Abstract
Background and Aims
Dormancy is a complex trait finely regulated by hormones and environmental factors. The phytochromes that sense red:far-red (R:FR) are the sole photoreceptors involved in the termination of dormancy and the induction of germination by light. The aims of this study were to identify and characterize loci controlling this process in seeds of Arabidopsis thaliana.
Methods
Recombinant inbred lines (RILs) derived from Landsberg erecta and Cape Verde Islands (Ler × Cvi), and Bayreuth and Shahdara (Bay-0 × Sha) were used to map loci related to light effects in seeds previously exposed to chilling and after-ripening periods.
Key Results
Substantial genetic variation was found between accessions of A. thaliana in the induction of germination by light. Twelve loci were identified under R, FR or darkness, some of which were novel loci: DOG8, DOG9, DOG13, DOG14 and DOG15 detected in the Ler × Cvi RIL population; and DOG10, DOG11 and DOG12 mapped in the Bay-0 × Sha RIL population. Furthermore, independent loci were mapped for the induction of germination by low fluence (DOG-LF1 and DOG-LF2) and very low fluence of light (DOG-VLF1) in the Ler × Cvi RIL population. Several loci were confirmed and characterized after different after-ripening and chilling treatments through near-isogenic lines (NILs) and heterogeneous inbred families (HIFs).
Conclusions
The results show that one group of loci act in a wide range of environmental scenarios, whereas a smaller group of loci are relevant only under a narrower set of conditions when the influence of the most-prevalent loci is reduced as a consequence of changes in the physiological status of the seeds. In addition, the identification of specific loci controlling the action modes of the phytochromes improves our understanding of the two independent signalling pathways that promote germination in response to light.
Key words: Arabidopsis thaliana, dormancy, germination, phytochromes, very-low-fluence response (VLFR), low-fluence response (LFR), natural genetic variation, quantitative trait loci (QTL), recombinant inbred line (RIL), near-isogenic line (NIL) heterogeneous inbred family (HIF)
INTRODUCTION
Dormancy is a most important seed characteristic since it defines the conditions required for germination (for thorough discussions of this concept see Baskin and Baskin, 2004, and Finch-Savage and Leubner-Metzger, 2006). It is modulated by a considerable number of factors acting from the early stages of seed development, and is eventually terminated by particular environmental cues. Since these cues are related to ecological scenarios likely to be favourable for the future development of the new individual, dormancy plays a crucial role in the adjustment of plant populations to their habitat (Benech-Arnold et al., 2000; Finch-Savage and Leubner-Metzger, 2006). There are usually large differences in the behaviour of different species, and even between populations of the same species established in diverse environments, and it is likely that adaptation has resulted in divergent responses to the environment (Donohue et al., 2005). Consequently, the induction and loss of dormancy following seed dispersal can be modulated by a variety of environmental factors acting through many apparently different physiological mechanisms or, at least, adaptations of the same mechanism controlled through natural allelic variation in key regulatory genes (Finch-Savage and Leubner-Metzger, 2006).
In natural populations, dormancy is commonly deep at the time of seed dispersal, thus severely restricting the possibilities of germination. After the seeds reach the soil, the dormancy can be progressively reduced under the influence of a variety of factors, among which temperature is of paramount importance, modulating seed responses to signals such as light or alternating temperatures that can terminate dormancy and initiate germination (Benech-Arnold et al., 2000). Depending on the conditions, the seeds can subsequently enter into secondary dormancy and so the possibilities of germination are again reduced (Finch-Savage and Leubner-Metzger, 2006). Available information on both physiological effects and gene expression patterns is consistent with the view that different environmental factors remove successive blocks to germination and need to be experienced by the seeds in the appropriate sequence (Cadman et al., 2006; Finch-Savage et al., 2007). Genetic variation in dormancy may thus result from different combinations of alleles affecting diverse stages in the processes leading to germination. Genetic variation for seed dormancy and germination has been studied for a long time in several crops (Foley and Fennimore, 1998) and wild populations of different species, such as Arabidopsis thaliana as a model system (Evans and Ratcliffe, 1972; Napp-Zinn, 1975; Ratcliffe, 1976). Understanding the variation is complicated by the interaction between environmental and genetic factors. Quantitative trait mapping is a useful approach to analyse such complex processes. Studies of dormancy quantitative trait loci (QTLs) using recombinant inbred lines (RILs) that originated from three independent populations of A. thaliana have identified several loci that produce small effects in populations of Ler × Col RIL (van der Schaar et al., 1997) and large effects in populations of Ler × Cvi and Ler × Sha RIL (Alonso-Blanco et al., 2003; Clerkx et al., 2004). The fine mapping of one of the delay of germination loci (DOG1) mapped in the RIL population of Ler × Cvi and the isolation of a loss-of-function dog1 mutant in the Col background, displaying reduced dormancy, has led to the identification and cloning for the first time of a seed-dormancy gene accounting for variation that occurs in natural populations (Bentsink et al., 2006).
The identification of several loci (DOG1 to DOG7) and the molecular characterization of DOG1 represent highly significant contributions that open the way for a substantial advance in our understanding of the genetic and molecular bases of seed dormancy. Nevertheless, it should be noted that in previous mapping studies with the Ler × Cvi RIL population, germination was tested by incubating the seeds under a long photoperiod of white light (Alonso-Blanco et al., 2003). Under natural conditions, low temperatures and light commonly act sequentially on dormancy. The low temperature effects are integrated over a relatively long time and increase the sensitivity of the seeds to light, a factor that terminates dormancy and initiates germination (Benech-Arnold et al., 2000; Finch-Savage and Leubner-Metzger, 2006). Therefore, to further advance our mechanistic understanding of the genetic architecture of dormancy, it is necessary to separately analyse the effects of low temperature and light in seed germination.
In the field, light cues elicit responses that reflect different environmental situations (Casal and Sánchez, 1998). Promotion of germination by light through phytochromes includes two types of responses or modes of action (Casal et al., 1998). First, germination can respond to the ratio between red (R, 600–700 nm) and far-red (FR, 700–800 nm) spectral bands, which is perceived mainly through phytochromes B and E: this is called the low-fluence response (LFR; Botto et al., 1995; Shinomura et al., 1996; Henning et al., 2002). The LFR modulates germination according to the density of the vegetation covering the soil. Opening a gap in the canopy increases the R/FR ratio that reaches the seeds and this light signal will promote germination in a site where the future seedling will not experience competition from larger, established plants (Deregibus et al., 1994; Insausti et al., 1995). Second, germination can also be promoted through phytochrome A (Botto et al., 1996; Shinomura et al., 1996) by a response requiring very small amounts of photons (millisecond exposures to sunlight): this is the very-low-fluence response (VLFR). The VLFR has been shown to participate in the stimulation of germination caused by soil disturbances such as those produced by mouldboard ploughing (Scopel et al., 1994; Botto et al., 1998a). The VLFR can be saturated by small amounts of Pfr (the active form of phytochrome) established by a saturating pulse of FR (Scopel et al., 1991; Botto et al., 1996). Whether a particular seed can be stimulated by a LFR or a VLFR depends on its dormancy status, which is affected by the conditions during burial in the soil (Botto et al., 1998b). Extreme sensitivity to light (the capacity to be induced to germinate by a VLFR) is found in seeds with shallower dormancy than those requiring larger Pfr levels and that are promoted by a LFR. The fact that each photoresponse is mediated by different photoreceptors suggests the possibility of additional sources of genetic variation, which therefore require investigation.
In this study, we report on the natural genetic variation for the light responses of seeds of Ler × Cvi and Bay-0 × Sha RIL populations and their interactions with after-ripening and incubation temperatures before light treatments. Twelve loci were identified under R, FR or darkness in both RIL populations. These include the previously identified DOG1, DOG2, DOG3 and DOG7 (Alonso-Blanco et al., 2003) and five additional novel loci (DOG8, DOG9, DOG13, DOG14 and DOG15) mapped in the Ler × Cvi RIL population, and three loci (DOG10, DOG11 and DOG12) identified in the Bay-0 × Sha RIL population. Moreover, DOG-LF1 and DOG-LF2 were mapped in the LFR promotion of seed germination, and DOG-VLF1 was mapped in the VLFR promotion of seed germination.
MATERIALS AND METHODS
Plant material and growth conditions
Ler and Cvi parental lines and a set of 162 RILs (Alonso-Blanco et al., 1998), and the Bay-0 and Sha parental lines and a set of 165 RILs (Loudet et al., 2002) were obtained from ABRC (University of Ohio). Plants of RILs and parental genotypes were cultivated in a growth chamber at 22°C under long-day conditions (16 h white light, PAR = 100 µmol m−2 s−1 and 8 h dark). Plants of different genotypes were grown together and their mature seeds were harvested on the same day at the moment that all siliques had senesced. Seeds of each genotype were harvested as single bulk samples of 3–6 plants and stored in open tubes inside a closed box containing silica gel at room temperature until used for germination experiments. DOG1 and DOG2 were confirmed and characterized in different light conditions using NIL DOG17-1 (generously provided by M. Koornneef, Wageningen University, The Netherlands) and NIL 42, NIL 45 and NIL-EDI (generously provided by C. Alonso- Blanco, Centro Nacional de Biotecnología, Madrid, Spain), respectively. In addition, we selected the dog1 mutant in the Col background from the Salk line 000867 (http://signal.salk.edu) previously isolated and characterized as a non-dormant line by Bentsink et al. (2006). To confirm the presence and the allelic effects of DOG10, DOG11 and DOG12, we developed HIFs from individual RILs that still segregate in a single marker and limited genomic region following protocols published previously (Tuinstra et al., 1997). DOG10 was confirmed using RIL61 (for marker MSAT2·36) and RIL84 (for marker MSAT2·41); DOG11 was confirmed using RIL350 (for marker MSAT5·9); and DOG12 was evaluated using RIL111 (for marker MSAT4·18) and RIL57 (for marker MSAT4·9). For each of these lines, 20 F7 seeds were germinated and the plants were genotyped individually, with two or three plants fixed with the Bay-0 or the Sha alleles at the respective segregating marker being selected. Harvested seeds from these plants were then used for the germination experiments.
Light conditions and germination experiments
Samples of 25 seeds were sown in clear plastic boxes (40 × 33 × 15 mm, width × breadth × height), each containing 3 mL of 0·8 % (w/v) agar in demineralized water. To avoid undesired effects of relatively high proportions of Pfr present in dry seeds, seeds imbibed for 2 h were irradiated with a saturated pulse of FR (15 min) to establish a minimum photoequilibrium into the seeds in order to reduce germination in darkness by as much as possible. The boxes were then wrapped with black plastic and incubated at 7°C in darkness for 0, 3 or 10 d. After that, the seeds were irradiated for 15 min with a saturated pulse of FR with a RG9 filter establishing a Pfr/P ratio of 0·03 (42 µmol m−2 s−1) or a saturated pulse of R establishing a Pfr/P ratio of 0·87 (35 µmol m−2 s−1; Casal et al., 1991). Intermediate ratios of Pfr/P were provided by different R + FR mixtures of a single saturated pulse of light, as described previously (Casal et al., 1991; Botto et al., 1996). Following the light treatments, the boxes containing the seeds were again wrapped in black plastic and incubated at 25°C for 3 d before germination was determined. The criterion for germination was the emergence of the radicle.
Seeds with high sensitivity to Pfr/P were obtained before the light treatments by incubation for 3 d at 7 °C followed by 8 h at 35 °C (Cone et al., 1985; Botto et al., 1996). The short heat treatment enhances the light sensitivity to the FR pulse without affecting the response to the R pulse (Cone et al., 1985; Botto et al., 1996), in contrast to the effect of imbibition of seeds for long periods at high temperatures that allows them to enter secondary dormancy (Finch-Savage and Leubner-Metzger, 2006). In this experiment, the classical low-fluence R/FR reversible response (i.e. LFR) was calculated as the difference between the percentage of germination in R minus the FR pulse, and the VLFR was calculated as the difference between the percentages of germination after the FR pulse minus germination in darkness (Casal et al., 1998).
Mapping and QTL analysis
Each QTL mapping experiment consisted of two (first experiment) or three (second and third experiments) randomized complete blocks (each block being a different day of sowing). The QTL analysis was based on the average percentage of germination for each RIL. The percentage of germination was probit-transformed to normalize the data (Cone and Kendrick, 1986). The S statistic Qstats package of QTL Cartographer (Wang et al., 2004) was used to test the normality of the distributions. ANOVA was used to partition variance into sources originating from after-ripening (A), chilling (C), light (L) and error. In this model, we excluded the RIL genotype factor in order to conduct a robust analysis for the three environmental factors. The general linear model module of the InfoStat statistical package was used (Grupo InfoStat, 2002).
Mapmaker/EXP 3·0 (Lander et al., 1987) was used to construct the linkage map. Linkage groups were verified with a minimum LOD = 3 and a maximum distance = 50 cM (Kosambi function). Marker segregation data for the Ler × Cvi (Alonso-Blanco et al., 1998) and the Bay-0 × Sha (Loudet et al., 2002) RIL populations were obtained at http://www.arabidopsis.org/. The composite interval mapping procedure of QTL Cartographer (Zeng, 1994) was used for QTL analysis. QTL co-factors were initially selected by using forward–backward stepwise multiple regression. Mapping was conducted with a walking speed = 0·5 cM and a window size = 3 cM. LOD thresholds for each trait were calculated with 5000 permutations (Doerge and Churchill, 1996) and ranged between 2·5 and 2·7 (P = 0·05).
Two-way interactions among the identified QTLs were tested by ANOVA using the corresponding two markers as random factors. The analysis was performed using the computer program EPISTAT (Chase et al., 1997) with LOD thresholds corresponding to a significant value of P < 0·001. Ten-thousand trials were used in Monte Carlo simulations performed with EPISTAT in order to establish the statistical significance of the LOD scores for the detected interactions (Chase et al., 1997).
For each putative QTL, the interaction QTL × E (environment) was tested by repeated-measures of two-way ANOVA using the segregation of the corresponding marker and the environmental conditions in which seeds were exposed as classifying factors (P < 0·005). The general linear model module of the InfoStat statistical package was used (Grupo InfoStat, 2002).
RESULTS
Phenotypic variation in responses of germination to light after different chilling and after-ripening periods
The induction of germination after a single R or FR pulse was analysed in 15- or 90-d after-ripened seeds of Ler and Cvi followed by 0, 3 or 10 d of incubation at 7 °C. Ler and Cvi seeds after-ripened for 15 d without chilling did not germinate (Table 1). Ler seeds incubated for 3 d at 7 °C germinated close to 75 % and 0 % after R and FR pulses, respectively. In contrast, Cvi seeds displayed a deep dormancy and a smaller response to the light stimulus than the Ler seeds (Table 1). When chilling was extended to 10 d, the difference in germination between R- and FR-treated seeds was reduced in both Cvi and Ler accessions (Table 1). Ler R-treated seeds with after-ripening for 90 d showed large germination values independent of the chilling period, and FR-treated seeds required 10 d of chilling to reach similar values compared with R (Table 1). For Cvi seeds, the promotion of germination required a period of chilling and the response was almost independent of the R and FR pulse (Table 1). Significant differences were not found between germination in darkness and after a FR pulse in seeds of Ler and Cvi accessions (data not shown).
Table 1.
Summary statistics for seeds of the Ler × Cvi RIL population that had been after-ripened for 15 or 90 d, then induced to germinate by chilling (0, 3 or 10 d) and then subjected to a saturated pulse of light (R or FR)
| After-ripening period |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 d |
90 d |
|||||||||||
| Red pulse |
Far-red pulse |
Red pulse |
Far-red pulse |
|||||||||
| Chilling (d) |
||||||||||||
| 0 | 3 | 10 | 0 | 3 | 10 | 0 | 3 | 10 | 0 | 3 | 10 | |
| Ler | 0 | 74 ± 9 | 97 ± 4 | 0 | 1 ± 1 | 79 ± 3 | 74 ± 9 | 77 ± 7 | 94 ± 4 | 1 ± 1 | 47 ± 9 | 79 ± 3 |
| Cvi | 0 | 13 ± 4 | 68 ± 2 | 0 | 0 | 60 ± 0 | 0 | 30 ± 12 | 68 ± 2 | 0 | 13 ± 4 | 60 ± 0 |
| RIL mean | 23 ± 2 | 64 ± 3 | 83 ± 2 | 0·5 | 11 ± 1 | 78 ± 3 | 33 ± 3 | 64 ± 3 | 83 ± 2 | 8 ± 1 | 11 ± 1 | 72 ± 2 |
| RIL max / min | 98 / 0 | 100 / 0 | 100 / 7 | 16 / 0 | 97 / 1 | 100 / 7 | 98 / 0 | 100 / 0 | 100 / 9 | 78 / 0 | 97 / 1 | 100 / 6 |
| Vg | 833·5 | 1024·8 | 443·8 | 3·5 | 282·5 | 588·4 | 961·7 | 1021·5 | 469·8 | 173·9 | 281·2 | 776·2 |
| Ve | 14·2 | 40·3 | 41·2 | 0·9 | 28·7 | 50·2 | 29·2 | 29·7 | 27·5 | 10·9 | 18·7 | 32·1 |
| H2 | 0·98 | 0·96 | 0·92 | 0·79 | 0·91 | 0·92 | 0·97 | 0·97 | 0·94 | 0·94 | 0·94 | 0·96 |
Vg, component of variance due to among-RIL variation.
Ve, component of variance due to within-RIL variation.
H2, broad-sense heritability calculated as the proportion of total variance (Vg + Ve) attributable to genotype (Vg).
Seed germination of Ler × Cvi RILs showed transgressive variation in both directions compared with the parental lines (Table 1). The heritability index was over 0·79, indicating that a high proportion of the phenotype was explained by the genotype in our experimental conditions (Table 1). Three-way ANOVA analysis showed significant effects for the environmental factors (i.e. after-ripening, chilling and light) in modulating the germination behaviour of the RILs (see Supplementary Information Table S1, available online). Significant interactions between chilling and light, and chilling and after-ripening were detected (Supplementary Information Fig. S1 and Table S1). Overall, the results indicate that enough genetic variation exists between RILs to map the loci controlling termination of dormancy by light after the seeds had experienced different after-ripening and chilling conditions.
Mapping loci for light induction of germination for seeds of Ler × Cvi RIL population after exposure to different chilling and after-ripening treatments
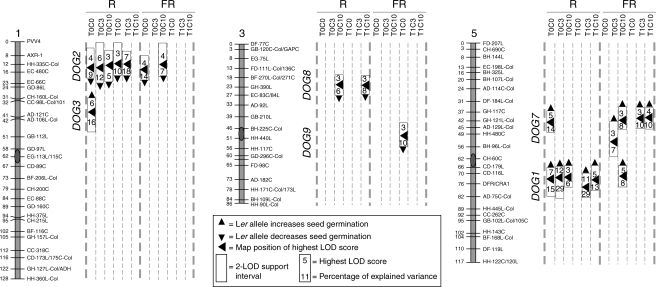
QTL analysis detected six loci in seeds after-ripened for 15 or 90 d and incubated for 0, 3 or 10 d at 7 °C and then exposed to a single pulse of R or FR. The loci have been named following the criteria of Alonso-Blanco et al. (2003). Four of them were located at the same positions and showed similar additive effects to those previously identified: DOG1, DOG2, DOG3 and DOG7 (Fig. 1, and Alonso-Blanco et al., 2003). DOG1, DOG2 and DOG7 were the most prevalent QTLs, affecting the light response of seeds in a large number of after-ripening and chilling conditions, whereas DOG3 was detected only in R-treated and non-chilled seeds after-ripened for 15 d (Fig. 1). The Ler alleles at DOG1 and DOG7 increased germination up to 36 % and 23 %, respectively, and the Cvi alleles at DOG2 increased germination up to 25 % (Table 2). Two novel QTLs were mapped at chromosome 3: DOG8 and DOG9 (Fig.1). DOG8 was detected in R-treated seeds incubated at 7 °C for 10 d, and DOG9 was mapped in non-chilled and FR-treated seeds after-ripened for 90 d. Cvi alleles at DOG8 and DOG9 loci increased germination (Fig. 1). DOG9 co-localized with DOG6, but they are probably distinct loci because they showed opposite allelic effects (Fig. 1, and Alonso-Blanco et al., 2003). The QTL mapping analysis explained up to 65 % or 34 % of the total phenotypic variation displayed in R and FR-treated seeds, respectively (Table 2).
Fig. 1.
QTL mapping for the induction of seed germination by a R or FR pulse in the Ler × Cvi RIL population. Seeds were stored at 25 °C for 15 d (T0) or 90 d (T1), and then incubated for 0, 3 or 10 d (C0, C3 or C10, respectively) at 7 °C followed by a pulse of R or FR. Only linkage groups with significant QTLs are shown. Rectangles show the two-LOD support interval; the arrowheads inside the rectangles point to the position of the highest LOD score; the numbers above the arrowheads indicate the LOD score; the numbers below the arrowheads indicate the percentage of explained variance for each QTL. The arrowheads outside the rectangles show the allelic effects of each QTL: the arrowheads point upwards when the Ler alleles increased the average value of the trait and downwards when the Cvi alleles increased the average value of the trait.
Table 2.
Summary of QTL detected for seeds in the Ler × Cvi RIL population that had been after-ripened 15 or 90 d, then induced to germinate by chilling (0, 3 or 10 d) and then subjected to a saturated pulse of light (R or FR)
| T0 = 15 d after-ripening |
T1 = 90 d after-ripening |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Red pulse |
Far-red pulse |
Red pulse |
Far-red pulse |
|||||||||||
| Chilling (d) |
||||||||||||||
| 0 | 3 | 10 | 0 | 3 | 10 | 0 | 3 | 10 | 0 | 3 | 10 | QTL × E | ||
| DOG1 (DFR) C5·72 cM | LOD | 6·75 | 11·5 | 3·4 | 5·26 | 11·26 | 4·91 | <0·0001 | ||||||
| Add % | 24 | 36 | 15 | 18 | 35 | 16 | ||||||||
| Var % | 15 | 29 | 6 | 8 | 29 | 13 | ||||||||
| Interval cM | 66–82 | 65–83 | 66–77 | 66–76 | 71–82 | 65–77 | ||||||||
| DOG2 (HH·335) C1·15 cM | LOD | 3·68 | 5·60 | 3·17 | 4·28 | 3·80 | 2·77 | 7·12 | <0·0001 | |||||
| Add % | −19 | −22 | −13 | −14 | −17 | −18 | −25 | |||||||
| Var % | 9 | 12 | 5 | 14 | 7 | 10 | 10 | |||||||
| Interval cM | 6–16 | 0–22 | 6–22 | 12–24 | 0–20 | 0–15 | 6–15 | |||||||
| DOG1 × DOG2 | Var % | 8 | 9 | 10 | 8 | 9 | 10 | |||||||
| DOG3 (AD·121) C1·38 cM | LOD | 5·76 | 0·0050 | |||||||||||
| Add % | 25 | |||||||||||||
| Var % | 16 | |||||||||||||
| Interval cM | 31–49 | |||||||||||||
| DOG7 (nga139) C5·42 cM | LOD | 5·38 | 3·27 | 3·64 | 3·50 | 4·26 | 0·0013 | |||||||
| Add % | 22 | 21 | 23 | 10 | 20 | |||||||||
| Var % | 14 | 7 | 8 | 10 | 10 | |||||||||
| Interval cM | 38–46 | 42–58 | 34–46 | 34–46 | 34–46 | |||||||||
| DOG1 × DOG7 | Var % | 12 | ||||||||||||
| DOG2 × DOG7 | Var % | 11 | 14 | 9 | 12 | 14 | ||||||||
| DOG8 (GH·390) C3·20 cM | LOD | 2·97 | 3·05 | NS | ||||||||||
| Add % | −14 | −11 | ||||||||||||
| Var % | 6 | 8 | ||||||||||||
| Interval cM | 16–27 | 20–27 | ||||||||||||
| DOG1 × DOG8 | Var % | 6 | ||||||||||||
| DOG9 (HH·440) C3·49 cM | LOD | 2·93 | NS | |||||||||||
| Add % | −11 | |||||||||||||
| Var % | 10 | |||||||||||||
| Interval cM | 43–54 | |||||||||||||
| Complete model | Var % | 65 | 60 | 40 | 14 | 19 | 33 | 10 | 47 | 39 | 10 | 22 | 34 | |
The closest marker to each QTL is shown in the first column, and its location is indicated by the linkage group followed by the map position. Additive effects are given as the difference between means of the two RIL genotypic groups (a positive value implies that Ler alleles induce germination more in comparison with Cvi alleles, and a negative value indicates the opposite). NS, not significant.
Analysis of QTL × E interactions showed that four of the six loci (DOG1, DOG2, DOG3 and DOG7) had significant effects depending on the after-ripening, chilling and light conditions experienced by the seeds (P < 0·005, Table 2). Interactions of these QTL with after-ripening confirm previous results (Alonso-Blanco et al., 2003).
Two-way interactions were analysed among the six QTL identified. DOG1 interacted significantly with DOG2, DOG7 and DOG8, while DOG2 interacted with DOG7 (P < 0·0005, Table 2). The interactions between DOG1, DOG2 and DOG7 alleles were also detected previously (Alonso-Blanco et al., 2003). A novel QTL × QTL interaction was detected between DOG1 and DOG8. The reaction norms of DOG1 × DOG8 interaction showed that the Cvi alleles at DOG1 and the Ler alleles at DOG8 decrease the light sensitivity to R (Supplementary Information Fig. S2).
Mapping loci for light induction of seed germination in the Bay-0 × Sha RIL population
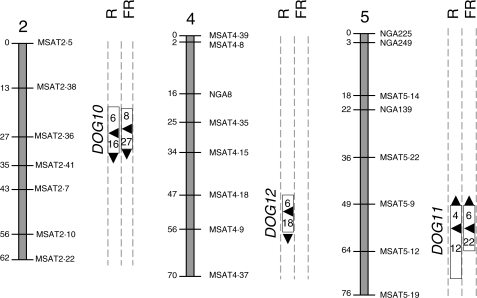
Ninety-day after-ripened seeds of Bay-0, Sha and their RILs were incubated for 3 d at 7 °C and then exposed to a R or FR pulse (i.e. identical environmental conditions to those for T1C3R and T1C3FR in Fig. 1). Bay-0 seeds showed a deep dormancy whilst Sha seeds germinated by 98 % and 15 % after a R or FR pulse, respectively (Supplementary Information Table S2). Thus, Bay-0 and Sha differed considerably in their seed dormancy status, displaying a wide genetic variation in their responses to light. The QTL analysis mapped DOG10 and DOG11 in R- and in FR-treated seeds with a higher contribution under FR and DOG12 in R-treated seeds (Fig. 2). The Sha alleles at DOG10 and DOG12 and the Bay-0 alleles at DOG11 promoted seed germination (Fig. 2 and Supplementary Information Table S3). The genetic component of the total phenotypic variance was similar between R and FR treatments (46 % and 49 %, respectively; Supplementary Table S3). Neither a significant two-way QTL × QTL interaction, nor a QTL × E interaction were detected.
Fig. 2.
QTL mapping for the induction of seed germination by a R or FR pulse in the Bay-0 × Sha RIL population. Seeds were stored at 25 °C for 90 d and then incubated for 3 d at 7 °C, followed by a pulse of R or FR. Only linkage groups with significant QTLs are shown. The arrowheads outside the rectangles show the allelic effects of each QTL: the arrowheads point upwards when the Bay alleles increased the average value of the trait and downwards when the Sha alleles increased the average value of the trait. For other details see Fig. 1.
An objective of this study was to evaluate the presence of common loci controlling the termination of dormancy by light in accessions of A. thaliana originating from diverse locations and displaying different levels of dormancy. Linkage map positions between Ler × Cvi and Bay-0 × Sha RIL populations were aligned with known physical positions for markers and with estimated physical positions for the QTLs. DOG1 and DOG11 at chromosome 5 overlapped between RIL populations for R-treated seeds incubated 3 d at 7 °C (Figs 1 and 2).
Mapping loci for LFR and VLFR on seed germination in the Ler × Cvi RIL population
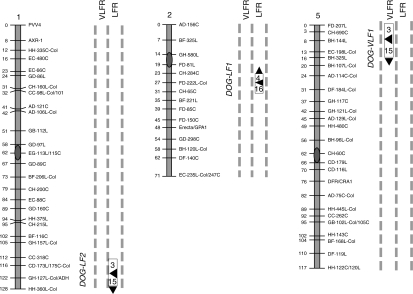
It is known that germination may be promoted by two different modes of action of the phytochromes depending on the seed sensitivity to R and FR. When germination is promoted by an R pulse and cancelled by a subsequent pulse of FR, the light response is R/FR reversible and classified as a low-fluence response (LFR). Under certain conditions, a pulse of FR can promote germination above the level of dark-imbibed seeds and the response induced is known as a very-low-fluence response (VLFR; Casal et al., 1998). To maximize the VLFR induction of seed germination, Ler, Cvi and RIL seeds after-ripened for 1 year were incubated for 3 d at 7 °C followed by 8 h at 35 °C, and then treated with R, FR or kept in darkness (Supplementary Information Table S4). Ler seeds displayed a larger LFR compared with Cvi seeds (59 vs. 39 %, respectively), and Cvi seeds showed an enhanced VLFR with respect to Ler seeds (14 vs. 3 %, respectively, Supplementary Information Table S4). The loci detected in the QTL mapping analysis were named delay of germination related with low-fluence (DOG-LF) or delay of germination related with the very-low-fluence (DOG-VLF). DOG-LF1, located to the 22–30 cM region of chromosome 2, and DOG-LF 2, located to the 115–125 cM region of chromosome 1, mapped for the LFR; and DOG-VLF1, located at the 0–18 cM of chromosome 5, mapped for the VLFR (Fig. 3 and Supplementary Information Table S5). DOG-VLF1 co-localized with DOG4 but it is likely to correspond to different loci because they showed opposite allelic effects (Fig. 3, and Alonso-Blanco et al., 2003). Neither a significant two-way QTL × QTL interaction among the three QTLs, nor a QTL × E interaction among modes of action were detected.
Fig. 3.
QTL mapping for the very-low-fluence response and the low-fluence response (VLFR and LFR, respectively) in the Ler × Cvi RIL population. Seeds were stored at 25 °C for 360 d, incubated for 3 d at 7 °C followed by 8 h at 35 °C, and then exposed to a pulse of R or FR, or kept in darkness. Only linkage groups with significant QTLs are shown. For details see Fig. 1.
Furthermore, the loci identified in R- and/or FR-treated seeds after-ripened for 15 or 90 d were not detected in seeds after-ripened for 360 d and incubated for 3 d at 7 °C followed by 8 h at 35 °C (compare R and FR conditions in Table 2 and Supplementary Information Table S5). DOG13 and DOG15 were mapped in R-treated seeds, suggesting that prolonged after-ripening and/or high incubation temperature are responsible for detection of different loci. In addition DOG14, mapped at the bottom of chromosome 2, contributed to the control of germination in darkness (Supplementary Information Table S5).
Genetic and physiological characterization of DOG1, DOG2, DOG10, DOG11 and DOG12
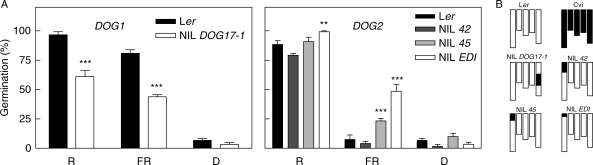
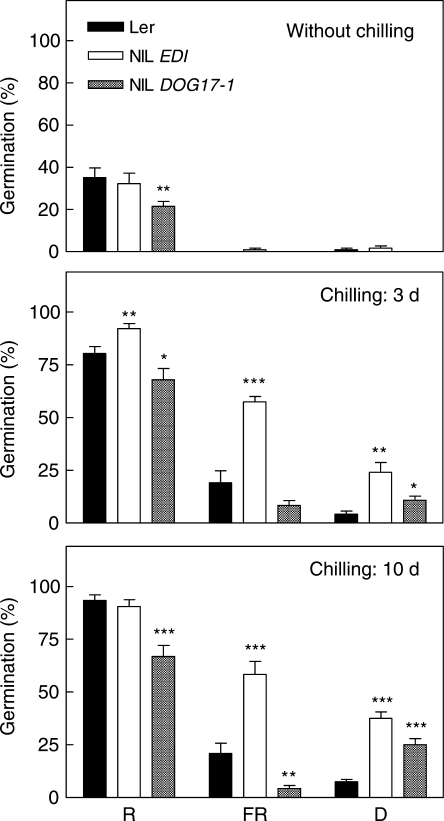
Seed germination was tested for NILs carrying a short DNA fragment of Cvi, inserted in the uniform background of Ler, in the region of DOG1 or DOG2 treated with R, FR or kept in darkness. NIL DOG17-1 seeds showed significantly reduced germination after R or FR, but not in darkness, compared with Ler seeds (P < 0·001, Fig. 4), confirming the participation of DOG1 in the termination of dormancy by light. The Cvi alleles enhanced germination at DOG2 in R (P = 0·0103 for NIL EDI) or FR-treated seeds (P = 0·0046 for NIL 45, and P = 0·0002 for NIL EDI, Fig. 4). These results show that DOG2 is contained in the shorter Cvi region included in the NIL EDI. To analyse the effects of chilling on the expression of DOG1 and DOG2, 120-d after-ripened seeds were incubated for 0, 3 or 10 d at 7 °C followed by a saturating pulse of R or FR, or kept in darkness (Fig. 5). The results showed that longer periods of chilling increased the Ler additive effects at DOG1 in seeds exposed to R and FR (Fig. 5). Significant effects of DOG1 alleles in seeds treated with a saturating pulse of FR were detected only when longer periods of chilling were used. This suggests that when the Pfr level of the seeds is very low the influence of DOG1 can be perceived if dormancy is substantially reduced. In contrast, seeds incubated in darkness showed a null or opposite contribution of the allelic effects, suggesting that this locus may be particularly linked to the promotion of germination by light (Fig. 5). The promotion of germination by Cvi alleles at DOG2 was manifested in seeds incubated at low temperatures and their effects were independent of the light treatment (Fig. 5).
Fig. 4.
(A) Confirmation and characterization of DOG1 and DOG2 in the promotion of germination by light. Seeds were stored at 25 °C for 90 d (DOG1) or 45 d (DOG2) and then incubated 3 d at 7 °C, followed by a pulse of R or FR, or kept in darkness (D). Seeds were harvested simultaneously from plants grown in identical environmental conditions, and therefore the NILs are directly comparable with the Ler control. Values are means of 3–6 replicates ± s.e. Only significant differences between means are shown: ***, P < 0·01; **, P < 0·05 (Student's t-test). (B) Graphical representation of the genotypes of the parental lines Ler, Cvi and the four NILs carrying a single Cvi introgression fragment.
Fig. 5.
Chilling effects on the promotion of germination by light of DOG1 and DOG2. Seeds were stored at 25 °C for 120 d and then incubated at 7 °C for 0, 3 or 10 d, followed by a pulse of R or FR, or kept in darkness (D). Seeds were harvested simultaneously from plants grown in identical environmental conditions, and therefore the NILs are directly comparable with the Ler control. Values are means of 6–9 replicates ± s.e. Only significant differences between means are shown: ***, P < 0·01; **, P < 0·05; *, P < 0·10 (Student's t-test).
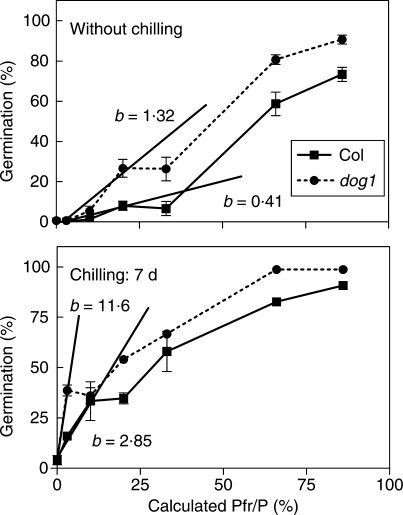
In addition, the role of DOG1 was studied in the promotion of germination by single light pulses that established a series of Pfr/P ratios. The germination of wild-type (WT, Columbia accession) and dog1 seeds plotted against Pfr/P showed two phases separated by a plateau (Fig. 6). The first phase, at lower Pfr/P, corresponds to the VLFR and the second one to the LFR. In seeds not exposed at 7 °C before the light pulse, the VLFR occurred below Pfr/P = 0·20 and the LFR was observed between Pfr/P = 0·33 and Pfr/P = 0·85. Increasing the light sensitivity of seeds by a previous incubation at 7 °C for 7 d reduced the VLFR range to Pfr/P = 0·03 and Pfr/P = 0·10 in dog1 and WT seeds, respectively (Fig. 6). The higher germination response of dog1 seeds compared with WT seeds was a consequence of the enhancement of the VLFR in the mutant genotype (Fig. 6). In dog1 seeds, the slope of the VLFR was 320–410 % higher than in the WT (Fig. 6).
Fig. 6.
Germination of wild-type (Columbia, Col) and dog1 mutant seeds plotted against different Pfr/P ratios provided by a light pulse. Seeds were stored at 25 °C for 50 d and then incubated at 7 °C for 0 or 7 d, followed by a pulse of light that established different calculated Pfr/P ratios. The slopes of the regression line for the VLFR are indicated with the letter b. Values are means of 3–6 replicates ± s.e.
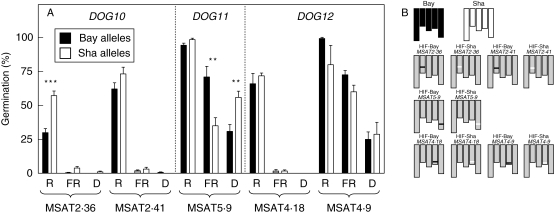
To confirm the position of the QTLs mapped in the Bay-0 × Sha RIL population, HIFs were developed from individual RILs that still segregate in a single marker of interest. DOG10 was confirmed at the MSAT2·36 marker. The Sha alleles increased germination by 27 % in R-treated seeds (P < 0·0001, Fig. 7). R- and FR-treated seeds of the same HIF exposed after different after-ripening periods confirmed the additive effects of Sha at the MSAT2·36 marker (data not shown). The promotion of germination by the Bay-0 alleles at DOG11 was associated with the MSAT5·9 marker and their effects were detected in FR-treated seeds (P < 0·0046) but not in seeds kept in darkness (Fig. 7). We could not confirm that DOG12 is associated with either the MSAT 4·18 or the MSAT4·9 markers (Fig. 7).
Fig. 7.
(A) Confirmation and characterization of DOG10, DOG11and DOG12 in the promotion of germination by light using heterogeneous inbred families (HIFs). Seeds were stored at 25 °C for 180 d (DOG10), 210 d (DOG11) and 300 d (DOG12), and then incubated for 3 d at 7 °C followed by a pulse of R or FR, or kept in darkness (D). Seeds were harvested simultaneously from plants grown in identical environmental conditions, and therefore the HIFs are directly comparable at the same marker. Values are means of 6–18 replicates ± s.e. Only significant differences between means are shown: ***, P < 0·01; **, P < 0·05 (Student's t-test). HIFs carried on Bay and Sha alleles at the indicated marker are shown. (B) Graphical representation of the genotypes of the parental lines Bay, Sha and the 5 HIFs segregating at the marker of interest as Bay (black) or Sha (white), and homozygous elsewhere (grey).
DISCUSSION
Light is a crucial factor that controls seed behavior, enhancing the chances of germination in environmental situations favourable for the future development of a new plant. We found substantial genetic variation between accessions of A. thaliana in the responses of germination to light. Seeds of Ler and Sha showed low levels of dormancy and a clear R/FR response, whereas Cvi and Bay-0 genotypes showed deeper dormancy. QTL mapping using recombinant inbred lines of Ler × Cvi and Bay-0 × Sha allowed us to identify 12 loci under R or FR light pulses, or under darkness (Figs 1, 2 and Supplementary Information Table S5). Interestingly, the loci identified in younger seeds after-ripened for no more than 90 d were not detected in older seeds incubated at 35 °C for 8 h after chilling, suggesting that prolonged after-ripening and/or high temperatures during incubation make the seeds sensitive to the influence of different light-related loci such as DOG13 and DOG15 (Supplementary Information Table S5). In addition, DOG-LF1, DOG-LF2 and DOG-VLF1 influenced the variation in LFR and VLFR, indicating specific and independent molecular points of control for the two modes of action of the phytochromes that promote seed germination (Fig. 3).
It was found that several loci mapped in a wide range of situations characterized by different combinations of after-ripening, low temperature and light, whereas a different set of loci influenced germination only within a narrow set of conditions and apparently when the influence of the most-prevalent QTLs was reduced. These results suggest that dormancy relief is a process finely regulated by a defined hierarchy of genes expressed in a temporal sequence associated with the physiological status of the seeds. DOG1 and DOG2 were prevalent loci mapped almost exclusively in seeds treated with R after a wide range of after-ripening and chilling conditions; whereas also DOG3 appeared in R-treated seeds but only in a narrow window of conditions such as short after-ripening and no chilling (Fig. 1, and Alonso-Blanco et al., 2003). Furthermore, a novel group of loci including DOG8 and DOG9 was also mapped in particular environments, suggesting that they act into specific signalling events induced by R or FR, respectively (Fig. 1). For example, DOG8 required a long incubation at low temperature to be influential (Fig. 1). Although environment-specific loci were found, we cannot discount the fact that those loci could display their actions in other environmental scenarios that determine a similar physiological status in the seeds (Finch-Savage et al., 2007). In fact, comparing global transcript expression patterns of Cvi seeds related with after-ripening, low temperature, nitrate and light stimuli, Finch-Savage et al. (2007) found that the gene expression profiles of seeds were grouped according to the depth of dormancy rather than to the dormancy-relieving treatments.
The identification of several loci inducing germination through the VLFR (DOG-VLF1) and LFR (DOG-LF1 and DOG-LF2) indicates that at least two independent signalling pathways control this process via light (Fig. 3). It is interesting to note that Yanovsky et al. (1997) observed significant allelic effects for the VLFR and LFR induction of seed germination between Col and Ler accessions close to DOG-LF1. This is consistent with the fact that each mode of action involves a different photoreceptor (phyB and phyE for LFR, phyA for VLFR) and, consequently, the signalling cascades are not identical. Moreover, each one participates in the perception of particular environmental cues that promote germination. The promotion of germination through the VLFR in response to very brief exposures to light during tillage in agricultural fields (Scopel et al., 1994; Botto et al., 1998a) or the perception of high a R/FR ratio associated with the opening of gaps in closed canopies leading to a LFR (Vázquez-Yanes and Orozco-Segovia, 1990; Deregibus et al., 1994; Insausti et al., 1995) are most likely regulated by different genes. It appears that each set of these loci play detectable functions only after proper changes in the light sensitivity of the seeds caused by factors such as after-ripening, chilling or high temperatures. It seems likely, then, that the expression of allelic variations in the VLFR and LFR loci identified in this study may be related to the seasonal patterns of light sensitivity observed in wild populations of seeds in the soil (Derkx and Karssen, 1994; Botto et al., 1998b).
DOG1 and DOG2 were confirmed and characterized by comparing the germination behavior of NIL seeds exposed to several chilling and light treatments. Whereas DOG2 allelic effects operated under light and darkness conditions, the inhibitory action of Cvi alleles at DOG1 was confined to seeds irradiated with a R and/or FR pulse (Figs 4 and 5). The results consistently showed that NIL DOG17-1 seeds germinated more than Ler seeds in darkness, suggesting an active role of DOG1 specifically in the induction of germination by light (Fig. 5). The apparent discrepancy of our data for seeds geminating in darkness with those of Bentsink et al. (2006) is likely to reflect differences in the quantity of the active form of phytochrome in seeds originated from plants grown in light environments with different R/FR ratios (McCullough and Shropshire, 1970; Wulff, 1995). Our experimental protocol substantially decreased the levels of Pfr derived from the light environment experienced by the seeds during ripening. In addition, we showed that DOG1 acts as a negative regulator of the VLFR induction of seed germination (Fig. 6). It has been reported that Col and Nossen accessions have reduced VLFR compared with the Ler accession (Yanovsky et al., 1997; Alconada-Magliano et al., 2005). Our data demonstrate that the loss-of-function of DOG1 might be one of the changes leading to the increase of the VLFR in nature.
Using heterogeneous inbred families, we were able to confirm the presence and expected additive effects of DOG10 and DOG11 located around the MSAT2·36 and MSAT5·9 markers in the QTL analysis of the Bay × Sha RIL population (Fig. 7). Meng et al. (2008) found QTLs named CDG-2 and CDG-5 that accounted for germination at low temperature in the dark, co-localizing with DOG10 and DOG11, respectively. In addition, DOG1 and DOG11 are loci linked at similar positions in the Ler × Cvi and Bay-0 × Sha mapping populations, respectively (Figs 1 and 2). In both RIL populations, DOG1 and DOG11 explained a significant fraction of the phenotypic variation and displayed opposite additive effects in darkness with respect to the light stimulus (Table 2, Supplementary Information Table S3, and Figs 5 and 7).
Considering the mapping intervals, PIL5 (At2g20180) appears as a strong candidate gene for DOG10 and/or DOG-LF1 (Figs 2 and 3). PIL5 protein, a basic helix–loop–helix transcription factor, is a negative regulator of phytochrome-mediated seed germination (Oh et al., 2004, 2006). When activated by light, the phytochromes accelerate the degradation of PIL5 in seeds, releasing the transcriptional repression of the GA3-oxidase genes and the transcriptional activation of the GA2-oxidase gene, leading to increases in the level of bioactive GA in seeds. Taking into account that there are sufficient polymorphisms at the PHYB gene to cause differential plant responses to light (Filiault et al., 2008), we cannot discount PHYB (At2g18790) as another candidate gene for DOG10 and/or DOG-LF1 into the mapping intervals, and neither can it be discounted that different genes might be responsible for the natural genetic variation at DOG10 and/or DOG-LF1 (i.e. PIL5 and PHYB).
CONCLUSIONS
Seed germination is determined by genetic control via complex regulatory networks that continuously integrate environmental signals. The goal of this paper was to explore the natural genetic variation in two populations of recombinant inbred lines in order to identify the loci controlling the induction of germination by light under different chilling and after-ripening treatments. The results presented here establish some novel conclusions. First, one group of loci act in a considerable number of different conditions, whereas a smaller group of light-related loci are effective only under a narrower set of environmental conditions, apparently when the influence of the most-prevalent QTLs is reduced as consequence of changes in the physiological status of the seeds. Second, the identification of specific loci associated with the two action modes of phytochromes that promote germination improves our understanding of the two independent signalling pathways that relate seed germination with specific environmental clues.
SUPPLEMENTARY INFORMATION
ACKNOWLEDGEMENTS
We thank Carolina Di Santo for performing the germination experiments of the Ler × Cvi RIL population under different after-ripening and chilling conditions, and Amy Austin for critically reading the manuscript. We also thank Maarten Koornneef and Carlos Alonso-Blanco for kindly providing the seeds of NILs. Financial support for J.F.B. came from the University of Buenos Aires (Grant G013) and the Agencia Nacional de Promoción Científica y Tecnológica (PICT 32137).
LITERATURE CITED
- Alconada-Magliano T, Botto JF, Godoy V, Symonds VV, Lloyd AM, Casal JJ. New Arabidopsis recombinant inbred lines (Ler/No-0) reveal natural variation in phytochrome-mediated responses. Plant Physiology. 2005;138:1126–1135. doi: 10.1104/pp.104.059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant Journal. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-Vries H, Koornneef M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164:711–729. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Baskin CC. A classification system for seed dormancy. Seed Science Research. 2004;14:1–16. [Google Scholar]
- Benech-Arnold RL, Sánchez RA, Forcella F, Kruk BC, Ghersa CM. Enviromental control of dormancy in weed banks in soil. Field Crops Research. 2000;67:105–122. [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proocedings of the National Academy of Science of the USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Sánchez RA, Casal JJ. Role of phytochrome B in the induction of seed germination by light in Arabidopsis thaliana. Journal of Plant Physiology. 1995;l46:307–312. [Google Scholar]
- Botto JF, Sánchez RA, Whitelam GC, Casal JJ. Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiology. 1996;110:439–444. doi: 10.1104/pp.110.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Scopel AL, Ballaré CL, Sánchez RA. The effect of light during and after cultivation with different tillage implements on weed seedling emergence. Weed Science. 1998;a 46:351–357. [Google Scholar]
- Botto JF, Sánchez RA, Casal JJ. Burial conditions affect the light responses of Datura ferox seeds. Seed Science Research. 1998;b 8:423–429. [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Sánchez RA. Phytochromes and seed germination. Seed Science Research. 1998;8:317–329. [Google Scholar]
- Casal JJ, Sánchez RA, Di Benedetto AH, de Miguel LC. Light promotion of seed germination in Datura ferox is mediated by a highly stable pool of phytochrome. Photochemistry and Photobiology. 1991;53:249–254. [Google Scholar]
- Casal JJ, Sánchez RA, Botto JF. Modes of action of phytochromes. Journal of Experimental Botany. 1998;49:127–138. [Google Scholar]
- Chase K, Adler FR, Lark KG. EPISTAT: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theorical Applied Genetics. 1997;94:724–730. [Google Scholar]
- Clerkx E, El-Lithy M, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SP, Vreugdenhil D, Koornneef M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accesions landsberg erecta and shakdara, using a new recombinant inbred line population. Plant Physiology. 2004;135:1–12. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone JW, Kendrick RE. Photocontrol of seed germination. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in plants. Dordrecht: Kluwer; 1986. pp. 443–465. [Google Scholar]
- Cone JW, Jaspers PAPM, Kendrick RE. Biphasic fluence- response curves for light-induced germination of Arabidopsis thaliana seeds. Plant Cell Environment. 1985;8:605–612. [Google Scholar]
- Deregibus VA, Casal JJ, Jacobo EJ, Gibson D, Kauffman M, Rodríguez AM. Evidence that heavy grazing may promote the germination of Lolium multiflorum seeds via phytochrome-mediated perception of high red/far red ratios. Functional Ecology. 1994;8:536–542. [Google Scholar]
- Derkx MPM, Karssen CM. Are seasonal dormancy patterns in Arabidopsis thaliana regulated by changes in seed sensitivity to light, nitrate and gibberellin? Annals of Botany. 1994;73::129–136. [Google Scholar]
- Doerge RW, Churchill GA. Permutations tests for multiple loci affecting quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, Schmitt J. Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution. 2005;59:740–757. [PubMed] [Google Scholar]
- Evans J, Ratcliffe D. Variation in ‘after-ripening’ of seeds of Arabidopsis thaliana and its ecological significance. Arabidopsis Information Service. 1972;9 [Google Scholar]
- Filiault D, Wessinger C, Dinneny J, Lutes J, Borevitz J, Weigel D, Chory J, Maloof J. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proceedings Academy of Sciences of the USA. 2008;105:3157–3162. doi: 10.1073/pnas.0712174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant Journal. 2007;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- Foley ME, Fennimore SA. Genetic basis for seed dormancy. Seed Sience Research. 1998;8:173–182. [Google Scholar]
- Grupo InfoStat. InfoStat. Córdoba, Argentina: Universidad Nacional de Córdoba; 2002. [Google Scholar]
- Henning L, Stoddart WM, Dieterle M, Whitelam GC, Schäfer E. Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiology. 2002;128:194–200. [PMC free article] [PubMed] [Google Scholar]
- Insausti P, Soriano A, Sánchez RA. Effects of flood-influenced factors on seed germination of Ambrosia tenuifolia. Oecologia. 1995;103:127–132. doi: 10.1007/BF00328433. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMARKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theorical and Applied Genetics. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- McCullough JM, Shropshire WJ. Physiological predetermination of germination responses in Arabidopsis thaliana (L.) Heynh. Plant Cell Physiology. 1970;11:139–148. [Google Scholar]
- Meng P-H, Macquet A, Loudet O, Marion-Poll A, North HM. Analysis of natural allelic variation controlling Arabidopsis thaliana seed germinability in response to cold and dark: identification of three major quantitative trait loci. Molecular Plant. 2008;1:145–154. doi: 10.1093/mp/ssm014. [DOI] [PubMed] [Google Scholar]
- Napp-Zinn K. On the genetical basis of the light requirement in seed germination of Arabidopsis. Arabidopsis Information Service. 1975;12 [Google Scholar]
- Oh E, Kim J, Park E, Kim J-I, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix–loop–helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung W, Choi G. Light activates the degradation of PIL5 protein to promote seed germination through giberellin in Arabidopsis. Plant Journal. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- Ratcliffe D. Germination characteristics and their inter- and intra-population variability in Arabidopsis. Arabidopsis Information Service. 1976;13 [Google Scholar]
- Scopel AL, Ballaré CL, Sánchez RA. Induction of extreme light sensitivity in buried weed seeds and its role in the perception of soil cultivations. Plant Cell and Environment. 1991;14:501–508. [Google Scholar]
- Scopel AL, Ballaré CL, Radosevich SR. Photostimulation of seed germination during soil tillage. New Phytologist. 1994;126:145–152. [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and phytochrome B-specific photoinduction of seed germination in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theoretical and Applied Genetics. 1997;95:1005–1011. [Google Scholar]
- van der Schaar W, Alonso-Blanco C, Léon-Kloosterziel KM, Jansen RC, Van Ooijen JW, Koornneef M. QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity. 1997;79:190–200. doi: 10.1038/hdy.1997.142. [DOI] [PubMed] [Google Scholar]
- Vázquez-Yanes C, Orozco-Segovia A. Ecological significance of light-controlled seed germination in two contrasting tropical habitats. Oecologia. 1990;83:171–175. doi: 10.1007/BF00317748. [DOI] [PubMed] [Google Scholar]
- Wang X, Korstanje R, Higgins D, Paigen B. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Research. 2004;14:1767–1772. doi: 10.1101/gr.2668204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff R. Environmental maternal effects on seed quality and germination. In: Kigel J, Galili G, editors. Seed development and germination. New York: Marcel Dekker; 1995. pp. 491–505. [Google Scholar]
- Yanovsky MJ, Casal JJ, Luppi JP. The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia dissect two branches of phytochrome A signalling pathways that correspond to the very-low fluence and high-irradiance responses of phytochrome. Plant Journal. 1997;12:101–109. doi: 10.1046/j.1365-313x.1997.00659.x. [DOI] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]