Abstract
Background and Aims
The aim of this work was the identification and molecular characterization of novel sugar beet (Beta vulgaris) repetitive sequences to unravel the impact of repetitive DNA on size and evolution of Beta genomes via amplification and diversification.
Methods
Genomic DNA and a pool of B. vulgaris repetitive sequences were separately used as probes for a screening of high-density filters from a B. vulgaris plasmid library. Novel repetitive motifs were identified by sequencing and further used as probes for Southern analyses in the genus Beta. Chromosomal localization of the repeats was analysed by fluorescent in situ hybridization on chromosomes of B. vulgaris and two other species of the section Beta.
Key Results
Two dispersed repetitive families pDvul1 and pDvul2 and the tandemly arranged repeat family pRv1 were isolated from a sugar beet plasmid library. The dispersed repetitive families pDvul1 and pDvul2 were identified in all four sections of the genus Beta. The members of the pDvul1 and pDvul2 family are scattered over all B. vulgaris chromosomes, although amplified to a different extent. The pRv1 satellite repeat is exclusively present in species of the section Beta. The centromeric satellite pBV1 by structural variations of the monomer and interspersion of pRv1 units forms complex satellite structures, which are amplified in different degrees on the centromeres of 12 chromosomes of the three species of the Beta section.
Conclusions
The complexity of the pBV1 satellite family observed in the section Beta of the genus Beta and, in particular, the strong amplification of the pBV1/pRv1 satellite in the domesticated B. vulgaris indicates the dynamics of centromeric satellite evolution during species radiation within the genus. The dispersed repeat families pDvul1 and pDvul2 might represent derivatives of transposable elements.
Key words: Beta vulgaris, dispersed repeats, satellite DNA, FISH
INTRODUCTION
Repetitive sequence families are major components of plant genomes (Heslop-Harrison, 2000). They are divided into tandemly arranged and dispersed sequences according to their genome organization (Schmidt and Heslop-Harrison, 1998). Tandem repeats include satellite DNA, micro- and minisatellites, telomeric repeats and ribosomal genes. Typical satellite repeating units are 160–180 bp or 320–360 bp long and are arranged in tandem arrays of hundreds of thousands copies (Hemleben et al., 2000; Macas et al., 2002), thus comprising a significant portion of the repetitive DNA. Satellites, together with transposable elements, are a major driving force for the variation in the plant genome regarding size and complexity (Heslop-Harrison, 1996, 2000). Dispersed elements are scattered over the genome and interspersed with other genomic sequences. Many of these repeats are derived from mobile DNA sequences, in particular from retrotransposons, as a result of an erroneous reverse transcription or divergence and rearrangements of integrated elements at the DNA level (Bennetzen et al., 1994; SanMiguel et al., 1996).
Repetitive DNA is predominantly localized in heterochromatic and subtelomeric regions of chromosomes (Heslop-Harrison, 1996). Particularly, plant centromeres, which are detectable as primary constrictions or as AT- or GC-rich heterochromatin, consist of various classes of repetitive sequences often separated by islands of higher complexity (Presting et al., 1998; Copenhaver et al., 1999; Heslop-Harrison et al., 1999; Gindullis et al., 2001a). Centromere-associated repeats have been described in various monocots (Dong et al., 1998; Nagaki et al., 1998; Hudakova et al., 2001) and dicots (Harrison and Heslop-Harrison, 1995; Schmidt and Heslop-Harrison, 1996; Brandes et al., 1997; Gindullis et al. 2001b).
The fast evolution of both tandemly arranged and dispersed repetitive DNA leads to changes in sequence and abundance (Schmidt and Heslop-Harrison, 1998) and often results in species-specifc repeat variants and/or the generation of novel sequence families. Therefore, comparative studies of plant repetitive sequences are useful for the investigation of evolutionary relationships between plant species (Kamm et al., 1995; Bennetzen, 2000; Ohmido et al., 2000; Nouzova et al., 2001).
Species of the genus Beta are grouped into four sections Beta, Corollinae, Nanae and Procumbentes. All cultivated beets (sugar, fodder, garden and leaf beet) belong exclusively to the section Beta. With approx. 20 closely and distantly related species and subspecies, the genus provides a suitable system for the comparative study of nuclear genome composition and evolution. Many genus-, section- or species-specific repetitive DNA sequences have been analysed from cultivated and wild Beta species (Schmidt and Metzlaff, 1991; Kubis et al., 1997, 1998; Gao et al., 2000).
The sugar beet (Beta vulgaris) genome is 758 Mbp in size (Arumuganathan and Earle, 1991) and is estimated to contain 63 % repetitive sequences (Flavell et al., 1974). In this paper, the genomic organization of novel dispersed and tandemly arranged repetitive sequences are described. In particular, analysis of BAC-end sequences revealed structural modifications of a centromere-specific satellite including the formation of a complex centromeric tandem array of unusual monomere size. The chromosomal organization of these repeat families was analysed by multicolour fluorescent in situ hybridization (FISH), and their distribution within the genus Beta was elucidated by comparative Southern hybridization.
MATERIALS AND METHODS
Plant material and DNA preparation
Plants of Beta vulgaris ssp. vulgaris ‘KWS 2320’ (sugar beet), ‘Brigadier’ (fodder beet), ‘Rote Kugel’ (garden beet) and ‘Lukullus’ (leaf beet), Beta vulgaris ssp. maritima, Beta vulgaris ssp. adanensis, as well as the species Beta patula, Beta macrocarpa, Beta corolliflora, Beta macrorhiza, Beta nana, Beta procumbens, Beta patellaris and Spinacia oleracea, were grown under greenhouse conditions. Genomic DNA was isolated from young leaves using the CTAB (cetyltrimethyl/ammonium bromide) standard protocol (Saghai-Maroof et al., 1984).
Isolation of repetitive DNA
For the isolation of repetitive DNA families, the plasmid library SGZR00841 from genomic Beta vulgaris DNA (genotype ‘KWS 2320’), consisting of 27 648 clones with an average insert size of 0·5 kb spotted in duplicate onto nylon membranes (Menzel et al., 2006), was subsequently probed with radiolabelled genomic DNA of B. vulgaris ‘KWS2320’ and a pool of all known radiolabelled repetitive B. vulgaris sequences. The overnight hybridizations were performed at 60 °C in 5× SSPE with 5× Denhardt solution and 0·2 % SDS. Post-hybridization washings were performed twice at 60 °C in 1× SSC / 0·1 % SDS for 10 min. The signals of both hybridization experiments were analysed with the software VisualGrid 3·4·1·1000 (www.gpc-biotech.com).
Plasmid DNA of randomly chosen clones not hybridizing to the pool of known B. vulgaris repeats was further purified with the GFX Micro Plasmid Prep Kit (GE Healthcare, Chalfont St Giles, UK), and sequenced with a CEQ 8000 capillary sequencer (Beckman Coulter, Fullerton, CA, USA) according to the manufacturers instructions. Sequences were aligned by the MegAlign option of the Lasergene 6·0 software (DNAStar, Madison, Wisconsin) using CLUSTAL with default parameters.
PCR conditions
To generate probes for Southern hybridization, PCR reactions with 50 ng template DNA and a final primer concentration of 0·5 µm were performed in a 50-μL volume containing 0·2 mm dNTPs, 50 mm KCl, 1·5 mm MgCl2, 10 mm Tris–HCl (pH 9·0) and 1 unit of Taq DNA polymerase (GE Healthcare). Standard PCR conditions were 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 49 °C to 55 °C for 45 s, 72 °C for 3 min 30 s and a final incubation at 72 °C for 10 min. For the amplification of a pDvul1 probe, the primers 5′-ACCCGTATTTCGTCTTTCTT-3′ and 5′-TCGCCCGTGTGATAATG-3′ were used for PCR with the clone 44K4 as template at 50 °C annealing temperature. A probe representing the pDvul2 repeat family was amplified from the plasmid DNA of clone 17H23 with the primers 5′-GGAAACTTTTCCCATACCA-3′ and 5′-GGGTTTTTTAGTTGAAATCT-3′ at 50 °C annealing. Hybridizations with pRv1 were performed with a probe amplified from the plasmid clone 46C21 with the primer pair 5′-CGGATTTTATTGATAGAATG-3′ and 5′-CTTAGATAAGACTATTCATGC-3′ at an annealing temperature of 50 °C. A pBV1 probe was amplified from a cloned monomer sequence (EMBL accession Z22849) with the primer pair 5′-TTCAATGCAACGCACTCG-3′ and 5′-ATCCTCTAGTTCTCGAGC-3′ using an annealing temperature of 54 °C.
After gel electrophoresis, PCR fragments were purified with the QIAquick gel extraction kit (Qiagen, Hilden, Germany).
Southern hybridization
For Southern hybridization, 10 µg genomic DNA was restricted with different enzymes, separated on 1·2 % agarose gels, and transferred onto Hybond-N+ nylon membranes (GE Healthcare) using alkaline transfer. Southern hybridizations using 32P-labelled probes were performed using standard protocols (Sambrook et al., 1989). Filters were hybridized at 60 °C and washed at 60 °C in 2× SSC/0·1 % SDS and 1× SSC/0·1 % SDS for 10 min each. Signals were detected by autoradiography.
FISH
The meristem of young leaves from B. vulgaris plants was used for the preparation of mitotic chromosomes. The material was macerated in an enzyme mixture and dropped onto slides as described by Schwarzacher and Heslop-Harrison (2000) with modifications (Desel, 2002). The probes were labelled with biotin-11-dUTP or digoxigenin-16-dUTP by PCR (Schwarzacher and Heslop-Harrison, 2000), and hybridization and detection were performed according to Schmidt et al. (1994). Signal amplification was performed with a Thyramide Amplification Kit according to manufacturer's instructions (Molecular Probes, Invitrogen). Chromosome preparations were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and mounted in antifade solution (CitiFluor).
Examination of slides was carried out with a Zeiss Axioplan2 fluorescence microscope equipped with filters 09 (FITC), 15 (Cy3) and 01 (DAPI). Images were acquired directly with the Applied Spectral Imaging v. 3·3 software coupled with the high-resolution CCD camera ASI BV300-20A. The images were contrast optimized using only functions affecting the whole image equally and printed using Adobe Photoshop 7·0 software.
RESULTS
Identification and sequence analysis of novel dispersed DNA families
To identify novel B. vulgaris repetitive DNA families, high-density filters of the plasmid library SGZR00841 containing approx. 15 Mbp were subsequently hybridized with genomic B. vulgaris DNA and most previously characterized B. vulgaris repeat families (Table 1). Comparing the number of hybridizing clones in each experiment, this differential hybridization approach revealed that 84 % of repetitive DNA sequences within the B. vulgaris genome had been characterized previously. Plasmid clones hybridizing strongly to genomic DNA, but not cross-reacting with known repeats were sequenced. Those with no significant similarities to other sequences in the EMBL database represented novel repeats and could be grouped into three families designated pDvul1, pDvul2 and pRv1.
Table 1.
Major tandemly arranged and dispersed B. vulgaris repeats
| Repeat | Type | Reference |
|---|---|---|
| pEV1 | Satellite DNA | Schmidt et al., 1991 |
| pBV1 | Satellite DNA | Schmidt and Metzlaff, 1991 |
| pXV | Satellite DNA | Schmidt et al., 1994 |
| pAV34 | Satellite DNA | Dechyeva and Schmidt, 2006 |
| pHC28 | Satellite DNA | Schmidt and Heslop-Harrison, 1993 |
| pSV1 | Satellite DNA | Schmidt et al., 1998 |
| pDRV1 | Dispersed DNA | Schmidt et al., 1998 |
| BNR1 | Non-LTR retrotransposon, partial | Schmidt et al., 1995 |
| Tbv1 | Ty1-copia retrotransposon, partial | Schmidt et al., 1995 |
| TPvul1 | En/Spm transposon, partial | Jacobs et al., 2004 |
| Vulmar1 | Tc1/mariner transposon | Jacobs et al., 2004 |
| VulMITE I | Stowaway miniature inverted-repeat transposable element | Menzel et al., 2006 |
| VulMITE II | Miniature inverted-repeat transposable element | Menzel et al., 2006 |
| VulMITE III | Miniature inverted-repeat transposable element | Menzel et al., 2006 |
For the pDvul1 repetitive family, five clones (EMBL accessions AM177315 and AM228861–AM228864) ranging in size from 226 bp to 656 bp and sharing similarity between 74·2 % and 76·3 % were identified. Among those, the repeat pDvul1-2 represents a full-length repetitive unit of 656 bp.
The six members of the pDvul2 family (AM177316, AM228865 and AM944727–AM944730) are 187–291 bp long and 75·5–94·4 % similar. The repeats pDvul2-5 and pDvul2-6 form a subgroup within the family by the presence of four positionally conserved insertions with an average length of 53 bp, 22 bp, 9 bp and 7 bp. The interspersion of pDvul1 and pDvul2 family members was observed on the plasmid clone 17H23, where the repeats pDvul1-1 and pDvul2-1 are spaced by a genomic region of 40 bp.
Structural variability and complexity of a centromeric satellite repeat
From the pRv1 repetitive family, 12 members (AM944555–AM944566) were identified. The repeats pRv1-1 to pRV1-12 have a similar length of 211–244 bp. The sequence similarity within the members of the pRv1 family ranges from 63·5 % to 93·2 %.
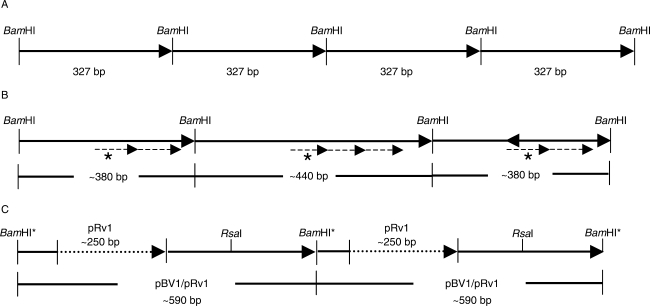
Sequence analyses of plasmid clones revealed the interspersion of pRv1 units with monomers of the satellite repeat pBV1. The tandem repeat pBV1 was first described by Schmidt and Metzlaff (1991) as a centromeric BamHI satellite with a monomer length of 327 bp (Fig. 1A). To unravel this organization of pBV1 and pRv1 units, the B. vulgaris entries in the EMBL database (McGrath et al., 2004) were screened using a pBV1 query, and subsequently compared. First, remarkable structural changes have been identified within pBV1 monomers, which can be assigned to three variants. Beside tandemly arranged pBV1 units of 327 bp (Fig. 1A), pBV1 monomers were observed containing an internal duplication of a 60-bp sequence motif, resulting in an increased repeat unit of 380 bp (Fig. 1B). Moreover, 380-bp pRv1 units were interspersed with monomers of approx. 440 bp, which are formed by a triple amplification of the 60-bp sequence motif (Fig. 1B). In addition to these observed variations in the pBV1 monomer structure, homology searches with a pRv1 query sequence to 5720 end sequences of a BAC library from the B. vulgaris cultivar ‘KWS 2320’ (Hohmann et al., 2003; D. Holtgräwe and B. Weisshaar, unpubl. res.) identified pBV1 units of 340 bp, each containing pRv1 monomers with an average length of 250 bp at conserved integration sites, thus suggesting the formation of a complex satellite unit of approx. 590 bp (Fig. 1C).
Fig. 1.
Schematic representation of the structural variations of the pBV1 satellite repeat in the B. vulgaris genome. (A) Tandemly arranged pBV1 monomeres of 327 bp. The BamHI restriction sites define the satellite monomeres according to Schmidt and Metzlaff (1991). (B) Size variations of pBV1 monomeres detected on a B. vulgaris bacterial artificial chromosome (BAC). The monomer length is increased to approx. 380 bp by an internal duplication (dashed arrow) of a 60-bp sequence motif (dashed arrows, labelled with an asterisk). An amplification of the 60-bp motif resulted in a monomer of approx. 440 bp. (C) Interspersion of a pRv1 unit of approx. 250 bp (dotted arrow) into pBV1 monomeres of approx. 340 bp. The combined pBV1/pRv1 unit observed on end-sequenced BACs and plasmid clones is approx. 590 bp in length and contains a unique RsaI restriction site, while the BamHI site is degenerated (indicated by an asterisk).
Genomic organization of repetitive families in B. vulgaris
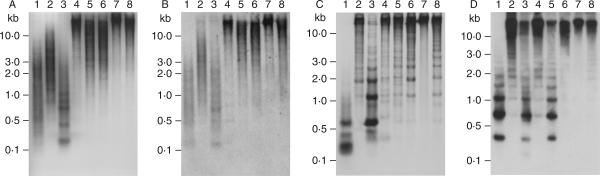
Genomic organization of the repetitive DNA in B. vulgaris was analysed by Southern hybridization using representative members of the pDvul1, pDvul2 and pRv1 families as probes (Fig. 2).
Fig. 2.
Southern hybridization of B. vulgaris repeats to genomic DNA of the B. vulgaris ssp. vulgaris cultivar ‘KWS 2320’, which was digested with AluI (lane 1), HaeIII (lane 2), RsaI (lane 3), EcoRI (lane 4), BamHI (lane 5), HindIII (lane 6), MspI (lane 7) and HpaII (lane 8). Blots were separately hybridized with pDvul1 (A), pDvul2 (B) and pRv1 (C). For comparison with the pRv1 pattern, a filter (D) was probed with a monomer sequence of the pBV1 satellite family (Schmidt and Metzlaff, 1991).
Probing of pDvul1-1 to genomic DNA digested with AluI, HaeIII, EcoRI, BamHI and HindIII detected a strong hybridization smear of a wide molecular weight range (Fig. 2A, lanes 1–2 and 4–6), reflecting a dispersed genomic organization of the pDvul1 family. In addition, the RsaI restriction revealed conserved fragments between 0·2 kb and 1·0 kb in size (Fig. 2A, lane 3). Hybridization to MspI- and HpaII-digested DNA indicated no significant different methylation at CNG sites of the pDvul1 repeat family (Fig. 2A, lanes 7 and 8).
Hybridization with pDvul2-1 resulted in a pattern of numerous fragments forming a smear, which is distributed over a molecular size range of 0·2 kb to above 10 kb (Fig. 2B, lanes 1–3), or a size above 1·0 kb (Fig. 2B, lanes 4–6). Additionally, in RsaI-digested genomic DNA, several faint, but conserved fragments in a size range of 0·2–1·0 kb are visible (Fig. 2B, lane 3). The hybridization pattern of MspI and HpaII digested genomic DNA suggests no differences of cytosine methylation of the pDvul2 family (Fig. 2B, lanes 7 and 8).
In contrast to the mostly dispersed patterns observed with pDvul1 and pDvul2, hybridization with a 170-bp pRv1-1 fragment produced distinct ladder-like patterns (Fig. 2C, lanes 1–6 and 8). In RsaI digested genomic DNA, strong hybridization signals consisting of a 0·6-kb monomer band and multimeric bands ranging up to approx. 10 kb (Fig. 2C, lane 3) suggests a satellite-like DNA organization. In contrast, weaker ladder-like banding patterns from 0·6 kb up to above 10 kb are visible in HaeIII, EcoRI, BamHI and HindIII restrictions (Fig. 2C, lanes 2–6). Furthermore, two strong AluI bands are localized at 0·2 kb and 0·6 kb, respectively (Fig. 2C, lane 1). Hybridization of MspI and HpaII digests shows significant differences in the cytosine methylation of pRv1 repeats (Fig. 2C, lanes 7 and 8).
Because of the interspersion of pRv1 in the BamHI satellite pBV1, a Southern comparative hybridization with a pBV1 probe was carried out (Fig. 2D). The AluI-, RsaI- and BamHI-restricted DNA reveals a typical satellite pattern including a monomere of approx. 0·35 kb and the corresponding di- and trimeres of 0·75 kb and 1·1 kb. Additionally, presence of weaker signals in the RsaI restriction forms an additional ladder with a monomere of 0·65 kb and a dimer of approx. 1 kb. Similar to pRv1, differences in methylation of the cytosine residues were observed by the occurence of the satellite hybridization pattern only in HpaII digested genomic DNA.
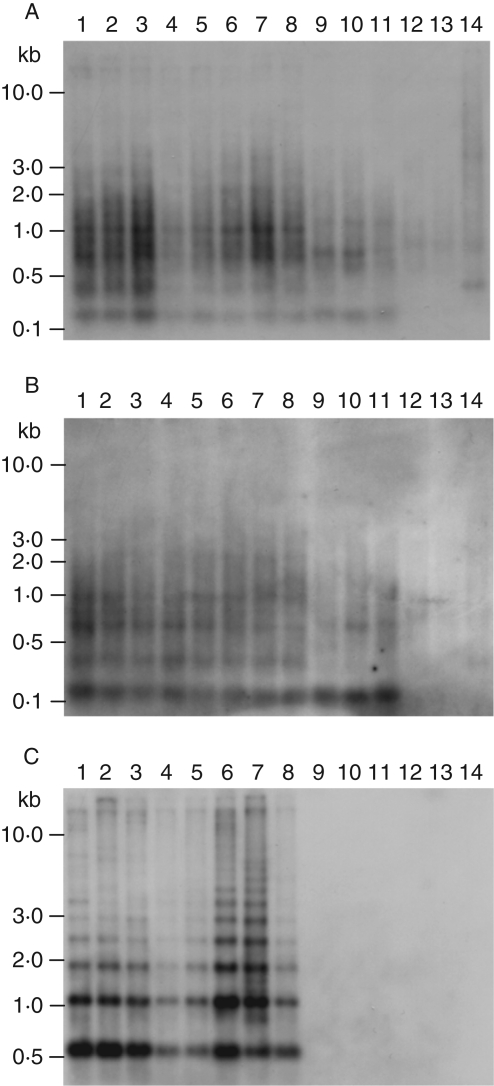
Distribution of repeat families within the genus Beta
The repeats distribution within the genus Beta was analysed by Southern hybridizations with RsaI-digested genomic DNA of representative species of the four Beta sections Beta, Corollinae, Nanae and Procumbentes. Spinach (Spinacia oleracea), which also belongs to the subfamily Chenopodiaceae, was chosen as an outgroup species.
Southern hybridization with the dispersed repeat pDvul1 revealed an almost identical hybridization pattern within the section Beta (Fig. 3A, lanes 1–8). In species of the section Corollinae (Fig. 3A, lanes 9 and 10) the weaker hybridization pattern appears to be almost similar to that in the section Beta, with the observed exception of two bands lacking at 0·4 kb and 1 kb, respectively. A similar but weaker pattern is visible in B. nana (Fig. 3A, lane 11). Genomic DNA of two species of the section Procumbentes (Fig. 3A, lanes 12 and 13) and the outgroup species S. oleracea (Fig. 3A, lane 14) shows only a faint hybridization.
Fig. 3.
Species distribution of B. vulgaris repeats was analysed by Southern hybridization to RsaI-digested genomic DNA of representative species of the four sections of the genus Beta. Species of the section Beta: sugar beet B. vulgaris ssp. vulgaris cultivar ‘KWS 2320’ (lane 1), fodder beet B. vulgaris ssp. vulgaris cultivar ‘Brigadier’ (lane 2), garden beet B. vulgaris ssp. vulgaris cultivar ‘Rote Kugel’ (lane 3), leaf beet B. vulgaris ssp. vulgaris cultivar ‘Lukullus’ (lane 4), B. vulgaris ssp. maritima (lane 5), B. vulgaris ssp. adanensis (lane 6), B. macrocarpa (lane 7), B. patula (lane 8); species of the section Corollinae: B. corolliflora (lane 9), B. macrorhiza (lane 10); species of the section Nanae: B. nana (lane 11); species of the section Procumbentes: B. procumbens (lane 12), B. patellaris (lane 13); outgroup species: S. oleracea (lane 14). Blots were separately hybridized with pDvul1 (A), pDvul2 (B) and pRv1 (C) probes.
Probing of a pDvul2 fragment resulted in a dispersed hybridization pattern (Fig. 3B, lanes 1–14), which appears to be almost similar to the pattern of the pDvul1 repeat family (Fig. 3A, lanes 1–14). Nevertheless, hybridization bands within the species of the four Beta sections are more distinct due to reduced background smear (Fig. 3B, lanes 1–11).
The satellite-like hybridization ladder formed by the strong 0·6-kb monomer band specific for the pRv1 family was exclusively identified in species of the section Beta. Multimeric bands extending to 3 kb were identified in all species of the section (Fig. 3C, lanes 1–8). Even after an extended time of exposure, no signals were visible in species of the sections Corollinae, Nanae and Procumbentes or in S. oleracea.
Chromosomal localization of dispersed repetitive families
The chromosomal organization of the repetitive families pDvul1 and pDvul2 on B. vulgaris mitotic chromosomes was analysed by FISH.
Probing of pDvul1 revealed dispersed fluorescent signals with banding of different intensity along all chromosome arms, while terminal euchromatin (indicated by arrows) was largely excluded from hybridization (Fig. 4A).
Fig. 4.
Fluorescent in situ hybridization on B. vulgaris mitotic chromosomes. In each panel, the DAPI-stained DNA (blue fluorescence) shows the morphology of the chromosomes. (A) pDvul1-1 (green) is dispersed along heterochromatin of all chromosomes with almost equal signal intensities, except for terminal euchromatin (indicated by arrows). (B) Hybridization with pDvul2-1 (red) revealed doublets of signals of varied strength in terminal (indicated by arrows), centromeric (arrowheads) and intercalary regions of all chromosomes. Scale bar in (B) = 10 µm.
Successful localization of pDvul2 on sugar beet chromosomes was compromised by its relatively low abundance in the genome. While pDvul1 produced >100 positives on a high-density filter of the plasmid library SGZR00841, there were only ten signals generated by pDvul2. To map pDvul2 physically by FISH, an extended hybridization time, adjusted stringency and signal amplification were applied. The repeat was found on all 18 B. vulgaris chromosomes producing doublets of signals of varying intensity. On four chromosomes it was found in the subtelomeric position (Fig. 4B, examples shown by arrows), on two pericentrically (Fig. 4B, arrowheads), and on the remaining chromosomes it was clustered in intercalary chromatin.
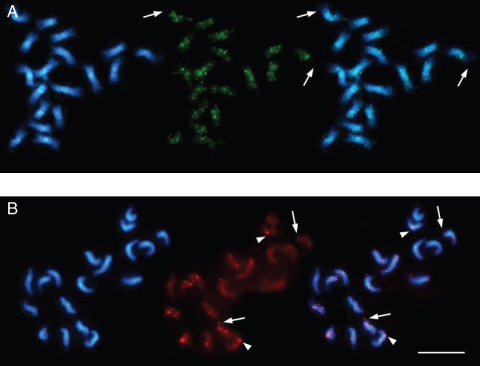
Centromeric co-localization of pRv1 and the pBV1 satellite family
The pBV1 repeat family, similar to pRv1, was shown to be exclusively present in species of the section Beta by Southern hybridization (Schmidt and Metzlaff, 1991). FISH on B. vulgaris metaphase chromosomes revealed that pBV1 is restricted to the centromeric regions on all 18 sugar beet chromosomes producing signals of different intensity (Fig. 5A, green fluorescence). In contrast, pRv1 hybridized clearly to 12 centromeric regions with signals of variable intensity (Fig. 5A, red fluorescence and overlay). The pRv1 signal strength falls into three classes of strong hybridization on four chromosomes, six centromeres with moderate intensity, and two chromosomes with weak signals (Fig. 5A). Moreover, signals of moderate strength could be confined to the chromosome pair I, which is recognizable by the presence of the terminal nucleolus organizer region (Fig. 5A, arrowed).
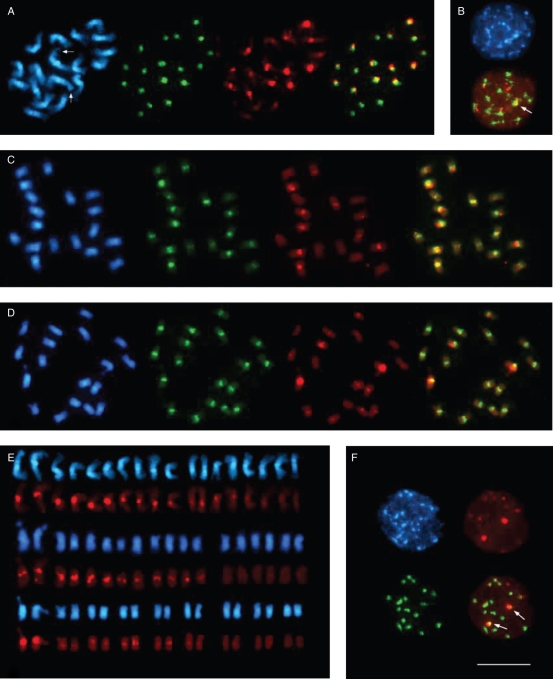
Fig. 5.
Fluorescent in situ hybridization to metaphase and interphase chromosomes of species of the section Beta. In each panel, the DAPI-stained DNA (blue fluorescence) shows the morphology of the chromosomes. The pBV1 hybridization signals are visible as green fluorescent signals, and red fluorescence indicates pRv1 hybridization. Comparison of the signal pattern of pBV1 and pRv1 probes hybridized to metaphase chromosomes of B. vulgaris (A), B. patula (C) and B. macrocarpa (D), indicating a species-specific centromeric localization of both repeat families. The B. vulgaris chromosome pair I is marked by arrows. The right picture in (A), (C) and (D) represents an overlay of the pBV1 and pRv1 hybridization. (B) Centromeric localization of pBV1 and pRv1 is visible as signals on chromocentres of B. vulgaris interphase nuclei. The embedment of pRv1 into areas of pBV1 is highlighted by an arrow in the overlay (bottom). (E) Arrangement of metaphase chromosomes from B. vulgaris (top), B. patula (middle) and B. macrocarpa (bottom) according to the observed signal strength of centromeric pRv1 hybridization. The pair of chromosome I for each species is placed on the left. (F) Centromeric localization of pBV1 and pRv1 signals in the centromeric heterochromatin of B. macrocarpa interphase chromosomes. The distinct cluster of pRv1 adjacent to a pBV1 area is visible as yellow fluorescence (indicated by arrows, bottom right). Scale bar (in F) = 10 µm.
The section Beta consists of the species B. vulgaris, B. patula and B. macrocarpa. To analyse the centromeric pRv1 and pBV1 organization within the section, comparative FISH on chromosomes of these species was performed. As in B. vulgaris, pBV1 hybridization in B. patula and B. macrocarpa is visible in the centromeric heterochromatin of all 18 chromosomes (Fig. 5C and D, green fluorescence). To accurately compare species-specific pRv1 hybridization patterns, the metaphase chromosomes stained with DAPI as well as the corresponding pRv1 hybridizations were arranged according to the classes of pRv1 signal intensity observed (Fig. 5E). In contrast to the three intensity classes detected on at least 12 B. vulgaris chromosomes (Fig. 5E, top), pRv1 hybridization strength in B. patula only falls into two classes of four strong and eight moderate centromeric signals, including the chromosome pair I (Fig. 5E, middle). In B. macrocarpa, the pair of chromosome I shows a strong pRv1 hybridization, while the centromeric regions of ten chromosomes reveal only weak signal intensities (Fig. 5E, bottom). FISH on B. vulgaris and B. macrocarpa show significant differences in hybridization pattern, suggesting a species- and chromosome-specific amplification and organization of pBV1/pRv1 units. Nevertheless, in overlays of pBV1 and pRv1 hybridization signals on metaphase chromosomes of both species, only the centromeric co-localization of both pBV1 and pRv1 signals was detected (Fig. 5A, C, overlays). To achieve higher resolution, pBV1 and pRv1 were hybridized to uncondensed interphase chromosomes of B. vulgaris and B. macrocarpa. The experiment showed a clear separation pRv1/pBV1 clusters from pBV1 arrays on chromocentres of both species (Fig. 5B, F, arrowed in overlay).
DISCUSSION
Repetitive sequences were estimated to form at least 63 % of the B. vulgaris genome (Flavell et al., 1974), with numerous families of repeats discovered by shot-gun cloning or targeted cloning of restriction satellites (Kubis et al., 1998; Dechyeva et al., 2003). Our differential hybridization experiments on high density filters, as well as the bioinformatic analysis of 54684 BAC end sequences indicates that the amount of repetitive DNA in the B. vulgaris genome might be even higher than 63 %.
From the fraction of B. vulgaris repetitive DNA, the novel families pDvul1, pDvul2 and pRv1 have been identified. Southern analysis with pDvul1 and pDvul2 showed a dispersed genomic organization of both repetitive families that was similar in the sections Beta, Corollinae and Nanae, but differs in the section Procumbentes. Such Procumbentes-specific absence or low abundance was also observed for Beta repeat families such as pHC, pRN, pSV and pDRV (Schmidt and Heslop-Harrison, 1993; Kubis et al., 1997; Schmidt et al., 1998). These results support the suggestion based on the analyses of ITS1 sequences, that species of Procumbentes might form a separate genus designated Patellifolia (Kadereit et al., 2006). In FISH on B. vulgaris metaphase chromosomes, pDvul1 is widely scattered including centromeres, while pDvul2 produced few pairs of signals dispersed in intercalary and terminal chromatin, indicating that pDvul2 is less abundant. A weak ladder-like pattern superimposed on a smear and additional irregular bands observed in Southern hybridization suggests an organization of pDvul2 in dispersed clusters of presumably small tandem arrays. Similarly, the AluI repeat pAp22 from the wild beet B. procumbens is dispersed with local amplification over the chromosomes, but is more abundant (Dechyeva et al., 2003). Dispersion along chromosomes is also typical for some Ty1-copia retrotransposons and LINEs (Katsiotis et al., 1995; Schmidt et al., 1995; Heslop-Harrison et al., 1997). Retrotransposons and transposons as highly amplified components of plant genomes play an important role in the evolution of the genome structure and function (Kumar and Bennetzen, 1999) and are a source of dispersed sequence families. In barley, dispersed repeats were identified as remnants of retrotransposon-specific reverse transcriptase fragments (Liu and Somerville, 1996). Transductions of genomic sequences by plant retroelements have been demonstrated for Bs1 from maize (Jin and Bennetzen, 1994), and the dispersed repeats pAp4 and pAp22 from B. procumbens are directly linked to Ty1-copia or env-like retrotransposons (Dechyeva et al., 2003). It is, however, unclear whether and by which mechanism mobile elements might have participated in the amplification of pDvul2 repeats. In fact, the repetitive families pDvul1 and pDvul2 exhibit no sequence homology to EMBL entries of conserved retrotransposon or transposon genes. Thus, pDvul1 and pDvul2 might be derivatives of untranslated transposon regions, including long terminal repeats, or of yet undiscovered B. vulgaris transposable elements. Otherwise, they might have an independent origin. Such dispersed repetitive families with no relationship to retrotransposon-like sequences have been observed in barley, rice and tobacco (Hueros et al., 1993; Kiefer-Meyer et al., 1996; Horakova and Fajkus, 2000).
Satellite DNA sequences comprise a large proportion of the repetitive plant genomic DNA. The differential hybridization presented in this work revealed that the majority of the previously described repetitive families belong to the group of satellite DNA. So far, 14 different satellite DNA families from the four sections of the genus Beta have been characterized (Dechyeva et al., 2003; Dechyeva and Schmidt, 2006). Most of these repetitive sequences were identified as restriction satellites with monomers falling into size classes of approx. 150–180 bp and 300–360 bp. The majority of the cloned Beta satellite monomers can be assigned to the group of 150- to 160-bp repeats, which presumably corresponds to the stretch of nuclear DNA wrapped around a single nucleosome core (Fischer et al., 1994; Vershinin and Heslop-Harrison, 1998; Heslop-Harrison, 2000). This length of satellite motifs may be favourable for chromatin packaging, thus undergoing selection resulting in accumulation of repeats of this size over evolutionary time scales. As satellite DNA represents a fast-evolving portion of plant genomes, the formation of larger and often complex repeats, including dimerization, is typical of satellite families and has been observed in many plant species (Ingham et al., 1993; Grebenstein et al., 1996; Simoens et al., 1988; Dechyeva et al., 2006). Among the Beta tandem repeats, the pRN1 satellite from B. corolliflora shows an unusual length of 202–233 bp (Kubis et al., 1997), and the 240 bp AluI satellite pAp11 from B. procumbens (Dechyeva et al., 2003) represents one and a half 160-bp monomers as a whole unit. By computational analyses of 27 342 end-sequenced B. vulgaris BACs, distinct variants have been observed with structural changes of the B. vulgaris satellite repeat pBV1, which was initially described as a BamHI satellite with a monomer size of 327 bp (Schmidt and Metzlaff, 1991). After the duplication of an internal 60-bp fragment, the pBV1 monomer size has increased up to 380 bp. Moreover, amplification of the 60-bp motif results in a pBV1 monomer unit of 440 bp. Tandemly arranged plant repetitive DNA often contains subrepeats of 60 bp, which are themselves built by two units of approx. 30 bp (Ingham et al., 1993). Such duplication was also described as the origin of the 60-bp subrepeat of pBV1 (Schmidt and Metzlaff, 1991). Thus, the stepwise duplication and subsequent amplification of short sequence motifs might be a common mechanism for the generation of plant satellite DNA.
Large arrays of tandemly repeated satellite DNA localized in centromeric heterochromatin are assumed to substantially contribute to centromere function, e.g. segregation of sister chromatids during mitosis and meiosis, by an interaction with the kinetochore protein CENH3 (Houben and Schubert, 2003). Despite their conserved function, centromeric satellites evolve more rapidly than other genomic sequences (Hall et al. 2003), most likely by the genetic mechanisms of unequal crossing over, replication slippage and unequal sister chromatid exchange (Smith, 1976; Dover, 1982). As a result, sequences of centromeric satellite repeats vary even between closely related species, often resulting in new centromer-specific satellite variations taking over centromere function (Ma et al., 2007). Thus, the localization of the satellite pBV1 in the centromeric heterochromatin on all chromosomes exclusively observed in the three species of the section Beta might be an indication of a section-specific role for pBV1 satellites in centromere function, although there is no experimental evidence for this assumption so far.
Beside the structural variations of the pBV1 satellite monomer, Southern hybridization experiments of RsaI-digested genomic B. vulgaris DNA probed with the pRv1 repeat revealed the amplification of a complex satellite unit composed of pBV1 and pRv1 units. In plants, insects and humans, tandem repeats were described representing combinations of closely related satellite subrepeats (Grebenstein et al., 1996; Pons et al., 2003; Rudd and Willard, 2004; Dechyeva and Schmidt, 2006). In contrast to pBV1, no pRv1 multimers were identified by sequence analysis of BAC ends and plasmid clones. Morever, no amplification of tandemly arranged pRv1 di- and multimeric products was observed in PCR analyses using pRv1-specific primers (data not shown). Therefore, the existence of pRv1 as an independant, separate Beta satellite can largely be excluded.
The chromosome specificity of centromeric satellite DNA has been observed only in few plant species like Brassica, sugarcane, Arabidopsis and the wild beet B. procumbens (Harrison and Heslop-Harrison, 1995; Nagaki et al., 1998; Kawabe and Nasuda, 2005; Dechyeva et al., 2003). In the three species of the section Beta, the interspersion of the complex pBV1/pRv1 with pBV1 satellite arrays was detected by FISH in the centromeric heterochromatin of up to 12 out of 18 chromosomes. The presence of pBV1/pRv1 arrays on the remaining three chromosome pairs is unlikely due to the conditions used in FISH. The high degree of sequence identity in the B. vulgaris-specific pBV1/pRv1 units analysed suggests a strong homogenization process during amplification, which is comparable to the homogenization of higher order satellite structures observed in humans and insects (Rudd and Willard, 2004; Kuhn et al., 2008). Therefore, a loss of pBV1/pRv1 arrays, which might have been present on all chromosomes of a progenitor of Beta section species, is unlikely.
The comparison of the species-specific pRv1 FISH signal intensities indicates a remarkable chromosome-specific amplification of pBV1/pRv1 units in the cultivated species B. vulgaris. It would be interesting to analyse the physical organization of pBV1/pRv1 units in an ancestor of cultivated beets, like the wild beet B. maritima, to unravel whether the observed dynamics of centromeric pBV1 satellite amplification has occured during domestication of sugar beet, which started in the late 18th century (Fischer, 1989).
ACKNOWLEDGMENTS
We thank Ines Walther for excellent technical assistance. This work is funded in part by the BMBF grant ‘Verbundprojekt GABI-Beet Physical map: Physikalische Genomkarte der Zuckerrübe zur Nutzung in der Pflanzenzüchtung’, sub-projects 0313127B and 0313127E.
LITERATURE CITED
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter. 1991;9:208–218. [Google Scholar]
- Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Molecular Biology. 2000;42:251–269. [PubMed] [Google Scholar]
- Bennetzen JL, Schrick K, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodifed and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- Brandes A, Tompson H, Dean C, Heslop-Harrison JS. Multiple repetitive DNA sequences in the paracentromeric regions of Arabidopsis thaliana L. Chromosome Research. 1997;5:238–246. doi: 10.1023/a:1018415502795. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, et al. Genetic defnition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- Dechyeva D, Schmidt T. Molecular organization of terminal repetitive DNA in Beta species. Chromosome Research. 2006;14:881–897. doi: 10.1007/s10577-006-1096-8. [DOI] [PubMed] [Google Scholar]
- Dechyeva D, Gindullis F, Schmidt T. Divergence of satellite DNA and interspersion of dispersed repeats in the genome of the wild beet Beta procumbens. Chromosome Research. 2003;11:3–21. doi: 10.1023/a:1022005514470. [DOI] [PubMed] [Google Scholar]
- Desel C. Chromosomale Lokalisierung von repetitiven und unikalen DNA-Sequenzen durch Fluoreszenz-in situ-Hybridisierung in der Genomanalyse bei Beta-Arten. Germany: Christian-Albrechts University Kiel; 2002. PhD Thesis. [Google Scholar]
- Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proceedings of the National Academy of Sciences of the USA. 1998;95:8135–8140. doi: 10.1073/pnas.95.14.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:430–440. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Fischer HE. Origin of the ‘Weisse Schlesische Rübe’ (white Silesian beet) and resynthesis of sugar beet. Euphytica. 1989;41:75–80. [Google Scholar]
- Fischer TC, Groner S, Zentgraf U, Hemleben V. Evidence for nucleosomal phasing and a novel protein specifically binding to cucumber satellite DNA. Zeitschrift für Naturforschung. 1994;49:79–86. doi: 10.1515/znc-1994-1-213. [DOI] [PubMed] [Google Scholar]
- Flavell RB, Bennett MD, Smith JB, Smith DB. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochemical Genetics. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- Gao D, Schmidt T, Jung C. Molecular characterization and chromosomal distribution of species-specific repetitive DNA sequences from Beta corolliflora, a wild relative of sugar beet. Genome. 2000;43:1073–1080. doi: 10.1139/g00-084. [DOI] [PubMed] [Google Scholar]
- Gindullis F, Dechyeva D, Schmidt T. Construction and characterization of a BAC library for the molecular dissection of a single wild beet centromere and sugar beet (Beta vulgaris) genome analysis. Genome. 2001;a 44:846–855. [PubMed] [Google Scholar]
- Gindullis F, Desel C, Galasso I, Schmidt T. The large-scale organization of the centromeric DNA in Beta species. Genome Research. 2001;b 11:253–265. doi: 10.1101/gr.162301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenstein B, Grebenstein O, Sauer W, Hemleben V. Distribution and complex organization of satellite DNA in sequences of Aveneae species. Genome. 1996;39:1045–1050. doi: 10.1139/g96-131. [DOI] [PubMed] [Google Scholar]
- Hall SE, Kettler G, Preuss D. Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Research. 2003;13:195–205. doi: 10.1101/gr.593403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GE, Heslop-Harrison JS. Centromeric repetitive DNA sequences in the genus Brassica. Theoretical and Applied Genetics. 1995;90:157–165. doi: 10.1007/BF00222197. [DOI] [PubMed] [Google Scholar]
- Hemleben V, Schmidt T, Torres-Ruiz RA, Zentgraf U. Molecular cell biology: role of repetitive DNA in nuclear architecture and chromosome structure. Progress in Botany. 2000;61:91–117. [Google Scholar]
- Heslop-Harrison JS. Comparative analysis of plant genome architecture. Symposia of the Society for Experimental Biology. 1996;50:17–23. [PubMed] [Google Scholar]
- Heslop-Harrison JS. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. The Plant Cell. 2000;12:617–635. doi: 10.1105/tpc.12.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Brandes A, Taketa S, Schmidt T, Vershinin AV, Alkhimova EG, et al. The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica. 1997;100:197–204. [PubMed] [Google Scholar]
- Heslop-Harrison JS, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. The Plant Cell. 1999;11:31–42. doi: 10.1105/tpc.11.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Jacobs G, Telgmann A, Gaafar RM, Alam S, Jung C. A bacterial artificial chromosome (BAC) library of sugar beet and a physical map of the region encompassing the bolting gene B. Molecular Genetics and Genomics. 2003;269:126–136. doi: 10.1007/s00438-003-0821-7. [DOI] [PubMed] [Google Scholar]
- Horakova M, Fajkus J. TAS49 – a dispersed repetitive sequence isolated from subtelomeric regions of Nicotiana tomentosiformis chromosomes. Genome. 2000;43:273–284. doi: 10.1139/g99-126. [DOI] [PubMed] [Google Scholar]
- Houben A, Schubert I. DNA and proteins of plant centromeres. Current Opinion in Plant Biology. 2003;6:554–560. doi: 10.1016/j.pbi.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Hudakova S, Michalek W, Presting GG, ten Hoopen R, dos Santos K, Jasencakova Z, et al. Sequence organization of barley centromeres. Nucleic Acids Research. 2001;29:5029–5035. doi: 10.1093/nar/29.24.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueros G, Loarce Y, Ferrer E. A structural and evolutionary analysis of a dispersed repetitive sequence. Plant Molecular Biology. 1993;22:635–643. doi: 10.1007/BF00047404. [DOI] [PubMed] [Google Scholar]
- Ingham LD, Hanna WW, Baier JW, Hannah LC. Origin of the main class of repetitive DNA within selected Pennisetum species. Molecular and General Genetics. 1993;8:350–356. doi: 10.1007/BF00291993. [DOI] [PubMed] [Google Scholar]
- Jacobs G, Dechyeva D, Menzel G, Dombrowski C, Schmidt T. Molecular characterization of Vulmar1, a complete mariner transposon of sugar beet and diversity of mariner- and En/Spm-like sequences in the genus Beta. Genome. 2004;47:1–10. doi: 10.1139/g04-067. [DOI] [PubMed] [Google Scholar]
- Jin YK, Bennetzen JL. Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. The Plant Cell. 1994;6:1177–1186. doi: 10.1105/tpc.6.8.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G, Hohmann S, Kadereit JW. A synopsis of Chenopodiaceae subfam. Betoidae and notes on the taxonomy of Beta. Willdenowia. 2006;36:9–19. [Google Scholar]
- Kamm A, Galasso I, Schmidt T, Heslop-Harrison JS. Analysis of a repetitive DNA family from Arabidopsis arenosa and relationships between Arabidopsis species. Plant Molecular Biology. 1995;27:853–862. doi: 10.1007/BF00037014. [DOI] [PubMed] [Google Scholar]
- Katsiotis A, Schmidt T, Heslop-Harrison JS. Sequences of Ty1-copia-like retrotransposon elements in genus Avena. Chromosome Research. 1995;3:52–53. [Google Scholar]
- Kawabe A, Nasuda S. Structure and genomic organization of centromeric repeats in Arabidopsis species. Molecular Genetics and Genomics. 2005;272:593–602. doi: 10.1007/s00438-004-1081-x. [DOI] [PubMed] [Google Scholar]
- Kiefer-Meyer MC, Reddy AS, Delseny M. Complex arrangement of dispersed repeated DNA sequences in Oryza officinalis. Genome. 1996;39:183–190. doi: 10.1139/g96-024. [DOI] [PubMed] [Google Scholar]
- Kubis S, Heslop-Harrison JS, Schmidt T. A family of differentially amplified repetitive DNA sequences in the genus Beta reveals genetic variation in Beta vulgaris subspecies and cultivars. Journal of Molecular Evolution. 1997;44:310–320. doi: 10.1007/pl00006148. [DOI] [PubMed] [Google Scholar]
- Kubis S, Heslop-Harrison JS, Schmidt T. Repetitive DNA elements as a major component of plant genomes. Annals of Botany. 1998;82:45–55. [Google Scholar]
- Kuhn GC, Sene FM, Moreira-Filho O, Schwarzacher T, Heslop-Harrison JS. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Research. 2008;16:307–324. doi: 10.1007/s10577-007-1195-1. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen J. Plant retrotransposons. Annual Review of Genetics. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Liu K, Somerville S. Cloning and characterization of a highly repeated DNA sequence in Hordeum vulgare L. Genome. 1996;39:1159–1168. doi: 10.1139/g96-146. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Shaw RS, de los Reyes BG, Weiland JJ. Construction of a sugar beet BAC library from a hybrid with diverse traits. Plant Molecular Biology Reporter. 2004;22:23–28. [Google Scholar]
- Ma J, Wing RA, Bennetzen JL, Jackson SA. Plant centromere organization: a dynamic structure with conserved functions. Trends in Genetics. 2007;23:134–139. doi: 10.1016/j.tig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Macas J, Meszaros T, Nouzova M. PlantSat: a specialized database for plant satellite repeats. Bioinformatics. 2002;18:28–35. doi: 10.1093/bioinformatics/18.1.28. [DOI] [PubMed] [Google Scholar]
- Menzel G, Dechyeva D, Keller H, Lange C, Himmelbauer H, Schmidt T. Mobilization and evolutionary history of miniature inverted-repeat transposable elements (MITEs) in Beta vulgaris L. Chromosome Research. 2006;14:831–844. doi: 10.1007/s10577-006-1090-1. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Tsujimoto H, Sasakuma T. A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Research. 1998;6:295–302. doi: 10.1023/a:1009270824142. [DOI] [PubMed] [Google Scholar]
- Nouzova M, Neumann P, Navratilova A, Galbraith DW, Macas J. Microarray-based survey of repetitive genomic sequences in Vicia spp. Plant Molecular Biology. 2001;45:229–244. doi: 10.1023/a:1006408119740. [DOI] [PubMed] [Google Scholar]
- Ohmido N, Kijima K, Akiyama Y, de Jong JH, Fukui K. Quantification of total genomic DNA and selected repetitive sequences reveals concurrent changes in different DNA families in indica and japonica rice. Molecular and General Genetics. 2000;263:388–394. doi: 10.1007/s004380051182. [DOI] [PubMed] [Google Scholar]
- Pons J, Bucur R, Vogler AP. Higher-order repeats in the satellite DNA of the cave beetle Pholeuon proserpinae glaciale (Coleoptera: Cholevidae) Hereditas. 2003;139:28–34. doi: 10.1111/j.1601-5223.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- Presting GG, Malysheva L, Fuchs J, Schubert I. A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. The Plant Journal. 1998;16:721–728. doi: 10.1046/j.1365-313x.1998.00341.x. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends in Genetics. 2004;30:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proceedings of the National Academy of Sciences of the USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. Variability and evolution of highly repeated DNA sequences in the genus Beta. Genome. 1993;36:1074–1079. doi: 10.1139/g93-142. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. High-resolution mapping of repetitive DNA by in situ hybridization: molecular and chromosomal features of prominent dispersed and discretely localized DNA families from the wild beet species Beta procumbens. Plant Molecular Biology. 1996;30:1099–1119. doi: 10.1007/BF00019545. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends in Plant Science. 1998;3:195–199. [Google Scholar]
- Schmidt T, Metzlaff M. Cloning and characterization of a Beta vulgaris satellite DNA family. Gene. 1991;101:247–250. doi: 10.1016/0378-1119(91)90418-b. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Jung C, Metzlaff M. Distribution and evolution of two satellite DNAs in the genus Beta. Theoretical and Applied Genetics. 1991;82:793–799. doi: 10.1007/BF00227327. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schwarzacher T, Heslop-Harrison JS. Physical mapping of rRNA genes by fluorescent in situ hybridization and structural analysis of 5S rRNA genes and intergenic spacer sequences in sugar beet (Beta vulgaris) Theoretical and Applied Genetics. 1994;88:629–636. doi: 10.1007/BF01253964. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Kubis S, Heslop-Harrison JS. Analysis and chromosomal localization of retrotransposons in sugar beet (Beta vulgaris L.): LINEs and Ty1-copia-like elements as major components of the genome. Chromosome Research. 1995;3:335–345. doi: 10.1007/BF00710014. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Kubis S, Katsiotis A, Jung C, Heslop-Harrison J. Molecular and chromosomal organization of two repetitive DNA sequences with intercalary locations in sugar beet and other Beta species. Theoretical and Applied Genetics. 1998;97:696–704. [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Practical in situ hybridization. Oxford: BIOS Scientific Publishers; 2000. [Google Scholar]
- Simoens CR, Gielen J, Van Montagu M, Inzé D. Characterization of highly repetitive sequences of Arabidopsis thaliana. Nucleic Acids Research. 1988;16:6753–6766. doi: 10.1093/nar/16.14.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Vershinin AV, Heslop-Harrison JS. Comparative analysis of the nucleosomal structure of rye, wheat and their relatives. Plant Molecular Biology. 1998;36:149–161. doi: 10.1023/a:1005912822671. [DOI] [PubMed] [Google Scholar]