize roots.
Methods
A custom μParaflo™ microfluidic array containing plant miRNA (miRBase: http://microrna.sanger.ac.uk) probes was used to explore differentially expressed miRNAs. Small RNAs from treated roots were hybridized with the microarray. The targets and their cis-acting elements of small RNA were predicted and analysed by RT-PCR.
Key Results
Microarray data revealed that the expression levels of 39 miRNAs from nine maize and some other plant miRNA families were significantly altered (P < 0·01). Four expression profiles were identified across different submergence time-points. The zma-miRNA166, zma-miRNA167, zma-miRNA171 and osa-miRNA396-like were induced in the early phase, and their target genes were predicted to encode important transcription factors, including; HD-ZIP, auxin response factor, SCL and the WRKY domain protein. zma-miR159, ath-miR395-like, ptc-miR474-like and osa-miR528-like were reduced at the early submergence phase and induced after 24 h of submergence. The predicted targets for these miRNAs were involved in carbohydrate and energy metabolism, including starch synthase, invertase, malic enzyme and ATPase. In addition, many of the predicted targets were involved in the elimination of reactive oxygen species and acetaldehyde. Overall, most of the targets of induced miRNAs contained the cis-acting element, which is essential for the anaerobic response or hormone induction.
Conclusions
Submergence-responsive miRNAs are involved in the regulation of metabolic, physiological and morphological adaptations of maize roots at the post-transcriptional level.
Key words: Anaerobic metabolism, Zea mays, gene expression, transcription factor, microRNA, flooding stress
INTRODUCTION
The regulation of gene expression in response to environmental cues is an important factor in plant survival and adaptability. Both transcriptional and post-transcriptional mechanisms have been implicated in the control of nuclear gene expression in response to submergence or flooding stress (Dennis et al., 2000; Dolferus et al., 2003). Transcriptome data have shown that the expression levels of a number of genes are altered in cells of submerged roots. These genes are involved in a broad spectrum of biochemical, cellular and physiological processes, including glycolysis, energy metabolism, lipid metabolism, signal transduction, DNA transcription, protein biosynthesis and digestion, cell components and photosynthesis (Klok et al., 2002; Liu et al., 2005; ZX Zhang et al., 2005, 2006). Many anaerobically induced genes share a consensus sequence called the anaerobic-responsive element (ARE), which is located in the promoter regions. The ARE is found to contain all the sequences necessary for both anaerobic induction and the binding of the transcription factors (TFs) of the Myb family (Walker et al., 1987; Dolferus et al., 1994; Hoeren et al., 1998). The data from the proteome and the bioactivity of functional proteins showed that the proteins efficiently synthesized in oxygen-deprived roots include enzymes involved in the breakdown of sucrose, glycolysis, ethanolic fermentation, aerenchyma formation and, especially, anaerobic polypeptides (Sachs et al., 1980, 1996; Andrews et al., 1994; Olson et al., 1995; Chang et al., 2000). A few of the protein products, especially some of the aerobic polypeptides (APs), did not increase while their mRNAs accumulated in oxygen-deprived cells. One potential cause of this is that the accumulated transcripts encoding these enzymes and anaerobic polypeptides are selectively translated, but not the APs. This indicates that the selected syntheses of anaerobic polypeptides are involved in transcriptional, as well as significant post-transcriptional regulation of gene expression (Fennoy and Bailey-Serres, 1995; Fennoy et al., 1998).
Plants exhibit a wide variety of anatomical, morphological and physiological responses to submergence in the roots or the whole plant. Some appear to have significant adaptability. The most conspicuous anatomical responses of maize roots to submergence, or anoxia, are the formation of an extensive aerenchyma system in the cortex and the development of adventitious roots in the vicinity of the cotyledonary nodes. These greatly facilitate gas transport in submerged root systems (Armstrong et al., 1971). Another response in leaves is stomatal closure. These changes can be mediated by diverse phytohormones, with ethylene and abscisic acid (ABA) playing prominent roles (Liao and Lin, 2001). Recently, Sub1A, a flooding-tolerance gene, was cloned in rice (Xu et al., 2006). The gene encodes a TF belonging to the B-2 subgroup of the ethylene-response factors, its expression increases the transcription of genes associated with ethanolic fermentation and represses the transcription of genes associated with cell elongation and carbohydrate catabolism. Regulation of genes involved in the morphological and physiological responses to oxygen deprivation, however, remains uncharacterized.
MicroRNAs (miRNAs) are approx. 21-nt-long, non-coding RNAs that play critical roles in the regulation of gene expression at the post-transcriptional level. Plant miRNAs regulate many genes involved in developmental control (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004), organ polarity (Eshed et al., 2001; McConnell et al., 2001; Kidner and Martienssen, 2004), development transitions (Aukerman and Sakai, 2003; Chen, 2004), leaf growth (Palatnik et al., 2003) and RNA metabolism (Xie et al., 2003). Recently, BH Zhang et al. (2005) found that >25 % of ESTs contain miRNAs in stress-induced plant tissues. It suggested that miRNAs may play an important role in plant responses to environmental cues. Several specific stress-responsive miRNAs have been identified under nutrient deficiency (Jones-Rhoades and Bartel, 2004; Fujii et al., 2005; Chiou et al., 2006), drought, cold and salinity stress (Sunkar and Zhu, 2004; Zhao et al., 2007), UV-B radiation (Zhou et al., 2007) and mechanical stress (Lu et al., 2005). Submergence is a serious abiotic stress to maize seedlings. In this study, a custom μParaflo™ microfluidic array (LC Sciences, Houston, TX, USA) containing version 10·0 plant miRNA probes was used to reveal the response of miRNAs to submergence-stress. The results indicated that these responsive miRNAs potentially play a vital role in the regulation of the morphological and metabolic adaptation response in cells of submerged roots of maize.
MATERIALS AND METHODS
Plant treatment and RNA isolation
Mo17, an inbred line of Zea mays sensitive to submergence, was germinated in an incubator at 30 °C. Uniformly germinated seeds were then sown in pots filled with vermiculite and grown until the seedlings developed three leaves in an incubator at 30 °C with a photosynthetic photon flux density of about 200 µmol m−2 s−1 immediately above the seedlings and 14/10 h light/dark cycles. During this period, the seedlings were irrigated every 2 d with 1× growth culture solution [Ca(NO3)2 820·7 mg L−1, KNO3 505·6 mg L−1, MgSO4·7H2O 616·2 mg L−1, KH2PO4 272·2 mg L−1, Fe-EDTA 13·02 mg L−1, H3BO3 2·860 mg L−1, MnSO4 1·015 mg L−1, CuSO4·5H2O 0·079 mg L−1, ZnSO4·7H2O 0·220 mg L−1, H2MoO4 0·090 mg L−1]. Seedlings with three leaves were used in all experiments. For the submergence treatment, seedlings were submerged with fresh culture solution, leaves were two-thirds underwater (i.e. the uppermost one-third of the shoot remained in air), and the roots were harvested at 12, 24 and 36 h post-treatment. Untreated seedlings were used as controls. About 2 g roots were collected from each treatment, washed free from vermiculite for approx. 30–60 s, immediately frozen in liquid nitrogen, and then ground into a fine powder. The total RNA from each sample was extracted using the Trizol reagent (Invitrogen, USA).
miRNA microarray and hybridization
Microarray assays were performed using a service provider by LC Sciences. A custom μParaflo™ microfluidic chip containing 623 unique plant probes from version 10·0 representing 799 miRNAs of 13 plant species miRNAs, with each chip containing five probe sets (Sanger Institute, Cambridge, UK; http://microrna.sanger.ac.uk/sequences/; Griffiths-Jones et al., 2006) was used. The 799 miRNAs consisted of 154 from Arabidopsis thaliana, 115 from Oryza sativa, 39 from Sorghum bicolor, 31 from Triticum aestivum, 10 from Saccharum, 43 from Zea mays and 407 from seven other plant species. PUC2-20B, a non-homologous nucleotide acid, and 5S rRNA served as positive controls. Blank cells and nucleic acid probes that were non-homologous with the sample RNA were also designed for the microarray and used as negative controls. No less than 5 µg of total RNA sample was used as the starting material for each assay. RNAs were size-fractionated using a YM-100 Microcon centrifugal filter (Millipore, Bedford, MA, USA), and the small RNAs (<300 nt) isolated were subsequently 3′-extended with a poly(A) tail using poly(A) polymerase. Among the control probes, PUC2PM-20B and PUC2MM-20B were a perfect match and single-base mismatch probe of a 20-mer RNA positive control sequence, respectively, which was spiked into the RNA samples prior to labelling. The oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining; two different tags were used for the two RNA samples used in the dual-sample experiments. Briefly, purified small RNAs were labelled with Cy3 or Cy5 fluorescent dyes (control and submergence-treatment, respectively) and then hybridized to the dual-channel microarray. The hybridization buffer was 100 µL 6× SSPE buffer (0·90 m NaCl, 60 mm Na2HPO4, 6 mm EDTA, pH 6·8) containing 25 % formamide at 34 °C. Microarray experiments were performed three times using distinct biological samples. Hybridization images were collected using a laser scanner (GenePix 4000B; Molecular Devices, Sunnyvale, CA, USA) and digitized using the Array-Pro image analysis software (Media Cybernetics, Bethesda, MD, USA). Data were analysed by first subtracting the background and then normalizing the signals using a LOWESS filter (locally weighted regression). In the two-colour experiments, the ratio of the two sets of detected signals was log2 transformed and balanced, and the P-values of the t-test were calculated. The difference of detected signals was considered significant at the 1 % level of significance.
miRNA target prediction and cis-acting element analysis
An miRNA is a transcript encoded by an miRNA gene. Theoretically, pre-miRNA sequences should be exact matches with specific maize genomic sequences. To determine the miRNA genes, each pre-miRNA sequence, which can be obtained by query at http://microrna.sanger.ac.uk/sequences/, was aligned to the maize genome sequence (maizesequence.org or maize.tigr.org). The DNA segment flanking queried sequences of not less than 10·0 kb was obtained, and used to predict the open reading frame using FGENESH (http://mendel.cs.rhul.ac.uk/). If the pre-miRNA sequence matched with a predicted gene sequence (including exon, intron and probable 5′- and 3′-terminal sequences), the DNA segment was regarded as an miRNA gene sequence. Since plant miRNAs recognize their target mRNAs by near-perfect base pairing, computational sequence similarity searches were used to identify potential targets. For the prediction of miRNA target mRNAs, a web-based integrated computing system, miRU, was queried using mature miRNA sequences, all potential complementary sequences containing limited mismatches were selected (Y Zhang, 2005). If miRNAs did not pair with any hit in miRU, then the miRNA and its reverse complementary sequence was used as a query in the public maize EST database (www.maizesequence.org). The number of mismatches was limited to less than three nucleotides; insertions or deletions were permitted to consist of no more than one nucleotide; and no more than five G-U pairs were admitted. The predicted target EST sequences were aligned with ClustalW. New contigs were then aligned and used for BLAST searching for functional annotation and for genomic sequence queries, respectively. The 1500-bp DNA sequence upstream of the transcription initiation site of each predicted miRNA gene and target gene was truncated to identify cis-acting motifs using PlantCARE (Rombauts et al., 1999; Lescot et al., 2002).
The expression assay of target genes
From each sample, 10 µg of total RNA was incubated with two units of RNase-free DNase I (New England BioLabs, Inc.) to remove DNA contamination from the RNA. An oligo(dT) DNA primer was then added to conduct the reverse transcription reaction as follows: 0·5 µL of total RNA and 1·75 µL of DEPC-free H2O were denatured for 5 min at 70 °C then incubated on ice for 3 min. The following reagents were then added in order: oligo(dT) DNA (0·25 µL of 2 µm), dNTP (0·5 µL of 10 mm each), RT buffer (1 µL of 5 × ), SuperScript II reverse transcriptase (0·25 µL of 200 U μL−1), RNase inhibitor (0·25 µL of 40 U μL−1) and DEPC-free H2O (0·5 µL). The reaction mixture was incubated for 60 min at 42 °C to synthesize cDNA. The specific target gene primers were then added to amplify the target genes from the cDNAs, and γ-tubulin was selected as the endogenous reference. The name, accession and primer sequences of target genes are listed in Table 1.
Table 1.
The accessions of target genes and primers for RT-PCR
| Target genes | Accession | Primer-sense | Primer-antisense |
|---|---|---|---|
| Acyl-CoA thioesterase | TC262881 | GCATCCATTCGTCAGCCTTC | TGATCCACGCTTCCCATCCC |
| AGO1-1 | TC255055 | AATGCCACAGTCATACCG | TAACTCCCTTGTTGCTTGT |
| Auxin response factor 12 | TC268913 | GGGCAAGTCCATCAGAAT | ACATCGTTCTCCCTGTCG |
| GAMYB | TC269992 | GTCAGCGTCCACCTTCTAC | CATTGAGGACCCACCACT |
| HD-zip | TC271068 | CGTAGCCCTGTTCCATTAGTT | TGTGCTGACAATCGCCTTTC |
| Phospholipase D | TC253142 | AGGGCTTACCTGCCTGTC | TGCTTCCTCCACCTCTGC |
| Serine/threonine protein phosphatase | TC270317 | CTGGACGCAATTTATTCAGC | CCCGACACCAACTACCTCTT |
| ZAG1 | TC274797 | ACGGCAGGATGTCTGAAACG | CTGAAGGAGGCACTGGAGCA |
| Gamma-tubulin | X83696 | TGTCGTCCAACCTTACAACTCACT | TCTCCAGGGTCCTCCATTCC |
RESULTS
Submergence-responsive miRNAs in roots of maize
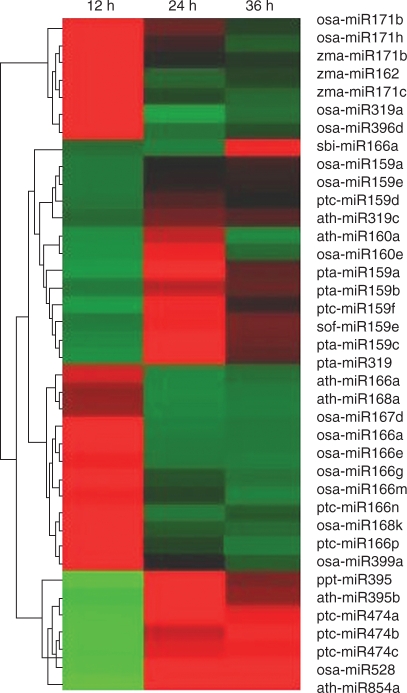
The microarray data showed that >100 expressed miRNAs had changed expression profiles owing to submergence treatment. These miRNAs were identified to be present in diverse families of plant species. In general, different plant species have similar mature miRNA sequences in the same miRNA families. They typically differ by only one to three nucleotides. For example, three members of pta-miR159 (pta-miR159a/b/c) from Pinus taeda have only one nucleotide difference with ptc-miR159d from Populus trichocarpa, and ptc-miR159d is homologous to zma-miRNA159c/d. The miRNA sof159e from Saccharum officinarum has two nucleotide differences with osa-miR159a from Oryza sativa which is homologous with zma-miRNA159a/b (Fig. 1). Thus, cross-hybridization among members from the same family and among orthologues across species could explain these microarray results. To refine the differentially expressed miRNAs, those miRNA members in the same family showing similar expression profiles were selected as the actual differentially expressed miRNAs. Four members of osa-miR166 (e, g, k and m) showed similar expression profiles. They were up-regulated after 12 h of submergence then down-regulated after 24 h of submergence. The miRNA osa-miR166 g is an orthologue of zma-miR166l and zma-miR166 m, osa-miR166 k is a homologue of zma-miR166j and zma-miR166 k. Thus it is suggested that the four identified zma-miRNAs were actually submergence-responsive miRNAs.
Fig. 1.
The similarity of sequences among miRNA members from different species. The miRNA in parentheses is the homologue of the miRNA before the parentheses. Shaded nucleotides show differences with zma-miRNA or its homologue. (–) indicates the absence of a nucleotide.
Although many differentially expressed miRNAs from arabidopsis and Oryza sativa have been identified, miRNAs are evolutionarily conserved throughout the plant kingdom (BH Zhang et al., 2006a), their microarray expression levels reflected the expression of their counterparts in maize. For example, the expression signal of pta-miR159a/b/c and sof-miR159e could have been attributed to hybridization with members of zma-miR159 and contributed to the change in the expression level of zma-miR159. In addition, non-homologous miRNAs with maize identified by microarray hybridization were suggested to be newly identified miRNAs in maize. A total of 39 miRNAs, about 6·26 % (39/623) of the probes on the microarray, were identified as submergence-responsive miRNAs at the 1 % level of significance (Table 2). Thirty out of the 39 miRNAs aligned with nine maize miRNA families, while the others corresponded to several members of miRNA families from three other plant species; these include ath-miR395, ath-miR854, ptc-miR474, osa-miR396 and osa-miR528.
Table 2.
Submergence-responsive miRNA families and members
| miR family | Members of identified miRNAs | Orthology miRNAs across species | Homologous miRNAs in Zea mays* |
|---|---|---|---|
| zma-miR159 | ptc-miR159d/f | zma-miR159c, zma-miR159d | |
| osa-miR159a/e | pta-miR159a/b/c, sof-miR159e | zma-miR159a, zma-miR159b | |
| zma-miR160 | ath-miR160a | zma-miR160a/b/c/d/e | |
| osa-miR160e | zma-miR160f | ||
| zma-miR162 | zma-miR162 | zma-miR162 | |
| zma-miR166 | ath-miR166a | ptc-miR166n/p | zma-miR166a |
| osa-miR166e/g/k/m | zma-miR166l/m, zma-miR166j/k | ||
| zma-miR166 | sbi-miR166a | zma-miR166b/c/d/e/f/g/h/i | |
| zma-miR167 | osa-miR167d | zma-miR167e/f/g/h/i | |
| zma-miR168 | osa-miR168a/b | ath-miR168a | zma-miR168a, zma-miR168b |
| zma-miR171 | osa-miR171b/h | zma-miR171d/e/i/j/h/k | |
| zma-miR171b/c | zma-miR171b/c | ||
| zma-miR319 | osa-miR319a | ath-miR319c, pta-miR319 | zma-miR319a/b/c/d |
| ptc-miR474 | ptc-miR474a/b/c | ||
| ath-miR395 | ath-miR395b | ppt-miR395 | |
| osa-miR396 | osa-miR396d | ||
| zma-miR399 | osa-miR399a | zma-miR399a/c | |
| osa-miR528 | osa-miR528 | ||
| ath-miR854 | ath-miR854a |
* Homology of identified miRNAs by microarray hybridization with maize miRNAs was obtained by searching in miRBase version 10·0 (Sanger Institute, Cambridge, UK; http://microrna.sanger.ac.uk/sequences).
At the three treatment time-points, the different miRNAs showed different expression levels, and four predominant expression profiles were detected using clustering analysis (Fig. 2). The expression of 19 miRNAs was up-regulated during the early stage (0–12 h) of submergence, then recovered to normal levels during later stages. The expression of 12 miRNAs, however, was down-regulated during the early stage, and increased after 24 h of submergence. Seven out of 39 miRNAs were dramatically induced between 24 h and 36 h of post-submergence, but sbi-miR166a from Sorghum bicolor showed a particular expression profile. It was down-regulated at the 24-h time-point, and then up-regulated at the 36-h time-point. These results indicated that the expression of miRNAs was regulated at different time-points during the submergence process.
Fig. 2.
The clustering of differentially expressed miRNAs for roots submerged for 12, 24 or 36 h relative to controls. miRNA from maize roots under normal growth condition was used as the reference. The colour saturation reflects the magnitude of the log2 expression ratio (Cy5/Cy3) for each transcript. Red colour means higher transcript levels than the reference, whereas green means lower transcript levels than the reference.
The potential target genes of submergence-responsive miRNAs
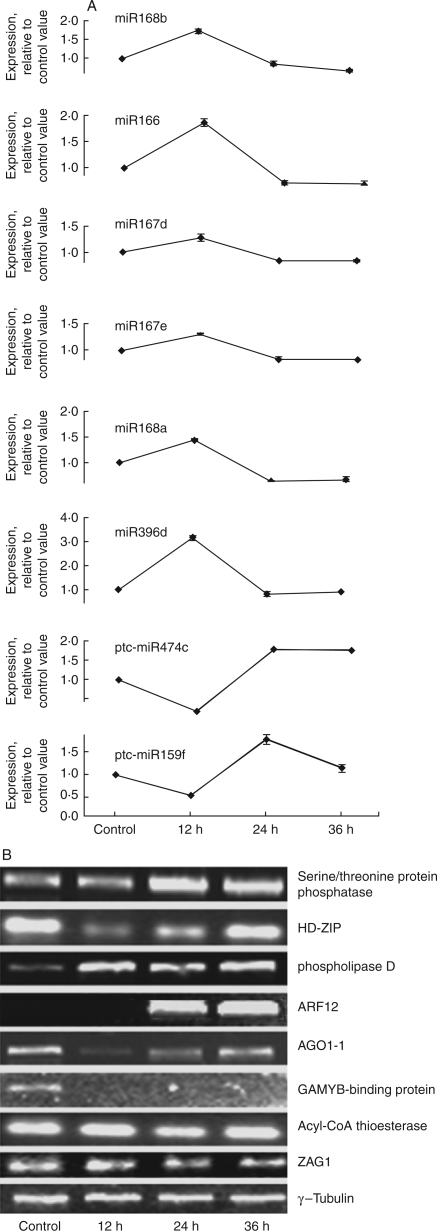
Previous studies have demonstrated that miRNAs regulate gene expression by binding to targeted mRNA sequences by a perfect or near-perfect hybridization (BH Zhang, 2005). This facilitates the prediction of plant miRNA targets using a homology search. A total of 38 potential target genes were identified. These potential target genes are involved in a broad spectrum of metabolite, cellular, and physiological processes, such as carbohydrate, lipid and energy metabolism, transcription, signal transduction, cell defence and differentiation. Of the predicted targets, the expression profiles of eight genes, including three genes encoding TFs (HD-ZIP, ZAG1 and GAMYB-binding protein), two signalling components (serine/threonine protein phosphatase, phospholipase D), an auxin response gene (ARF12), a key player in microRNA pathways (AGO1-1) and a metabolic gene (Acyl-CoA thioesterase) were confirmed by RT-PCR. Their expression profiles showed the expected negative regulation pattern of the target gene by the miRNAs (Fig. 3).
Fig. 3.
RT-PCR of representative target genes of miRNAs at different time-points in maize roots. (A) The expression of miRNAs associated with targets, expressed relative to the control value. (B) RT-PCR of representative targets of miRNAs. RT-PCR was performed with primers specific for each of the target genes. RT-PCR of γ-tubulin cDNA was used as a positive control.
Based on their biological function, the predicted target genes could be classified into three categories. First, many miRNAs directly targeted various TFs involved in plant development and organ formation. Most of the TFs were described previously as known targets of miRNAs in arabidopsis, Oryza sativa and Zea mays (Llave et al., 2002; Juarez et al., 2004; Kidner and Martienssen, 2004). For instance, ZAG1, an AGAMOUS-like gene in maize, was detected as the target of zma-miR159c/d. HD-ZIP and the scarecrow-like family (SCL) mRNA were, respectively, predicted as the target of zma-miR166 and zma-miRNA171. The expression levels of the three TFs were negatively correlated with those of corresponding zma-miRNAs (Fig. 3). Some of the transcription factor/activator mRNAs, such as heat shock factors, ethylene response element-binding proteins, MADS-box proteins, AP2 domain proteins, leucine zipper and zinc finger proteins, were up-regulated in response to various regimes of oxygen deprivation in arabidopsis and maize (Klok et al., 2002; Gonzali et al., 2005; ZX Zhang et al., 2006). It was found that these TFs were also predicted targets of miRNAs (Table 3). Secondly, several targets of miRNAs, such as GAMyb and auxin response factors (ARF12, ARF17 and ARF25), are involved in phytohormone signal cascades (Table 3). Thirdly, several targets of miRNAs encode proteins that function in diverse metabolic pathways and are involved in various physiological processes. Interestingly, many targets were involved in the biosynthesis of cell wall metabolites and ATP, such as β-d-xylosidase, glycoside hydrolase, carbonate dehydratase, starch synthase, malic enzyme, invertase, ATPase and ATP sulfurylase (Table 3). Of these metabolic-associated targets, the anaerobic-induced expression of starch synthase, malic enzyme and invertase were confirmed on the transcriptional and protein level in maize roots (reviewed by Drew, 1997; Vartapetian and Jackson, 1997), indicating that submergence-responsive miRNAs may be involved in regulating the pathway of anaerobic metabolism.
Table 3.
Predicted target genes and motif of target genes in 5′ upstream sequences
| MiR ID | Accession of miRNA targets* | Protein annotation | e-value | Predicted motif in 5′ upstream of target gene† |
|---|---|---|---|---|
| Zma-miR159a, b | AC186310·3 | Serine/threonine protein phosphatase | 4e-67 | ARE, GARE-motif, HSE, LTR, MBS, TGA |
| AC199576·2-Contig94 | GAMYB | 4e-170 | ABRE, ARE, LTR, MBS | |
| zma-miR159c/d | AC204211·2-Contig32 | Ubiquitin isopeptidase | 1e-78 | ABRE, ARE, GC-motif, LTR, MBS, TCA, TGA |
| AC194290·2-Contig69 | V-ATPase subunit D | 6e-98 | ARE, MBS, WUN | |
| AC188029·2 | ZAG1, AGAMOUS | 3e-45 | ABRE, ARE, GARE, GC-motif, MBS, TCA | |
| zma-miR160f | AZM5_84507 | Alpha/beta fold family protein | 1e-122 | ABRE, ACE, ARE, LTR, MBS |
| zma-miR162 | AC191256·2 | DICER-LIKE1 | 4e-54 | ABRE, DRE, LTR, TGA |
| zma-miR166l/m/j/k | AC191059·3 | Rolled leaf1, HD-ZIP | 0·0 | ABRE, EIRE, GC-motif, LTR, MBS, plant AP-2 |
| zma-miR167e/f/g/h/i | AC191413·2-Contig32 | Auxin response factor (12, 17, 25) | 0·0 | ARE, HSE, MBS, TCA, WUN |
| AC194260·2-Contig18 | Neutral invertase | 0·0 | ABRE, GARE, MBS, MBS, TCA, TGA | |
| zma-miR168a/b | AC194028·2-Contig34 | Serine/threonine-protein phosphatase | 4E-80 | ABRE, GARE-motif, TCA |
| AC190578·3-Contig63 | Leucine-rich repeat family protein | 0·0 | GARE, LTR, MBS, TCA, TGA | |
| AC199001·2-Contig25 | AGO1-1 | 2e-93 | ABRE, ARE, GARE, TGA | |
| AC194028·2-Contig34 | Putative tonneau 2 | 0·0 | ARE, EIRE, GARE, TCA | |
| zma-miR171c | AC205908·1-Contig87 | Scl1 | 7e-161 | ARE, GARE, HSE, LTR, MBS, P-box, TCA |
| AC202909·2-Contig32 | WRKY transcription factor | 4e-20 | ARE, EIRE, ERE, HSE, MBS | |
| zma-miR319 | AC189090·2-contig34 | Peptide transporter | 3e-122 | ARE, HSE, MBS, MRE, TCA |
| zma-miR399a/b | AC209708·1-Contig49 | Hypothetical protein | 1e-162 | ABRE, ARE, GC-motif, TGA |
| AC212680·1 Contig186 | Aminotransferase | 2e-26 | GARE, HSE, MBS, TCA | |
| AC190908·0-cotig39 | Granule-bound starch synthase | 0·0 | ABRE, ARE, GC-motif, LTR, TGA, plant-AP2 | |
| ath-miR395b | AC186565·2-Contig10 | Beta-d-xylosidase | 0·0 | ABRE, GC-motif, MBS, plant-AP2 |
| AC199419·3-Contig42 | NADP-dependent malic enzyme | 0·0 | ARE, GC-motif, HSE, LTR, MBS, TGA | |
| AC194854·2-Contig67 | ATP sulfurylase | 0·0 | ABRE, GC-motif, HSE, MBS, SARE | |
| AC206431·1-Contig49 | Glycoside hydrolase | 5e-88 | ABRE, ARE, AuxRR-core, GARE, HSE, MBS, TCA | |
| ath-miR854a | AC186510·2-Contig29 | Water stress-induced protein | 3e-6 | ABRE, ARE, GC-motif, MBS, TCA |
| AC196385·2-Contig31 | Dof zinc finger protein | 2e-68 | ABRE, ARE, GC-motif, MBS | |
| AC194714·3-Contig17 | ZmSIG6 | 2e-168 | ARE, C-repeat/DRE, MBS, GC-motif, TGA | |
| ptc-miR474a | AZM5_99082 | NAD-dependent malic enzyme | 2e-143 | LTR, MBS, MRE, TCA |
| AC208836·1 | Pumilio/Mpt5 family | 2e-69 | ERE, GARE, HSE | |
| ptc-miR474b | AZM5_12664 | Putative protein kinase | 0·0 | ARE, GC-motif,MRE, TGA |
| AZM5_102471 | Carbonate dehydratase | 2e-86 | ARE, GC-motif, HSE, LTR, TCA | |
| ptc-miR474c | AC199387·3-Contig31 | Acyl-CoA thioesterase | 3e-151 | Unknown |
| osa-miR396d | AC209432·1-Contig40 | Growth-regulating factor 2,6 | 1e-170 | Box-W1, HSE,P-box, TC-rich, TCA, Wun |
| AC190640·3-Contig19 | GAMYB | 0·0 | MBS, TCA | |
| Osa-miR528 | AC177830·3-Contig89 | Cu,Zn-superoxide dismutase | 6e-50 | ABRE, AuxRR-core, HSE, TCA, Wun |
| AC197013·3-Contig16 | Hypothetical protein | 5e-21 | ABRE, GC-motif, LTR, MBS, TGA | |
| AC208577·1-Contig2 | bHLH transcription factor | 2e-73 | ARE, AuxRR-core, GARE, LTR, MBS | |
| AC189099·3-Contig25 | Aldehyde dehydrogenase | 0·0 | ABRE, ERE, GC-motif, plant-AP2 |
* The mature miRNA sequence was used to maize or rice mRNA database using miRU, a plant microRNA potential target finder (http://bioinfo3.noble.org/miRNA/miRU.htm; Zhang, 2005). If miRNAs did not pair with any hit in miRU, then the miRNA and its reverse complementary sequence were used as a query in the public maize EST database (www.maizesequence.org). The number of mismatches was limited to no more than three nucleotides, insertions or deletions were permitted to consist of no more than one nucleotide, and no more than five G-U pairs were admitted.
† About 1500 bp of DNA sequence upstream of the transcription initiation site of each predicted target gene were used to identify cis-acting motifs using PlantCARE (Rombauts et al., 1999; Lescot et al., 2002). ARE, Anaerobic-responsive element; ABRE, ABA-responsive element; AuxRR, auxin-responsive element; ERE, ethylene-responsive element; GARE, gibberellin-responsive element; GC-motif, enhancer-like element involved in anoxic-specific inducibility; HSE, hot-shock element; LTR, low temperature-responsive element; MBS, MYB binding site; TCA/SARE-element, salicylic acid-responsive element; WUN, wound-responsive element.
Key cis-acting element within miRNAs and target genes
Using the PlantCARE motif-finding algorithm, significant matches with sequence motifs in the 5′-upstream of eight miRNA genes were found (Table 4). These cis-acting elements include; the ARE (anaerobic response element), GC-motif, HSE (hot shock element), LTR (low temperature-responsive element), ABRE (ABA-responsive element), GARE (gibberellin-responsive element) and ERE (ethylene-responsive element). Each of these miRNAs has more than one stress-responsive cis-acting element, implicating that these miRNAs are involved in response to various biotic, abiotic and phytohormone stimuli. As shown in Table 4, some of the submergence up-regulated miRNA genes have anaerobic response motifs in their promoters. Especially, seven out of eight miRNAs genes contained ARE and/or GC-motifs, which were found in the 5′-upstream regions of ADH1 (Walker et al., 1987; Dolferus et al., 1994; Hoeren et al.,1998), and specifically in response to anaerobic stress.
Table 4.
Mainly predicted motif in 5′ upstream of microRNA gene
| miRNA | Accession* | Predicted motif in 5′ upstream of microRNA gene† |
|---|---|---|
| zma-miR159c | AC194022·2-Contig40 | GARE, MBS, SARE, TCA |
| zma-miR159d | AC189038·2-Contig53 | GARE, GC-motif, MBS, TCA |
| zma-miR160f | AC191030·1-Contig73 | ARE, TGA |
| zma-miR166l | AZM5_6103 | ABRE, ARE,GARE, LTR, MBS, TCA |
| zma-miR166j | AC196406·3-Contig18 | ABRE, ARE, GARE, TGA |
| zma-miR168a | AC209687·1-Contig53 | ARE, GARE,HSE,LTR,MBS |
| zma-miR168b | AC209687·1-Contig53 | ARE,GARE, HSE, LTR, MBS |
| zma-miR319a | AZM5_88200 | ARE, GC-motif |
* The pre-miRNA sequence was used to query the maize genome sequence (maizesequence.org or maize.tigr.org). The DNA segment flanking queried sequences of not less than 10·0 kb was used to predict the ORF using FGENESH (http://mendel.cs.rhul.ac.uk/).
† About 1500 bp DNA sequence upstream of the transcription initiation site of each predicted target gene was used to identify cis-acting motifs using PlantCARE (Rombauts et al., 1999; Lescot et al., 2002). ARE, Anaerobic-responsive element; ABRE, ABA-responsive element; GARE, gibberellin-responsive element; GC-motif, enhancer-like element involved in anoxic specific inducibility; MBS, MYB-binding site; TCA/SARE, salicylic acid-responsive element; HSE, hot-shock element; LTR, low temperature-responsive element.
The 38 predicted target genes also contained various cis-elements involved in response to diverse stimuli. The prevalent cis-elements were: ARE, GC-motif, ABRE, GARE, HSE, LTR and TC-rich repeats (Table 3). The ARE and/or GC-motif was present in the 5′-upstream region of 28 out of 38 targets. In addition, the targets without ARE or GC-motifs had one or more specific cis-elements involved in response to GA or ABA or diverse abiotic stresses. This suggested that the stimuli inducing the target genes as well as the miRNAs at least partially overlap. The miRNAs are involved in the feedback loop control mechanism to attenuate the expression of the target genes.
DISCUSSION
miRNAs respond specifically or non-specifically to submergence stress in maize root cells
Recent studies have shown that ath-miR395 and ath-miR399 were, respectively, up-regulated under low-sulphate and low-phosphate levels in the media (Jones-Rhoades and Bartel, 2004; Fujii et al., 2005). The present microarray data revealed that expressions of ath-miR395b, ppt-miR395 and osa-miR399a were also altered in roots during submergence. Ath-miR395b and ppt-miR395 were down-regulated at the early submergence stage (0–12 h), but were significantly induced after 24 h. Osa-miR399a was dramatically up-regulated at the early stage, and then weakly down-regulated after 24 h, indicating that the three miRNAs may have been regulated by nutrient stress as well as submergence stress. The eight members from the miR159 family across species showed a similar expression profile. They were significantly down-regulated at the 12 h time-point, and recovered to normal levels at the later stage. Because these miRNAs are homologous with four members of zma-miR159 (miR159a/b/c/d), it is suggested that zma-miR159a/b/c/d may be part of the submergence stress response in maize roots.
The miRNAs identified in this study also included a few members of miRNA160, miR166, miR168 and miR171 of maize (Table 2). Sequence analysis of a stress-treated arabidopsis small RNA library indicated that ath-miR393, ath-miR397b and ath-miR402 were up-regulated by ABA, cold, dehydration and salt stress, whereas ath-miR398a was down-regulated (Sunkar and Zhu, 2004; Sunkar et al., 2006). The expression patterns of the four miRNAs, however, did not change in this study, indicating that these miRNAs in maize are not involved in response to submergence stress. Stress-specific regulated miRNAs were also observed in previous studies. The ath-miR319c was specifically up-regulated by cold but not by ABA, dehydration or salt (BH Zhang et al., 2006b). Osa-miR169 g was confirmed as the only member induced by drought in rice (Zhao et al., 2007). The expression profile of osa-miR169 g, however, did not change in cells of submerged maize roots (present study). The differences between miRNAs identified under adverse abiotic stresses with those identified under submerged root cells showed that submergence probably induced a set of specific miRNAs.
The 96 zma-miRNAs on the microarray constituted a very small sample of approx. 500 miRNA genes relative to the >50 000 protein-coding genes in the maize genome (Messing et al., 2004; Fu et al., 2005). In addition, the differentially expressed miRNAs identified in this study constituted only about 6·26 % of the predicted miRNAs. Furthermore, it is known that dramatic metabolic and physiological alterations occurred in maize roots after 12 h of submergence. Metabolic pathways rapidly switched from aerobic metabolism to fermentation (Drew, 1997). In addition, aerenchyma and adventitious roots develop for adaptation to the low-oxygen environment (Kawase, 1981). Amino-cyclopropane carboxylic acid (ACC) synthase, a critical enzyme in ethylene biosynthesis, was also induced (Olson et al., 1995). These metabolic and physiological alterations implicated changes in the global gene expression profiles. If miRNAs are truly involved in the regulation of those alterations, a set of specifically submergence-regulated miRNAs should be expected.
Submergence-responsive miRNAs potentially regulate metabolic and morphological adaptations facilitating survival of maize seedlings under submergence conditions
Plants respond to oxygen limitation with a significant reprogramming of gene expression. Numerous studies have shown that a number of anaerobic polypeptides are induced during low oxygen treatment, while normal APs are dramatically repressed (Sachs et al., 1980, 1996; Chang et al., 2000). Many of the induced proteins were subsequently identified as enzymes of carbohydrate and lipid metabolism, and their accumulation facilitated the supply of energy for normal physiological processes. Interestingly, several mRNAs encoding anaerobic-responsive proteins are also targets of differentially expressed miRNAs. The present microarray data showed that the expression levels of miR162 and miR168 changed as the result of anaerobic induction. The direct targets for these were, respectively, DCL1 and AGO1-1. Both are key proteins during mature miRNA formation in the plant kingdom (Bartel, 2004; Vaucheret et al., 2004). In addition, the accumulation of miR159, ath-miR395-like, ptc-miR474-like and osa-miR528-like were reduced in the early stage of submergence in root cells. Vacuolar-ATPase, ATP sulfurylase, granule bound starch synthase and malic enzyme are important anaerobic proteins. Their mRNAs also are potential targets of miR159, miR399, ath-miR395-like and ptc-miR474-like, respectively (Table 3). The down-regulation of zma-miR159, ath-miR395-like and ptc-miR474-like suggested the occurrence of an accumulation of vacuolar-ATPase, ATP sulfurylase and malic enzyme, while the up-regulation of miR399 could result in a decrease in granule-bound starch synthase and amino-transferase. The down-regulation of ath-miR395-like and ptc-miR474-like resulted in accumulations of sucrose degradation-associated proteins in the cell and enhanced carbohydrate breakdown to supply the substrate for glycolysis to produce ATP. In addition, the accumulation of acyl-CoA thioesterase due to a decrease of ptc-miR474c-like might also ensure a high level of β-oxidation of fatty acids resulting in the release of free coenzyme A and subsequently stored energy.
In addition to energy metabolism, miRNAs might also be involved in other aspects of cellular metabolism. As examples, accumulations of superoxide dismutase and aldehyde dehydrogenase, two targets of osa-miR528-like miRNA, would also aid the elimination of reactive oxygen species and acetaldehyde produced by either a plasma membrane NAD(P)H oxidase and/or mitochondria. This supports the view of a regulated response to oxygen deprivation (Bailey-Serres and Chang, 2005; Agarwal and Grover, 2006).
Accompanied by the metabolic adaptations mentioned above, some miRNAs, including miR166, miR167, miR171, miR399 and osa-miR396-like, were induced in the early phase. It directly altered the level of transcripts encoding TFs such as AGAMOUS and HD-ZIP. Rolled leaf 1 (rld1) of maize is a homologue of the arabidopsis HD-ZIP gene and can be cleaved by miR166 (Juarez et al., 2004). Previous studies showed that miR166-mediated cleavage of HD-ZIP was found to regulate the pattern of vasculature and the establishment or maintenance of abaxial–adaxial polarity in lateral organs (Emery et al., 2003; Kim et al., 2005; Prigge et al., 2005). The accumulation of zma-miR166 under submergence stress indicated that root meristem cell differentiation probably caused the formation of the vascular system with expanded xylem tissue. This hypothesis was confirmed by the induction of ACC synthase and xyloglucan endo-transglycosylase (Olson et al., 1995; Sachs et al., 1996).
Recently, several independent groups of workers have observed that MiR159 regulated key components of auxin signaling pathways and further regulated adventitious root formation and lateral root development (Achard et al., 2004; Mallory et al., 2005; Sorin et al., 2005). It the present study it was found that three ARF transcripts (ARF12, ARF17 and ARF25) and AGO1 in maize were predicted as the targets of zma-miR167 and zma-miR168, respectively. The changes in the expression profiles of both zma-miR167 and zma-miR168 at the early submergence stage can explain the accumulation of ARF transcripts. This event would result in the formation of adventitious roots in the hypocotyl of submerged maize seedlings. It is suggested that this mechanism of morphological adaptation to the anaerobic environment was associated with the regulation of the auxin signal pathway by the miRNA-directed cleavage of ARF mRNA.
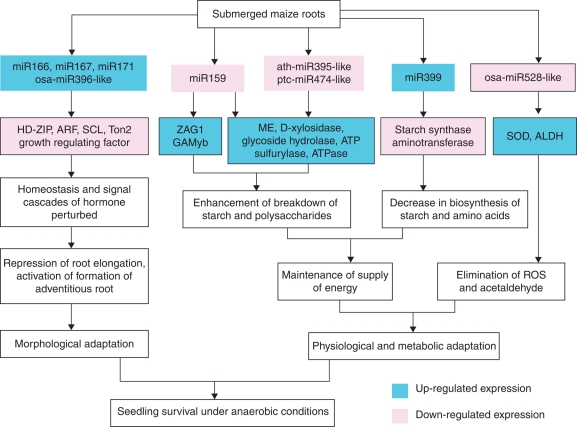
Based on the miRNA expression profiles and the functions of miRNA targets, the possible molecular responses and physiological changes that occurred in root cells of maize when submerged have been outlined (Fig. 4). The present study demonstrated the potential role of submergence-responsive miRNAs at the post-transcriptional level in the regulation of the metabolic, physiological and morphological adaptations of maize seedling survival under oxygen-deficient conditions.
Fig. 4.
The potential regulating network of submergence-responsive miRNAs in roots cell. The stress-responsive miRNAs could be involved in three pathways to reprogramme complex procedures of metabolism and physiology. (1) Up-regulated miR399 directly targeted cleavage of mRNAs of starch synthase and amino-transferase, prevented metabolic intermediates and glucose from synthesizing starch and amino acid. Accumulated low molecular weight carbohydrate could act as substrate of glycolysis pathway. In addition, the down-regulated miR159, ath-miR395-like and ptc-miR474-like could enhance accumulation of many anaerobic-responsive enzymes involved in carbohydrate and energy metabolism; these accumulated enzymes enhance glycolysis and thus ATP supply. The aerobic metabolism in roots cells would be shifted into anaerobic metabolism during the early submergence phase. (2) Accumulation of SOD and ALDH due to decreasing of osa-miR528-like would aid elimination of reactive oxygen species and acetaldehyde, and thus survival of root cells. (3) The accumulated miR166 and miRNA167 together with down-regulated miR159 could modulate hormone homeostasis via regulating transcripts of HD-ZIP, ARF and GAMYB and then trigger adventitious root formation and lateral root development. The plant has acquired morphological characteristics to adapt to the low-oxygen environment.
ACKNOWLEDGEMENTS
We are grateful to Dr Qiulei Lang (LC-Bio,Hangzhou,China) for technical assistance. We are also grateful to Dr Lifang Zhang (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York) and Dr Bailin Li (Dupont Pioneer USA) for critically revising the manuscript. This work was supported by the Hi-Tech Research and Development Program of China (grant numbers 2008AA10Z112 and 2006AA100103), National Natural Science foundation of China grant numbers 30571171 and 30771352.
LITERATURE CITED
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Grover A. Molecular biology, biotechnology and genomics of flooding-associated low O2 stress response in plant. Critical Reviews in Plant Sciences. 2006;25:1–21. [Google Scholar]
- Andrews DL, Drew MC, Johnson JR, Cobb BG. The response of maize seedlings of different ages to hypoxic and anoxic stress. Plant Physiology. 1994;105:53–60. doi: 10.1104/pp.105.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong W. Radial oxygen losses from intact rice roots as affected by distance from the apex, respiration and waterlogging. Physiologia Plantarum. 1971;25:192–197. [Google Scholar]
- Aukerman M, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R. Sensing and signalling in response to oxygen deprivation in plants and other organisms. Annals of Botany. 2005;96:507–518. doi: 10.1093/aob/mci206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identify important target genes. Proceedings of the National Academy of Sciences of the USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment and identification of proteins by mass spectrometry. Plant Physiology. 2000;122:295–317. doi: 10.1104/pp.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. Regulation of phosphate homeostasis by microRNA in Arabidopsis. The Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren F, et al. Molecular strategies for improving waterlogging tolerance in plants. Journal of Experimental Botany. 2000;51:89–97. [PubMed] [Google Scholar]
- Dolferus R, Peacock WJ, Dennis ES. Differential interactions of promoter elements in stress responses of the Arabidopsis ADH gene. Plant Physiology. 1994;105:1075–1078. doi: 10.1104/pp.105.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Klok EJ, Delessert C, Wilson S, Ismond KP, Good AG, et al. Enhancing the anaerobic response. Annals of Botany. 2003;91:111–117. doi: 10.1093/aob/mcf048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:223–250. doi: 10.1146/annurev.arplant.48.1.223. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum S, Perea J, Bowman J. Establishment of polarity in lateral organs of plants. Current Biology. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- Fennoy SL, Bailey-Serres J. Post-transcriptional regulation of gene expression in oxygen-deprived roots of maize. The Plant Journal. 1995;7:287–295. doi: 10.1046/j.1365-313X.1998.00249.x. [DOI] [PubMed] [Google Scholar]
- Fennoy SL, Nong T, Bailey-Serres J. Transcriptional and posttranscriptional processes regulate gene expression in oxygen-deprived roots of maize. The Plant Journal. 1998;15:727–735. doi: 10.1046/j.1365-313X.1998.00249.x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, et al. Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proceedings of the National Academy of Sciences of the USA. 2005;102:12282–12287. doi: 10.1073/pnas.0503394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Novi G, Poggi A, Alpi A, Perata P. The use of microarrays to study the anaerobic response in arabidopsis. Annals of Botany. 2005;96:661–668. doi: 10.1093/aob/mci218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. Evidence for a role of AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics. 1998;149:479–490. doi: 10.1093/genetics/149.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Kawase M. Anatomical and morphological adaptation of plants to waterlogging. HortScience. 1981;16:30–34. [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, et al. microRNA directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. The Plant Journal. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RB, Somerville SC, et al. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. The Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau K, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CT, Lin CH. Physiological adaptation of crop plants to flooding stress. Proceedings of the National Science Council of ROC, Part B. 2001;25:148–157. [PubMed] [Google Scholar]
- Liu F, Van Toai T, Moy LP, Bock G, Linford LD, Quackenbush J. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiology. 2005;137:1115–1129. doi: 10.1104/pp.104.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa, that are absent from Arabidopsis. The Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J, Emery J, Eshed Y, Bao N, Bowman J, Barton M. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, et al. Sequence composition and genome organization of maize. Proceedings of the National Academy of Sciences of the USA. 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DC, Oetiker JH, Yang SF. Analysis of LE-ACS3, a 1-aminocyclopropane-1-carboxylic acid synthase gene expressed during flooding in the roots of tomato plants. Journal of Biological Chemistry. 1995;270:14056–14061. doi: 10.1074/jbc.270.23.14056. [DOI] [PubMed] [Google Scholar]
- Palatnik J, Allen E, Wu X, Schommer C, Schwab R, Carrington J, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. The Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van montaqu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Research. 1999;27:295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM, Freeling M, Okimoto R. The anaerobic proteins of maize. Cell. 1980;20:761–767. doi: 10.1016/0092-8674(80)90322-0. [DOI] [PubMed] [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN. Anaerobic gene expression and flooding tolerance in maize. Journal of Experimental Botany. 1996;47:1–15. [Google Scholar]
- Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. The Plant Cell. 2005;17:1343–1359. doi: 10.1105/tpc.105.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. The Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartapetian BB, Jackson MB. Plant adaptations to anaerobic stress. Annals of Botany. 1997;79(Suppl. A):3–20. [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Development. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Howard EA, Dennis ES, Peacock WJ. DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase-1 gene. Proceedings of the National Academy of Sciences of the USA. 1987;84:6624–6628. doi: 10.1073/pnas.84.19.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau K, Carrington J. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Current Biology. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNA using EST analysis. Cell Research. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. The Plant Journal. 2006;a 46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Developmental Biology. 2006;b 89:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Research. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZX, Tang WH, Tao YS, Zheng YL. cDNA microarray analysis of early response to submerging stress in Zea mays roots. Russian Journal of Plant Physiology. 2005;52:43–49. [Google Scholar]
- Zhang ZX, Zou XL, Tang WH, Zheng YL. Revelation on early response and molecular mechanism of submergence tolerance in maize roots by microarray and suppression subtractive hybridization. Environmental and Experimental Botany. 2006;58:53–563. [Google Scholar]
- Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, et al. Identification of drought-induced microRNAs in rice. Biochemical and Biophysical Research Communications. 2007;354:585–90. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Molecular Systems Biology. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]