Abstract
Background and Aims
Platanaceae is an old family of angiosperms extending back to the Early Cretaceous but consisting of a single extant genus, Platanus. Species of Platanus have long been known to hybridize, and the London plane, Platanus × hispanica, is a well-known example of a hybrid species that formed in historical times. In addition, morphological studies have suggested past interspecific or interlineage hybridization and reticulation as possibly important factors in the evolution of the genus. This study aims at unravelling the complex evolutionary information contained in internal transcribed spacer (ITS) sequences.
Methods
The ITS of the 35S nuclear ribosomal DNA are biparentally inherited, multi-copy markers with a high potential for resolving intrageneric relationships especially when ancient hybridization (reticulation) is involved. Phylogenetic trees, splits graphs and motif analysis are used to extract phylogenetic information from 223 cloned ITS sequences, representing ten species and varieties of Platanus. Non-pseudogenous and pseudogenous sequence motives are assessed to explain how different evolutionary modes contribute to possibly conflicting character state patterns in the ITS.
Key Results
It was found that putative non-functional ITS copies (‘pseudogenes’) form distinct groups in phylograms and splits graphs, and that pseudogenous lineages reflect ancient hybridization events conserved in the ITS. Specifically, pseudogenous clones of an ‘Atlantic’ North American clade share sequence motives with non-pseudogenous clones of the western (‘Pacific’) North American P. racemosa species aggregate. In addition, evidence was found for recent lateral gene flow as a possible factor in the evolution of the central Mexican P. rzedowskii.
Conclusions
Broad ITS data sets that cover intra- and interindividual variability reveal past and ongoing speciation processes in Platanus. Evolutionary pathways can be visualized with splits graphs, but not with bifurcating trees.
Key words: Ancient hybridization, bipartition networks, evolutionary patterns, internal transcribed spacer, intraindividual variability, motif analysis, phylogenetic networks, Platanus, pseudogenes
INTRODUCTION
The northern hemispheric genus Platanus (plane tree, sycamore) comprises ten species and varieties in two subgenera and is the only extant representative of the family Platanaceae (Nixon and Poole, 2003). While the family has traditionally been placed within the order Hamamelidales (Cronquist, 1981; Takhtajan, 1987), recent phylogenetic studies suggest that it belongs to Proteales (including Platanaceae, Proteaceae and Nelumbaceae), which are part of a basal grade within the eudicots (e.g. Drinnan et al., 1994; Doyle and Endress, 2000; Angiosperm Phylogeny Group, 2003; Soltis et al., 2003). Platanaceae has an excellent fossil record dating back to the Early Cretaceous and showing that the family was much more diverse during the Cretaceous and Early Tertiary (Friis et al., 1988; Crane et al., 1993; Kvaček et al., 2001; Maslova, 2003).
Modern species of Platanus show a considerable amount of overlapping morphological variability, and morphological characters are difficult to use for phylogenetic reconstructions (Nixon and Poole, 2003). One exception is the distinct morphology of the south-east Asian species P. kerrii (subgenus Castaneophyllum) that differs from the remaining species by its simple, evergreen leaves, the more complex inflorescence axis and pollen morphology (Nixon and Poole, 2003; Denk and Tekleva, 2006). The species of the second modern subgenus within Platanus, subgenus Platanus, have been subject to a major morphological revision by Nixon and Poole (2003), who recognized two species groups. One group consists of the western North American taxa P. racemosa (varieties racemosa and wrightii) and P. gentryi, and the European–Middle Eastern P. orientalis. The second group is purely American comprising P. mexicana (varieties mexicana and interior), P. rzedowskii and P. occidentalis (varieties palmeri and occidentalis) along a south–north gradient (Nixon and Poole, 2003; see Table 1 for current taxonomy of Platanus). The pattern of character variation among Platanus species and the high level of interfertility among some taxa point to past interspecific or interlineage hybridization and reticulation as possibly important factors in the evolution of the genus (Nixon and Poole, 2003).
Table 1.
Taxonomic concept of Platanus adopted for the present study and geographical ranges of species and varieties (from Nixon and Poole, 2003)
| Taxon (Nixon and Poole, 2003) | Variety | Distribution | Notes* |
|---|---|---|---|
| Platanus kerrii Gagnep.† | Vietnam | ||
| P. orientalis L. | South-eastern Europe, south-western Asia | ||
| P. racemosa Nutt. | var. racemosa | California, Baja California | 1 |
| P. racemosa Nutt. | var. wrightii (S.Wats.) Benson | Arizona, New Mexico, Chihuaha | 1 |
| P. gentryi Nixon & Poole | Sonora, Sinaloa, Chihuaha | 1 | |
| P. occidentalis L. | var. occidentalis | Eastern Canada, eastern USA | 2 |
| P. occidentalis | var. palmeri (Kuntze) Nixon & Poole ex Geerinck | Texas, Coahuila | 2 |
| P. rzedowskii Nixon & Poole | Nuevo Leon, Tamaulipas | 2 | |
| P. mexicana Moric. | var. mexicana | Veracruz, Puebla, Oaxaca, Chiapas | 3 |
| P. mexicana Moric. | var. interior Nixon & Poole | San Luis Potosi, Queretaro | 3 |
* Notes: 1, P. racemosa species aggregate; 2, P. occidentalis species aggregate; 3, P. mexicana s.l.
† Subgenus Castaneophyllum; all the remaining species belong to subgenus Platanus.
In a recent molecular marker study, Feng et al. (2005) found limited evidence for reticulate evolution in Mexican Platanus, because a single sample of P. rzedowskii grouped with P. occidentalis var. occidentalis in the internal transcribed spacer (ITS)-based tree, while the same species grouped with P. mexicana var. mexicana in a tree based on data of the nuclear encoded low-copy LEAFY gene (3′ part of second intron). The nuclear-encoded ribosomal (r) DNA ITS1 and ITS2 of the 18S-5·8S-25S rDNA cistron (35S rDNA) provide a powerful tool to recognize lateral gene flow (reticulation) in flowering plants, including the detection of ancient hybridization events because ITS is a biparentally inherited multi-copy marker (Manen, 2004; Razafimandimbison et al., 2004; Denk and Grimm, 2005; Denk et al., 2005; Besnard et al., 2007; this study). Cloning of PCR products makes it possible to detect polymorphism in the ITS sequences of species and even individual plants (e.g. Baldwin et al., 1995; Grebenstein et al., 1998; Jobst et al., 1998; Denduangboripant and Cronk, 2001; Denk et al., 2002; Grimm, 2003; Won and Renner, 2005) including pseudogenous ITS sequence data (e.g. Samuel et al., 1998; Mayol and Rosselló, 2001, Quercus; Razafimandimbison et al., 2004, Naucleeae s.l.; this study). These polymorphisms can be used to detect ITS motives shared by taxa that had sympatric areas in the geological past enabling gene flow between them (ancient hybridization), but may be allopatric at present and only distantly related [e.g. Denk and Grimm (2005) for Zelkova; Denk et al. (2005) for Fagus; Besnard et al. (2007) for Olea europaea].
In the present study, cloned ITS sequences of 66 plants (223 clones) of all living taxa of Platanus, collected mostly from natural stands, were used to address the following questions: Does ‘ancient hybridization’ play a major role in the evolution of Platanus? Is there clear evidence for P. rzedowskii being a (young) hybrid between P. mexicana and P. occidentalis? What are appropriate phylogenetic models to visualize the evolutionary mode in Platanus when ITS data are analysed?
MATERIALS AND METHODS
Sampling and ITS sequencing
Most of the plant material was collected from natural stands in western Eurasia and North America (see Supplementary Information 1, available online, for voucher information and gene bank accession numbers). North American taxa have been sampled with a high geographical coverage including very rare types such as P. gentryi and P. mexicana var. mexicana from Oaxaca (‘P. oaxacana Standl.’). Varieties of P. mexicana and P. occidentalis have been sampled from their entire distribution range to capture possible geographical signals in the ITS. Specimens of P. kerrii were obtained from three cultivated trees in the greenhouses of the Komarov Botanical Institute, St. Petersburg, Russia. Additionally, samples of P. × hispanica Mill. ex Muenchh. [syn. P. × aceroides (Ait.) Willd.], a hybrid between P. occidentalis and P. orientalis that possibly originated in the 17th century (cf. Besnard et al., 2002), were collected in Austria and Sweden.
Total DNA was isolated following a modified cethyltrimethylammonium bromide (CTAB) protocol (Gebhardt et al., 1989). PCR was carried out with primers ITS-A and ITS-D (Denk et al., 2002; modified after Jobst et al., 1998) and Vent® polymerase (high proofreading capacity; Roche Diagnostics GmbH, Mannheim, Germany) for amplification of the 3′ end of 18S rDNA, ITS1, 5·8S rDNA, ITS2, and the 5′ end of 25S rDNA; the amplified fragment was ligated and transformed using a pUC18 vector Escherichia coli strain DH5α system (for details refer to Denk et al., 2002). Up to ten positive clones per individual were cultivated overnight, mixed 1 : 1 with glycerine, and stored at –70 °C for plasmid isolation and documentation. Cultures are available upon request. Plasmid DNA of up to ten clones was isolated and purified (High Pure Plasmid Isolation® Isolation Kit; Roche) and up to seven sequenced on an ABI Prism® automated sequencer with universal primers M13rev and M13uni or sent to a professional laboratory. Sequences were edited and aligned with Chromas® V.1·45 (Technelysium Pty, Tewantin, Australia) and SeqMan II® plus MegAlign® (DNAStar; Madison, USA with Clustal V algorithm implemented), and manually adjusted at two positions with minor length polymorphism. All sequences have been submitted to EMBL nucleotide data base.
Phylogenetic analyses
Phylogenetic trees were obtained under likelihood (ML) and parsimony (MP) optimality criteria. Distance and tree-based phylogenetic networks (splits graphs) relied on algorithms implemented in SplitsTree 4·6 (Huson and Bryant, 2006). The analyses were based on six matrices (annotated NEXUS-files available from GWG) with rRNA-coding positions generally excluded: (1) matrix ‘OMall’ with all original data (223 cloned ITS accessions, 553 characters); (2) matrix ‘OM+ps’ (215 accessions, 553 characters) without pseudorecombinant accessions (see below); (3) matrix ‘OMred’ (193 accessions, 553 characters) without pseudogenous and pseudorecombinant accessions; (4) matrix ‘CMall’ of strict consensus sequences (see Grimm et al., 2006) for all taxa (species/variety; Table 1) and distinct ITS variants (homoeologues in the case of one P. rzedowskii individual, distinct pseudogenous lineages and unique pseudogenous clones; 31 taxa, 553 characters; see also Supplementary Information 3 and 4, available online); (5) same matrix as in (4) without pseudorecombinant data, ‘CM+ps’ (26 taxa, 553 characters); (6) same matrix as in (4) but without pseudogenous and pseudorecombinant data, ‘CMred’ (15 taxa, 553 characters).
Maximum likelihood analyses were performed with RAxML-VI HPC (Stamatakis, 2006), using two runs, each with 100 inferences on the original alignment, and each starting from a randomized parsimony tree. Node support was assessed with non-parametric bootstrapping (BSML; Felsenstein, 1985). For the original data (matrices OMall/+ps/red), BSML is based on six times 167 (1002) replicates calculated in parallel on the computer cluster of the Cyber-Infrastructure for Phylogenetic Research project (CIPRES, www.phylo.org) using RAxML. For the consensus data with fewer operational taxonomic units, additional node support could be established with Bayesian inference using MrBayes 3·1 (CMall/+ps/red; Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) to calculate posterior probabilities (PP; Rannala and Yang, 1996; Huelsenbeck et al., 2001). MrBayes used one ‘cold’ and no heated chain in five parallel runs, each with 2 000 000 generations, following the manual's specifications for few-taxon matrices; trees in every 200th chain cycle were saved. Convergence was identified by converging log-likelihoods of the five runs and relative stationarity of substitution parameters of each run. For computation of posterior probabilities (PP), all pre-convergence trees were discarded. For the matrix CMall, PP are based on 42 615 Bayesian inferred saved trees (BIST) from the five parallel runs; PP for matrix CM+ps are based on 49 600 BIST and for matrix CMred on 49 915 BIST. A Bayesian information criterion (BIC) approach (DT-ModSel; Minin et al., 2003), pairwise likelihood ratio tests (LRT), and the Akaike information criterion (AIC; MrModeltest 2·2; Nylander, 2004) were used to select the best-fitting substitution model. The three approaches prefer either the HKY + Γ (BIC) or the GTR + Γ (AIC, LRT) model for the original data (matrix OMred). MrBayes and RAxML were hence allowed to estimate nine free parameters over the course of the runs.
ITS divergence was accessed using pairwise genetic distances (matrix OMall/red; with PAUP*; Supplementary Information 2, available online) under the best fit model HKY + Γ selected by DT-ModSel; parameters' values were selected according to AIC (MrModeltest).
Parsimony analyses were carried out with PAUP 4·0 beta 10 (Swofford, 2002) with the ‘branch-and-bound’ algorithm to find the most-parsimonious tree-like phylogenetic solution for the consensus data (matrices CMall, CM+ps and CMred). Consensus networks (Holland and Moulton, 2003; using SplitsTree) were used to investigate competing topologies of most-parsimonious trees. Node support was assessed with non-parametric bootstrapping (BSP; PAUP*, matrices CMall/+ps/red). Using a simple addition tree as a start, which was modified by tree bisection reconstruction, and holding one tree in memory, 100 000 bootstrap replicates were computed (Müller, 2005).
Phylogenetic splits (bipartitions) occurring in >5 % of ML bootstrap replicates, parsimony bootstrap replicates (P-BR), and BIST (Bayesian inferred saved trees) were investigated using bipartition networks (Grimm et al., 2006). For a bipartition network, the bipartition tables calculated with MrBayes, PAUP* and RAxML, are the input matrix for SplitsTree; the weight of each split is given by the frequency of the according bipartition in percentage of ML and parsimony bootstrap replicates and BIST. Thus, the support of alternative bipartitions that compete to define nodes in phylogenetic trees can be visualized.
The neighbour-net (NN) algorithm (Bryant and Moulton, 2002, 2004) was used to compute splits graphs based on uncorrected pairwise (p) distances based on the original data (matrices OMall, OM+ps and OMred; using SplitsTree). NN bootstrapping (BSNN) was based on 1000 replicates.
Pseudogenes, pseudorecombinants and homoeologues
Pseudogenous clones (called ‘ITS pseudogenes’ hereafter) are characterized by (a) decreased CG content, (b) increased pairwise distances (inter- and intra-taxonomic), (c) occasional deletions in genes and spacers, (d) non-synonymous mutations in the otherwise almost identical rRNA-coding regions (generally 0–1 nt difference in the 5·8S rDNA), and (e) ‘aberrant’ behaviour in phylogenetic analyses as compared with non-pseudogenous accessions (see below). In addition, pseudogenous sequence portions were observed in putative recombinant (chimeric) clones, called ‘pseudorecombinants’. These are sequences that contain pseudogenous and non-pseudogenous parts, and are obtained from samples yielding pseudogenous and non-pseudogenous clones (except for three clones of two individuals of P. mexicana var. mexicana represented only by non-pseudogenous clones). Recombination occurred in generally conserved ITS portions, which could be susceptible for crossing-over. Alternatively, the pseudorecombinants could be PCR-mediated artefacts (cf. Cronn et al., 2002). Such artefacts have been documented for Acer (Grimm et al., 2007). Polyploid hybrids as well as diploid hybrids are characterized by two or more homoeologous (‘paralogous’) nuclear genomes, i.e. at least two sets of chromosomes (and genes) that have an independent evolutionary origin. The resultant ITS homoeologues induce phylogenetic paralogy (ITS paralogues; Buckler et al., 1997; Leitch and Bennett, 1997; Cronn et al., 2002; Bailey et al., 2003). In contrast to ‘true’ gene paralogues and pseudogenes, ITS homoeologues within an individual may be orthologous: They do not necessarily differ in function, and distinct arrays of 35S rDNA may undergo concerted evolution, (epi-)genetic silencing, as well as intragenomic recombination (e.g. Arnheim et al., 1980; Chen and Pikaard, 1997; Pikaard, 2001; Cronn et al., 2002; Komarova et al., 2004; Volkov et al., 2007).
Motif analysis
Complete ITS1 and ITS2 sequences were partitioned into series of logically dependent (linked) mutations (‘motives’) as elaborated in previous studies on Acer, Fagus and Zelkova (Denk and Grimm, 2005; Denk et al., 2005; Grimm et al., 2006, 2007). For Platanus, ten motives exhibiting mainly site variability (motives I to X; Supplementary Information 3, available online) were distinguished. The motif variants detected are organized as a differentiation framework following the principles of parsimony and likelihood (Grimm, 2003; Denk and Grimm, 2005). This framework is used to assess alternative evolutionary hypotheses. Motives can be assigned to the c- and v-regions originally described by Hershkovitz and Lewis (1996) for ITS1 and ITS2; in the case of ITS2, conserved and variable regions can be correlated to hypothetical secondary structure elements (Hershkovitz and Zimmer, 1996).
For detailed investigation of pseudogenous data, all mutations were compiled (Supplementary Information 4, available online) and compared with non-pseudogenous motif sequences of the source taxa and the remaining taxa analysed.
RESULTS
General ITS variability
Length polymorphisms were restricted to a few sequence portions and/or individuals of a specific taxon and all accessions could be unambiguously aligned. ITS divergence varied significantly between clones (Supplementary Information 2). Platanus mexicana, P. occidentalis, P. rzedowskii and P. racemosa var. wrightii (one clone) displayed markedly increased intra- and interindividual variability due to pseudogenous and pseudorecombinant ITS clones (Table 2 and Supplementary Information 2 and 4).
Table 2.
ITS variability in modern Platanus
| Interclonal distances* |
||||||||
|---|---|---|---|---|---|---|---|---|
| Number of clones |
Complete data |
Only non-pseudogenous data |
||||||
| Used for calculation | Aberrant clones† | Mean | Maximal | Minimal | Mean | Maximal | Minimal | |
| P. kerrii | 13 | None | 0·006 | 0·012 | 0·000 | N/A | ||
| P. mexicana s.l. | ||||||||
| Total | 40 | 5 | 0·020 | 0·163 | 0·000 | 0·003 | 0·014 | 0·000 |
| var. interior | 10 | 3 | 0·040 | 0·087 | 0·000 | 0·001 | 0·002 | 0·000 |
| var. mexicana | 30 | 2 | 0·011 | 0·095 | 0·000 | 0·004 | 0·014 | 0·000 |
| P. occidentalis species aggregate | ||||||||
| Total | 65 | 21 | 0·056 | 0·232 | 0·000 | 0·005 | 0·016 | 0·000 |
| including rz-6525, rz-6531, and rz-6537‡ | 68 | 21 | 0·045 | 0·232 | 0·000 | 0·007 | 0·025 | 0·000 |
| P. rzedowskii | 25 | 7 | 0·065 | 0·232 | 0·000 | 0·004 | 0·010 | 0·000 |
| including rz-6525, rz-6531, and rz-6537‡ | 28 | 7 | 0·063 | 0·232 | 0·000 | 0·006 | 0·024 | 0·000 |
| var. occidentalis | 19 | 7 | 0·048 | 0·142 | 0·000 | 0·007 | 0·014 | 0·000 |
| var. palmeri | 21 | 7 | 0·055 | 0·136 | 0·000 | 0·005 | 0·010 | 0·000 |
| P. racemosa species aggregate | ||||||||
| Total | 48 | 1 | 0·005 | 0·069 | 0·000 | 0·003 | 0·012 | 0·000 |
| P. gentryi | 26 | None | 0·001 | 0·008 | 0·000 | N/A | ||
| var. racemosa | 11 | None | 0·003 | 0·006 | 0·000 | N/A | ||
| var. wrightii | 11 | 1 | 0·015 | 0·069 | 0·000 | 0·003 | 0·010 | 0·000 |
| P. orientalis s.l. | ||||||||
| Total | 40 | 0 | 0·008 | 0·018 | 0·000 | N/A | ||
| P. orientalis | 28 | None | 0·008 | 0·016 | 0·000 | N/A | ||
| P. × hispanica | 12 | None | 0·007 | 0·014 | 0·000 | N/A | ||
For each taxonomic entity (‘species’, ‘variety’, species aggregate), the mean (arithmetic), maximal and minimal genetic distance between pairs of clones is listed, including and excluding ITS pseudogenes and pseudorecombinants.
* Calculated using the substitution model favoured by DT ModSel and AIC; based on 209 clones (clones with missing data in either ITS1 or ITS2 excluded).
† ITS pseudogenes and pseudorecombinants (listet in Supplementary Information 4).
‡ Clones representing the P. mexicana homoeologue of the potentially hybrid individual Denk 2004093-2 S.
CG contents of ITS pseudogenes and pseudorecombinants of Platanus are comparable to non-pseudogenous ITS accessions of other tree genera (≥56 % in ITS1 and ITS2; various gene bank data), but are consistently lower than CG contents of non-pseudogenous accessions of Platanus. CG contents of non-pseudogenous ITS1 and ITS2 of Platanus (ITS1, 65–68 %; ITS2, 66–69 %) are relatively high for northern hemispheric woody angiosperms; e.g. Fagus (Fagaceae; 64–64·5 %/65–66 %) and Acer Section Acer (Sapindaceae: 60–66 %/59·5–65·5 %; Grimm, 2003). Basal eudicots with increased CG contents (ITS1/ITS2) are found among Proteaceae, the sister family of Platanus: Protea (70 %/72 %), Faurea (63 %/71 %), Adenanthos spp. and Isopogon spp. (>65 %; gene bank data of Barker et al., 2002). Potential functionality of rRNA genes was assessed by comparison with proposed secondary structures of plant rRNA (CRW database; Cannone et al., 2002).
Lower CG contents of ITS pseudogenes and pseudorecombinants are caused by numerous G → A and C → T transitions throughout the gene and spacer regions (see ‘Motif analysis’) accounting for markedly higher mean distances and up to 45 times higher maximal distances within a taxon (Table 2).
The alignment and pairwise distance distribution (greyish zones in Supplementary Information 2) readily distinguished pseudogenous and non-pseudogenous clones of the same taxon. Pseudogenous and non-pseudogenous clones were placed separately in an ML phylogram (Fig. 1) and an NN splits graph (below). ITS pseudogenes were encountered in Platanus wrightii (one clone; specimen 2003029 S), P. mexicana var. interior (four clones; 2004086-1 S, 2004072-1 S), P. rzedowskii (eight out of 11 clones from five specimens), P. occidentalis var. palmeri (five out of 12, three specimens), and var. occidentalis (four out of 14, three specimens). Pseudorecombinants were encountered in P. mexicana var. mexicana (li-3108; li-3109; li-3809; 2004011-3 S, 2004032–3 S, no additional pseudogenes were obtained) and P. occidentalis s.l. (oc-804, oc-806, oc-908, pa-2204, pa-2311; in addition to pseudogenes and non-pseudogenous sequences).
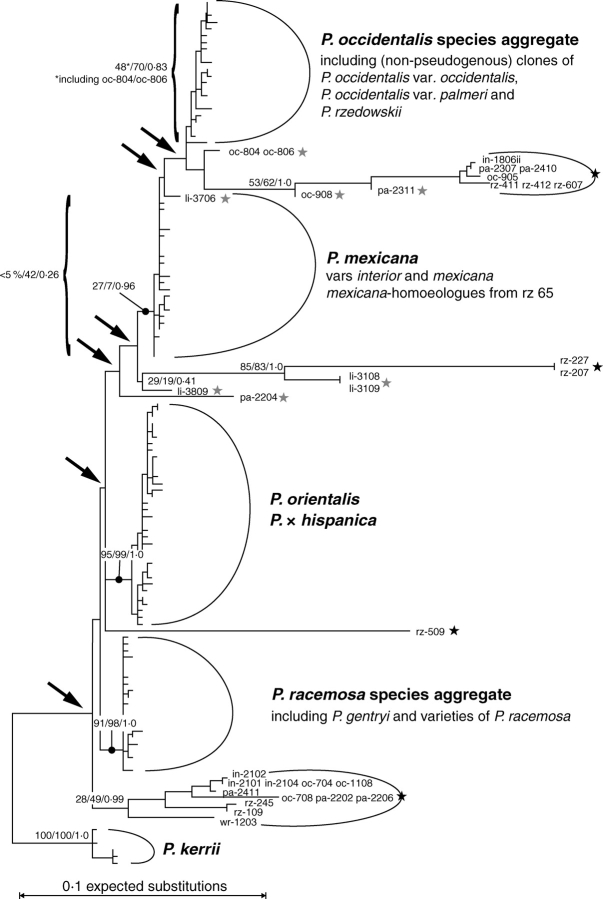
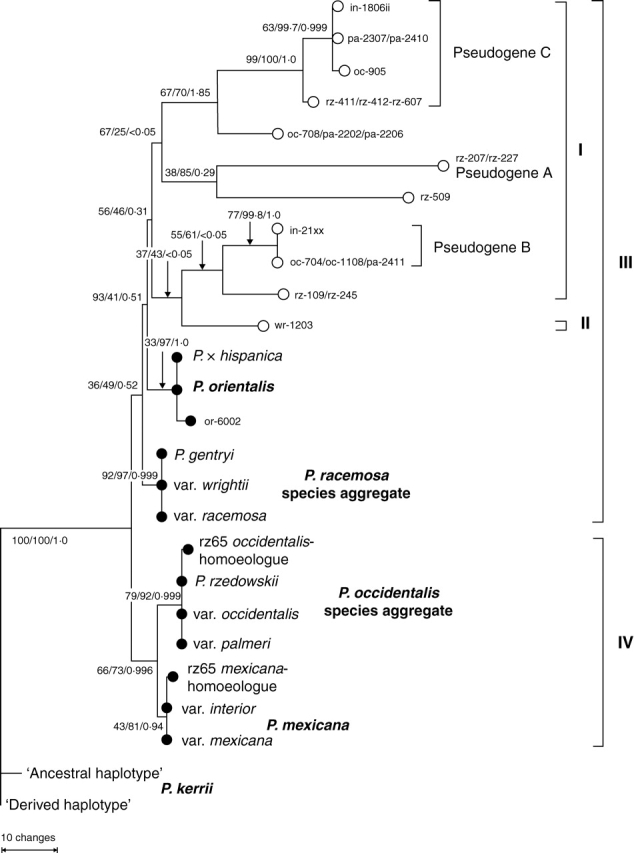
Fig. 1.
ML phylogram based on the original data matrix OMall (223 cloned ITS accessions), rooted with P. kerrii. Arrows indicate nodes affected by pseudogenous and pseudorecombinant sequences. Black stars, ITS pseudogenes; grey stars, pseudorecombinants. Node support (BSML/BSP/PP) is indicated for selected nodes; BSP and PP refer to analyses of consensus data (matrix CMall). A clade comprising only P. mexicana, P. occidentalis species aggregate and oc-8xx pseudorecombinants received BSP = 31.
When pseudogenous and pseudorecombinant sequences were excluded, inter- and intrataxonomic divergence was low but taxonomically significant (Table 2). Platanus kerrii was most distinct compared with all other taxa. Highest overall sequence similarity was found among P. gentryi, P. racemosa and P. wrightii (Fig. 1 and Table 2), called ‘P. racemosa species aggregate’ hereafter, and among P. mexicana var. interior and var. mexicana. Increased intra- and interindividual divergences in P. orientalis (Table 2) are diagnostic at the species level (six single nucleotide polymorphisms; one 2 nt-long duplication). One clone, individual Ori-0407 S, showed a 16-nt-long duplication. Platanus × hispanica fell within the natural variability of P. orientalis, i.e. only P. orientalis ITS homoeologues were obtained. Non-pseudogenous clones of P. rzedowskii, P. occidentalis var. palmeri and P. occidentalis var. occidentalis differed mainly at four sites but the respective point mutations were not found in all ITS copies because of incomplete concerted evolution. The resulting site variability is diagnostic (specific) for all three taxa, referred to as ‘P. occidentalis species aggregate’ in the following. Overall, individuals of P. occidentalis s.l. were more variable than those of P. rzedowskii (Table 2). One individual of P. rzedowskii (2004093–2 S) exhibited possibly homoeologous ITS variants, namely a P. mexicana and a P. occidentalis species aggregate homoeologue (Table 2, last three columns). In addition, this individual showed some morphological features not typical of P. rzedowskii (e.g. abaxial leaf surface becoming glabrous as in P. occidentalis var. palmeri).
Phylogenetic analyses (ML, MP and NN)
Platanus kerrii (subgenus Castaneophyllum) was used as outgroup to the remaining species following Feng et al. (2005), because ITS1 and ITS2 sequences of Platanus and Proteaceae differ greatly in length and could not be unambiguously aligned.
Low overall sequence divergence, significant intraindividual/intraspecific site variability, and incompatible phylogenetic signals accounted for the weak resolution in phylogenetic trees, especially when ITS pseudogenes and pseudorecombinant accessions were included (Fig. 1). In trees rooted on P. kerrii, the P. racemosa species aggregate was placed basally among non-pseudogenous lineages in subgenus Platanus. Exclusion of pseudorecombinants resulted in better resolved and less ambiguous topologies: Under parsimony, pseudogene lineages formed a discrete clade that was placed as sister to P. orientalis, both of which were sister to the P. racemosa species aggregate. Support of backbone nodes of subtrees with pseudogenous terminals was low and varied under different optimality criteria (ML and MP) and methods (bootstrapping, Bayesian analysis; Fig. 2). ML using original data and Bayesian analysis of consensus data favoured different topologies (not shown) concerning the organization of the pseudogene subtree (I + II in the MP tree; Fig. 2): Affected were the placement of accessions that had not been assigned to one of the pseudogenes A, B and C as labelled in Fig. 2 (Supplementary Information 4; cf. ‘Motif analysis’), and the relationship of pseudogenes A, B and C to each other. This topological conflict was caused by incompatible signals in the data and also reflected by equally low BS/PP support for alternative nodes (details not shown). For example, pseudogene rz-509 was placed as sister to all other pseudogenes [BSML (original data) = 42; PP (consensus data) = 0·58] under ML. The topological alternative favoured by MP (rz-509 sister to pseudogene A, as shown in Fig. 2) received less support under ML (BSML = 38, PP = 0·29). When pseudogenes and pseudorecombinants were excluded, high BS and PP (BSML ≥ 83, original data, BSP ≥ 90, PP ≥ 0·97, consensus data; Figs 3 and 4) supported the following clades: Platanus kerrii (both haplotypes), P. racemosa species aggregate, P. orientalis and P. × hispanica, and P. occidentalis species aggregate. Platanus mexicana (var. mexicana and interior) received low support based on the original data (46 % BSML) but high support based on the consensus data (BSP = 81 %, PP = 0·93). ITS homoeologues of P. rzedowskii 2004093–2 S grouped with either P. mexicana or P. occidentalis species aggregate. Two main genetic lineages could be recognized within subgenus Platanus:
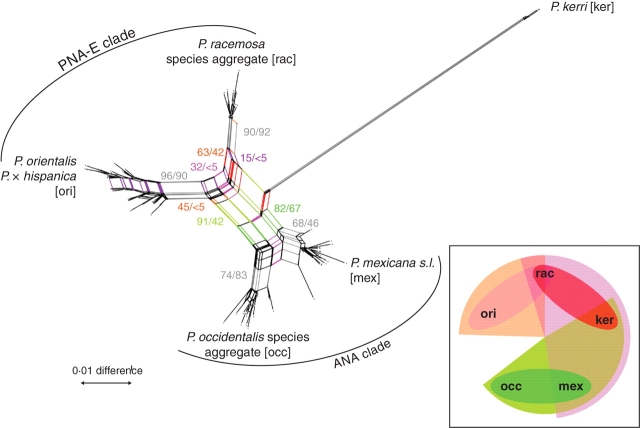
An eastern North American/Central American clade (ANA clade) comprising P. mexicana and P. occidentalis species aggregate. The ANA clade was supported by tree-building methods (Fig. 3; BSML = 67, BSP = 64, PP = 0·98), and well documented in the NN splits graph (Fig. 4) and bipartition networks (not shown).
A Pacific North American/European clade (PNA-E clade) comprising P. orientalis (+ P. × hispanica) and P. racemosa species aggregate. The PNA-E clade received less support by tree-building methods (Fig. 3; BSML = 53, BSP = 56, PP = 0·41) due to an incongruent phylogenetic split between P. racemosa species aggregate + P. kerrii (BSML = 42, BSP = 40, PP = 0·51; red edges in Fig. 4) and the remaining taxa. Strongest evidence for the PNA-E clade came from the NN splits graph (BSNN = 91; Fig. 4) and motif analyses (see below).
The Central American ITS types (cf. Figs 1, 3 and 4), or a consensus of them, possibly retain the basic motives of all extant types (illustrated in the splits graph Fig. 4 and inset in same figure). Bundles of shared edges connected P. kerrii to both P. racemosa species aggregate and P. mexicana, but not to their respective sister groups P. orientalis (plus P. × hispanica) and P. occidentalis species aggregate (Fig. 4). The ‘splits rose’ (inset in Fig. 4) shows the derived position of P. occidentalis s.l. and P. orientalis within the genus. In contrast, P. racemosa species aggregate and P. mexicana form a link between the two major clades, and are assumed to have an ITS type that is closer to a hypothetical ancestor of subgenus Platanus than the ITS of any other modern taxon of Platanus.
Fig. 2.
One of the 234 most-parsimonious trees inferred from the consensus data excluding pseudorecombinants (matrix CM+ps), rooted with P. kerrii. The pseudogenes (white dots) obtained from P. mexicana s.l. and P. occidentalis species aggregate (I, together with the single pseudogene obtained from P. racemosa var. wrightii, II) fall in a clade comprising P. racemosa species aggregate and P. orientalis (III) but not with their non-pseudogenous counterparts (IV). Node support (BSML original data, matrix OM+ps/BSP /PP consensus data, matrix CM+ps) is indicated for selected nodes.
Fig. 3.
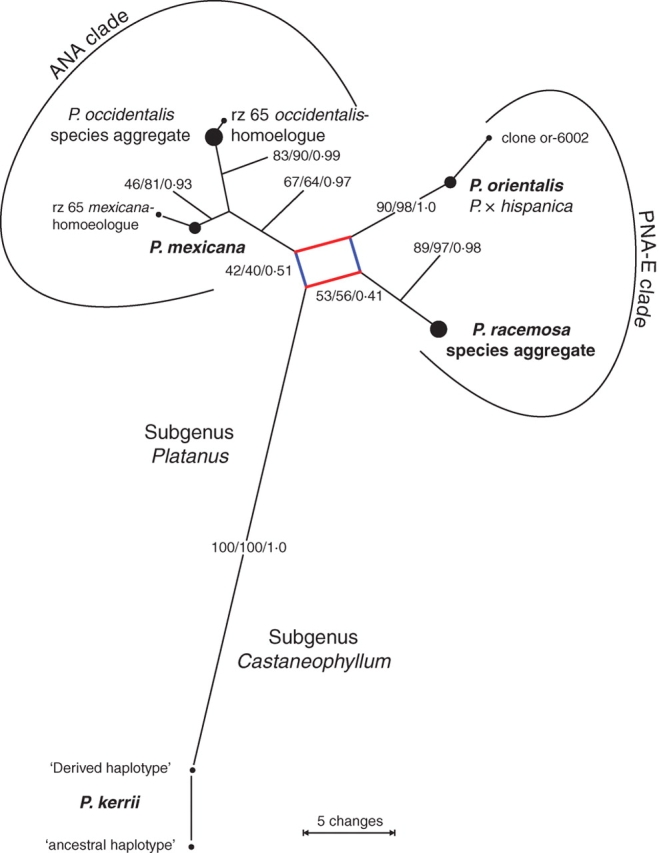
Consensus network of the 156 most-parsimonious trees (MPT) inferred from matrix CMred (consensus data, only non-pseudogenous data; edge lengths equal the according mean branch lengths among all 156 MPT). Node support (BSML-original data, matrix OMred/BSP-/PP-consensus data, matrix CMred) is indicated along edges. Note, that BS and PP equally support two alternative relationships (red and blue edges) considering the position of the P. racemosa species aggregate.
Fig. 4.
NN splits graph based on uncorrected p-distances inferred from the original data; only non-pseudogenous data are included (matrix OMred). Support (BSNN, based on 1000 replicates/BSML of potential nodes in an ML tree) is indicated for the competing central edges (coloured). Inlet, a ‘splits rose’ illustrating the competing relationships. Note the pink semicircle corresponding to the pink-shared edges in the splits graph that interconnects all major groups in Platanus.
Except for a single P. racemosa clone (var. wrightii), ITS pseudogenes were exclusively obtained from members of the ANA clade. Pseudogenes interfered with tree-building methods (Figs 1 and 2 vs. Fig. 3) because pseudogenous signals (Supplementary Information 4) were often incompatible with non-pseudogenous phylogenetic signals. Incompatibility was also encountered with pseudorecombinants as they have chimeric sequences. NN splits graphs were used to further group pseudogenous data in a phylogenetic context. As in the MP analysis (Fig. 2), pseudogenes formed a highly differentiated cluster nested in the PNA-E clade after exclusion of pseudorecombinants from the analysis, supported to some degree by ML bootstrapping (Fig. 5). The topology of the NN remained otherwise unchanged with respect to the placement of non-pseudogenous data (cf. Fig. 4). When including pseudorecombinants (graph not shown) the signal of the ‘typical’ non-pseudogenous ANA clade sequence portions disconnected the pseudogenes from P. orientalis (PNA-E clade): The pseudogenous cluster (as in Fig. 5) remained but was placed between P. racemosa species aggregate (PNA-E clade) and P. mexicana s.l. (ANA clade) instead of being placed next to P. occidentalis species aggregate. The position of pseudogenes in phylograms (Figs 1 and 2) and phylogenetic networks (Fig. 5) indicate that pseudogenes of the ANA clade are more closely related to (ancestors of) the PNA-E clade, which is further supported by detailed sequence inspection (see following section).
Fig. 5.
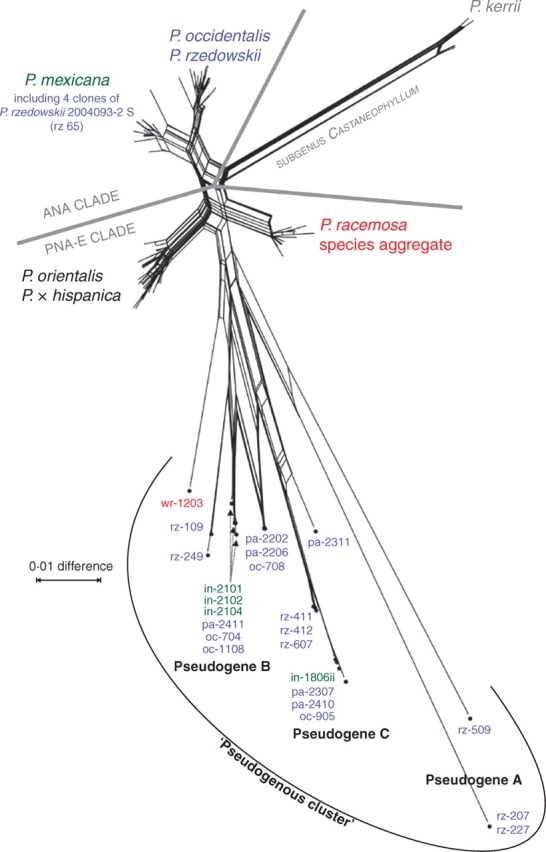
NN splits graph based on uncorrected p-distances inferred from the original data, with pseudogenes included and pseudorecombinants excluded (matrix OM+ps). Pseudogenous lineages obtained from ANA clade individuals (except for clone wr-1203) occur separated from their non-pseudogenous accessions (blue and green fonts) and close to the PNA-E clade taxa P. orientalis and P. racemosa species aggregate. Pseudogenous clones (e.g. wr-1203, rz-109) are coloured according to the source specimen's taxonomic identity (Supplementary Information 1, available online) and are grouped (Pseudogene A, B and C; Fig. 2 and Supplementary Information 4).
Motif analysis
Ten motives (I–X; Fig. 6 and Supplementary Information 3 – only non-pseudogenous data) were defined to summarize phylogenetically informative portions of the complete ITS1 and ITS2. Motif variants of P. kerrii were most distinct from variants of the remaining species (one to five mutations are needed to derive them from P. kerrii). Closely related taxa shared identical motives, which were diagnostic at the species level. Identical motives were further found in members of the same clade (ANA clade vs. PNA-E clade) and accounted for the moderate support of according nodes in phylogenetic trees. In the case of more than one motif type within an individual or species, the according types were highly similar and could be derived from each other by a single mutation (mainly transitions). This points to ongoing molecular differentiation, i.e. gradual genetic evolution and has a direct negative effect on phylogenetic tree building and support. For motives differing among clade members (species, varieties), the following could be observed:
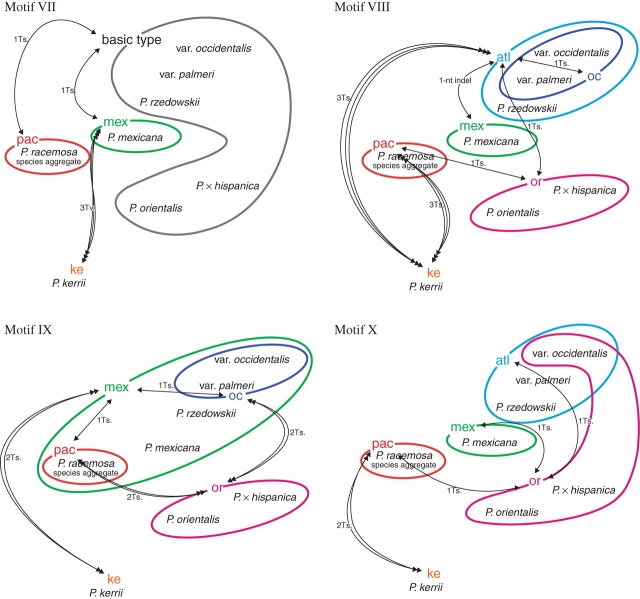
Fig. 6.
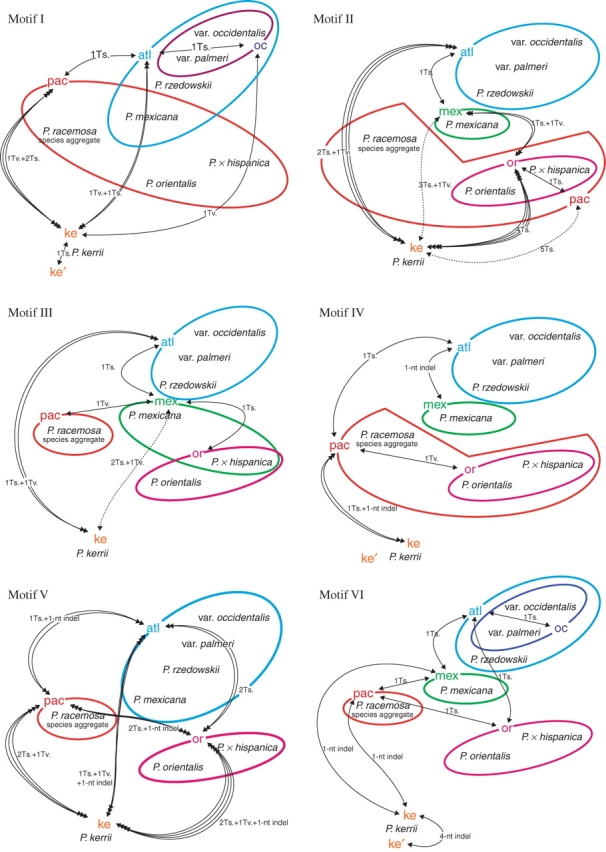
Distribution of ITS motif types (roman numerals, cf. Supplementary Information 3) in Platanus. Detected motif variants (‘atl’, ‘mex’, ‘ke’, ‘oc’, ‘or’ and ‘pac’; Supplementary Information 3) are organized in a framework showing the minimum number of fixed mutations to derive one variant from another. Coloured circles refer to motif types (co-)occurring in modern taxa of Platanus; dotted lines indicate alternative derivation pathways. Abbreviations: Ts., transition; Tv., transversions.
Species of the ANA clade showed a genetic pattern that interconnected them along a south–north gradient (P. mexicana s.l. ↔ P. rzedowskii ↔ P. occidentalis s.l.). The increased intraindividual variability in P. occidentalis s.l. distinguished it from P. rzedwoskii. Motives that were polymorphic in representatives of P. occidentalis always comprised the type also found in P. rzedowskii. In all cases, a single mutation was needed to change the P. occidentalis-unique type into the shared type [labelled as ‘atlantic’ type (atl) in Fig. 6 and Supplementary Information 3]. In the same way, all types detected in P. mexicana differed in only one mutation from the ‘atlantic’ types, and strongly confirmed the ANA clade. General similarity between motif types and the occasional co-occurrence of different motif types within individuals/taxa resulted in low support for the according nodes in phylogenetic trees. Nevertheless, P. mexicana ITS variants could be unambiguously distinguished from P. occidentalis species aggregate ITS variants. The motif types exhibited by the PNA-E clade taxa P. racemosa species aggregate and P. orientalis (including P. × hispanica) were less similar in several cases than those in the ANA clade; derivation involved two or three changes instead of one minimum fixed mutational change (motives III, V and IX vs. II, IV, VI, VII, VIII and X; Fig. 6).
Motif analysis further showed that P. mexicana (in the ANA clade) and P. racemosa species aggregate (in the PNA-E clade) typically share motif elements with the other major clade (Fig. 6) indicating their basal positions. Shared motif types of P. mexicana and the PNA-E clade (motives I, II, III, VI, IX together with P. rzedowskii) form a ‘phylogenetic bridge’ between the ANA and PNA-E clades. In the case of motives IV and VIII, P. racemosa and/or P. rzedowskii link the two clades. This is in accordance with the assumption (see above) that the Central American ITS types (‘pac’ and ‘mex’ in Fig. 6, ‘atl’ to some degree), or a consensus of them, represent the basic motives of all extant types and resemble a hypothetical ITS ancestor (illustrated in the splits graph Fig. 4 and inset in same figure). This ancestral ITS type may identify the root of the subgenus Platanus in a more appropriate way than by rooting the phylograms with the distantly related P. kerrii. Such potentially plesiomorphic motives contain phylogenetic signals that are per se incompatible with signals supporting the ANA and PNA-E clades. Furthermore, the motif analysis revealed general mutational trends (e.g. towards T → C transitions; Fig. 6, positions 99, 165, 523, 567 and 602) occurring independently in the unrelated species P. occidentalis and P. orientalis or P. kerrii, and in some pseudogenes (Supplementary Information 4 and Table 3). The resultant incompatible splits hampered phylogenetic tree reconstructions and accounted for lower node support (cf. Figs 2 and 3).
Table 3.
Summary of sequence characters of pseudogenes (Supplementary information 3)
| 3′ 18S rDNA | ITS1 | 5·8S | ITS2 | 5′ 25S rDNA | In total | |
|---|---|---|---|---|---|---|
| Total number of mutations linked to pseudogenization | 4 | 55 | 28 | 38 | 4 | – |
| Consensual C → T | 2 | 19 | 13 | 15 | 1 | 50 |
| Consensual G → A | 0 | 17 | 8 | 3 | 1 | 29 |
| Consensual G → A at CG site (potentially affected by C methylation*) | 2 | 13 | 6 | 13 | 2 | 36 |
| Consensual G → T, C → A | 0 | 6 | 1 | 4 | 0 | 11 |
| Deletion/sequence modified in comparison to the genus' consensus | 0 | 0 | 0 | 3 | 0 | 3 |
| Total number of sequence characters that could indicate ancesty | 0 | 13 | 1 | 11 | 0 | – |
| Same character state as otherwise typical (autapomorphic) for sequences of P. kerrii | 0 | 4 | 0 | 4 | 0 | 8 |
| Same (potentially plesiomorphic) state as found in ‘pac’ motives | 0 | 1 | 0 | 2 | 0 | 3 |
| Same state as in some P. gentryi clones | 0 | 0 | 0 | 1 | 0 | 1 |
| Same (symplesiomorphic) state shared by P. kerrii and ‘pac’ motives | 0 | 4 | 1 | 0 | 0 | 5 |
| Same (symplesiomorphic) state shared by ‘pac’ and ‘mex’ motives | 0 | 3 | 0 | 0 | 0 | 3 |
| Same state as in P. orientalis | 0 | 0 | 0 | 1 | 0 | 1 |
| Reflecting a consensual state, not the according taxon's typical (autapomorphic) state | 0 | 1 | 0 | 3 | 0 | 4 |
| Total number of unique (‘lost’ lineage) mutations | 0 | 5 | 1 | 3 | 0 | |
| G → C | 0 | 2 | 0 | 1 | 0 | 3 |
| T → C | 0 | 1 | 1 | 1 | 0 | 3 |
| Other sequence modifications (including 1nt-long length polymorphism) | 0 | 2 | 0 | 1 | 0 | 3 |
Pseudogenes and pseudogenous portions of pseudorecombinants are compared to consensus states for the genus and to states in non-pseudogenous sequences.
* See, for example, King et al. (1993) and Keller et al. (2007).
Identification of ancestral states is important to evaluate the evolutionary significance of ITS pseudogenes and to discuss their placement in phylogenetic reconstructions (Figs 1, 2 and 5). ITS pseudogeny in Platanus stemmed from two major mutational trends (Table 3; for details see Supplementary Information 3 and 4): (1) the replacement of the genus' consensual ‘G's or ‘C's by ‘A's and ‘T's (standard pattern of pseudogenization; cf. Mayol and Rosselló, 2001; Bailey et al., 2003; Manen, 2004) reflected by long edges in phylogenetic reconstructions; and (2) a retention of putatively ancestral nucleotides and according sequence motives. The latter is highly characteristic: ITS pseudogenes lacked diagnostic sequence motives of either the P. occidentalis species aggregate or the entire ANA clade (including P. mexicana). Instead, pseudogenous clones exhibited motif variants also found in P. kerrii (subgenus Castaneophyllum) and/or members of the PNA-E clade, or shared by P. racemosa species aggregate and P. mexicana. In pseudogenous sequences (sequence portions), small indels and several G → C or T → C point mutations were found (Table 3). These mutations counteract the general increase of ‘A's and ‘T's in the course of pseudogenization. Moreover, many mutations did not occur randomly, but were confined to specific sites (Supplementary Information 4). Geographically isolated specimens representing morphologically distinct taxa (Figs 1, 2 and 5) showed very similar or identical ITS pseudogenes (including pseudogenous sequence portions of pseudorecombinants; Supplementary Information 4). Pseudogene A (Figs 2 and 5) was obtained from two specimens (P. mexicana var. mexicana 2004011-3 S; P. rzedowskii 2004095-1 S; pseudogenous parts of clones li-3108/-3109; clones rz-207/-227), pseudogenes B and C (Figs 2 and 5) from P. mexicana var. interior (B, 2004086-1 S; C, 2004072-1 S), P. rzedowskii (C, 2004088-4 S and 2004092-4 S), and both variants of P. occidentalis s.l. (var. palmeri, B and C, from four out of five specimens analysed; var. occidentalis, B and C, three of five). Three clones of P. occidentalis s.l. appeared to be a recombinant or intermediate of pseudogenes B and C (oc-708, pa-2202, pa-2206). In addition to the main pseudogenes A, B and C shared by several specimens and taxa, other pseudogenous variants were obtained from P. rzedowskii: Clones rz-109 (2004097–6 S) and rz-245 (2004095-1 S), while almost identical to each other, displayed partly (predominantly within ITS2) the same pseudogenous mutations as in pseudogene C; clone rz-509 (2004089–1 S) was unique (Figs 2 and 5 and Supplementary Information 4). The only PNA-E pseudogenous clone obtained (P. wrightii 2003029 S; one out of three clones sequenced) was closer to pseudogene C than to any other lineage (Fig. 5 and Supplementary Information 3), but exhibited a number of unique mutations not found in ANA clade ‘pseudogenes’ (Supplementary Information 4).
DISCUSSION
Phylogenetic structure and taxonomy of Platanus
Two distinct species groups within the subgenus Platanus (an eastern North American and a Pacific North American–European group) and the isolated position of P. kerrii have already previously been suggested (Hsiao, 1973). The present and a number of other recent accounts (Nixon and Poole, 2003; Feng et al., 2005) recognized the same pattern. As far as they overlap in taxon sampling, the present results agree with the three-gene analysis by Feng et al. (2005), which recognized the same relationships as we do based on ITS data only. To accommodate possible past reticulation (Nixon and Poole, 2003), use was made of the multicopy biparentally inherited ITS. Indeed, incomplete concerted evolution and lateral gene flow in Platanus are reflected in the intraindividual ITS variability observed, and in particular, in the presence of ITS pseudogenes and homoeologues within the ANA clade.
The present results show that in extant Platanus ancestral, modern (rate of new mutations is higher than level of homogenization; Baldwin et al., 1995), and introgressive ITS variants (hybridization, lateral gene flow, polyploidization; e.g. Bailey et al., 2003; Draper et al., 2007) co-occur. An ancestral ITS variant and its derivate(s) are homologous and have a common origin, while introgressive ITS variants are homoeologous to the according domestic ITS variants. The co-occurrence of ancestral and derived variants induces incompatible split patterns and results in low node support (Grimm et al., 2006, 2007; this study). Low node support has traditionally been considered as ‘no resolution’ or even ‘polyphyly’ as opposed to a common origin. Using phylogenetic networks and motif analysis (Figs 3–6), low node support can be understood in an evolutionary context: For example, the low node support for the PNA-E clade (see Phylogenetic analysis) can be ascribed to a single alternative phylogenetic split grouping the P. racemosa species aggregate with the outgroup (P. kerrii), and hence separating it from P. orientalis (Figs 3 and 4). Plesiomorphic character states in ITS sequences of the P. racemosa species aggregate, which are absent in P. orientalis, account for this signal (Fig. 6; see ‘Motif analysis’).
About 50 clones of the three western North American taxa, P. racemosa var. racemosa, P. racemosa var. wrightii and P. gentryi, did not reveal intertaxonomic diagnostic sequence motives, which calls into question the recognition of P. gentryi as a distinct species (Nixon and Poole, 2003). Furthermore, overlapping patterns of nucleotide variability found in P. occidentalis species aggregate could reflect ongoing gene flow between P. rzedowskii and P. occidentalis. Additional morphological and molecular analyses (including data from other biparentally inherited markers) will clarify the final taxonomic status of the taxa accommodated into species aggregates in the present study.
Recent hybrids
Platanus × hispanica clearly falls within the ITS variability of the western Eurasian P. orientalis. This is in accordance with Hsiao and Li (1975) using phenol composition and Besnard et al. (2002) using RAPD fragments and (PCR-)RFLP analyses of 5S-rDNA intergenic spacers, plastid and mitochondrial DNA to explore the hybrid status of P. × hispanica. Besnard et al. (2002) ascribed the more important contribution of the P. orientalis genome to the hybrid to more frequent backcrosses by P. orientalis than by P. occidentalis in Europe. The elimination (complete loss) of one parental 35S rDNA array (no P. occidentalis homoeologues detected) has been shown in the case of the allopolyploid tetraploid Nicotiana tabacum (Volkov et al., 1999; Lim et al., 2000) and other plant models (reviewed in Pikaard, 2001).
In the study by Feng et al. (2005), a single sample of P. rzedowskii grouped with P. occidentalis var. occidentalis in the ITS tree, but was placed with accessions of P. mexicana var. mexicana in the LEAFY tree. Based on the present results the following statements can be made. (a) Most of the 29 clones of P. rzedowskii grouped with P. occidentalis (both varieties) in the ML tree and splits graphs. No geographic pattern was seen in the sequence data between and within these species except that P. occidentalis s.l. exhibited increased intraindividual ITS variability. Sequences of P. mexicana var. interior and var. mexicana (49 clones) showed highly overlapping variability that is diagnostic at the species level. (b) Motif analysis showed that, although P. rzedowskii shares most motif variants with P. occidentalis, some sequence patterns link it more closely to P. mexicana. All motives of the ANA clade can be derived from P. mexicana variants (they differ in 0 to 1 fixed mutations, mostly T ↔ C transitions, two transitions in motif X). (c) A single individual from Nuevo Leon yielded P. occidentalis s.l. and P. mexicana s.l. homoeologues that grouped accordingly in the phylogenetic trees and splits graphs. (d) Pseudogenes restricted to P. rzedowskii (i.e. not found in P. occidentalis s.l.) grouped with pseudogenous clones of P. mexicana and the PNA-E clade (e.g. pseudogene A; Figs 1 and 5). Overall, this suggests that ancestors of the ANA clade were more similar to P. mexicana than to P. occidentalis. Platanus rzedowskii and P. occidentalis appear to be derived from P. mexicana with P. rzedowskii connecting eastern North American populations to their Central American ancestors. Platanus mexicana and P. occidentalis homoeologues in a single individual of P. rzedowskii suggest that hybridization and interbreeding played a role in the formation of the modern species P. rzedowskii that clearly has evolved more closely towards P. occidentalis in more recent times [cf. Besnard et al. (2007) for Olea, and Besnard et al. (2002) for a similar scenario for P. × hispanica]. Gene flow from P. rzedowskii to P. mexicana could account for the placement of according accessions in the LEAFY tree by Feng et al. (2005, Fig. 3C vs. Fig. 3A and B).
Pseudogenes and ancient hybridization
In addition to ITS variants that represent potentially functional 35S rDNA, the ITS pseudogene lineages in the ANA clade constitute sets of non-functional 35S rDNA arrays. Simultaneous phylogenetic analyses of potentially functional and pseudogenous variants have a potential for evolutionary considerations that goes far beyond the resolution of standard phylogenetic methods and multigene analyses. However, the largely incompatible signal from possibly artificial (PCR-mediated) pseudorecombinants (Cronn et al., 2002; Grimm et al., 2007; this study) and pseudogenes hinders phylogenetic tree-building and can induce potential long-branching artefacts (LBA; Sanderson et al., 2000; but see Razafimandimbison et al., 2004). In Platanus, the negative affect of ITS pseudogenes and pseudorecombinants on tree-building phylogenetic methods and node support is due to three general phenomena. (1) Pseudorecombinants (Fig. 1) combine pseudogenous and non-pseudogenous sequence portions. In the non-pseudogenous portions, diagnostic sequence features of the respective taxa (ANA clade) were encountered. (2) ITS pseudogenes of P. rzedowskii and P. occidentalis s.l. are more similar (pairwise genetic distances; Supplementary Information 2; cf. Figs 2 and 5) to non-pseudogenous P. mexicana s.l., P. racemosa species aggregate and P. kerrii (Table 2 and Supplementary Information 4) than to non-pseudogenous sequences of P. rzedowskii and P. occidentalis s.l. (3) Pseudogenous mutations exceed non-pseudogenous mutations (cf. branch lengths; Figs 1 and 2 and Table 3).
ANA group pseudogenes form a sister clade to P. orientalis (Fig. 2) in phylogenetic trees independent of the optimality criterion used. Low node support does not contradict a possible common origin of the pseudogene clade and the non-pseudogenous ITS variants of P. orientalis. However, ITS sequences of P. orientalis can be considered to be ‘derived’ (Table 2 and Figs 4 and 6) and the position of pseudogenes in optimal tree-like reconstructions could be due to LBA. Considering all the taxa analysed, ITS sequences of P. kerrii are most distinct (see above). If the position of pseudogenes is highly affected by LBA, one could expect that the pseudogene cluster would fall between P. kerrii and P. orientalis in the NN splits graph, which is not the case (Fig. 5). Instead, the pseudogenes are placed between the ‘primitive’ P. racemosa species aggregate and the more derived P. orientalis (see ‘Motif’ analysis). This further supports the hypothesis that ITS pseudogenes (mainly from the ANA clade) are most closely related to non-pseudogenous ITS variants of the PNA-E clade. Accordingly, non-pseudogenous and pseudogenous 35S rDNA arrays in the ANA clade may have an independent evolutionary origin (see Fig. 7).
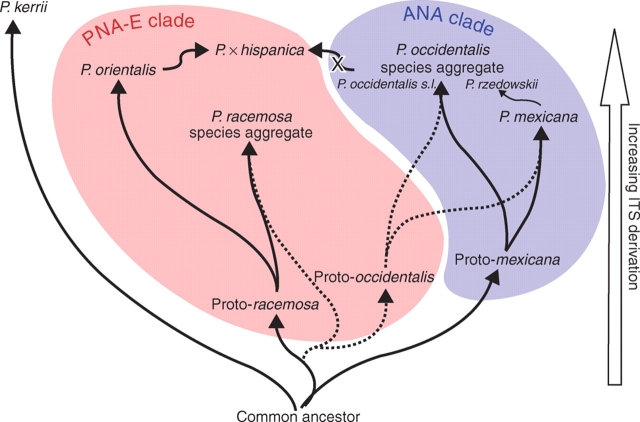
Fig. 7.
ITS evolution mirroring ancient reticulation processes leading to modern taxa of Platanus. Solid arrows reflect pathways based on non-pseudogenous data, dotted arrows refer to pseudogenous data. The P. occidentalis parental lineage (grey arrow) has been eliminated in P. × hispanica. The thin black arrow indicates recent introgression from P. mexicana into the P. occidentalis species aggregate via P. rzedowskii.
The most prominent evolutionary pattern revealed by ITS pseudogenes is the retention of ancestral sequence characteristics (Table 2 and Supplementary Information 4). Pseudogene lineages share significantly more elements with non-pseudogenous lineages of P. mexicana (basal in the ANA clade), P. racemosa species aggregate (basal in the PNA-E clade) and P. kerrii, than with their non-pseudogenous counterparts of the ANA clade (Tables 3 and 4 and Supplementary Information 3 and 4). The single pseudogenous accession obtained from the PNA-E clade also showed the general plesiomorphic pattern, placing this sequence further apart from its non-pseudogenous counterparts and closer to the ANA clade pseudogenes (clone wr 1203 in Figs 1, 2 and 5). In conclusion, ITS pseudogenes point towards ancient hybridization and/or introgression among lineages ancestral to the major modern clades in subgenus Platanus (Fig. 7 and Table 4).
Table 4.
Length, consistency and retention index (under parsimony), and likelihood of constrained topologies
| Length*,† | CI*,† | RI*,† | ln LhLRT*,‡ | ln Lmean Bayes*,§ | |
|---|---|---|---|---|---|
| Under no constraint | 385 | 0·81 | 0·8 | −1992·59 | −2017·67 |
| Pseudogenes originated outside of modern (sub)genus Platanus | 400 | 0·78 | 0·76 | −2022·64 | −2056·68 |
| Pseudogenes originated from a lineage ancestral to PNA-E clade | 399 | 0·78 | 0·76 | −2021·88 | −2054·92 |
| Pseudogenes originated from a lineage ancestral to P. racemosa | 399 | 0·78 | 0·76 | −2021·88 | −2054·92 |
| Pseudogenes A and B + C originated from two different lineages | 405 | 0·77 | 0·75 | −2023·14 | −2057·94 |
| Pseudogenes originated from a lineage ancestral to P. mexicana | 410 | 0·76 | 0·74 | −2038·59 | −2075·68 |
| Reticulation limited to the ANA clade; pseudogenes originated from lineage(s) ancestral to P. occidentalis species aggregate | 417 | 0·75 | 0·72 | −2067·05 | −2108·99 |
| No reticulation: pseudogenes are non-functional substitutes | 500 | 0·62 | 0·49 | −2359·95 | −2453·24 |
The basic topology is defined by the results from phylogenetic analyses of non-pseudogenous data (see text); the position of accessions representing pseudogenes A, B and C has been constrained.
* Using a NJ inferred (constrained) topology based on uncorrected p-distances as basis for calculation.
† Total number of steps (length), consistency (CI) and retention index (RI) under parsimony.
‡ Model parameters set according to the results of the hierarchial LRT and AIC (MrModeltest; matrix OMred).
§ Model parameters set according to the mean values inferred from the not-discarded BIST (MrBayes; matrix CMred).
In this context, several possible reticulation scenarios need to be discussed. The pseudogenes of the modern ANA clade taxa may have originated from hybridization of an extinct lineage related to the (modern) PNA-E taxa (scenario 1) with an ANA clade ancestor, or with the ancestor of the P. racemosa species aggregate (clone wr-1203; Supplementary Information 4). Alternatively (scenario 2), hybridization and introgression could have occurred only among ancestors of the ANA clade and P. racemosa species aggregate. That pseudogenous data were not obtained from most individuals of the PNA-E clade (support for scenario 1) does not necessarily indicate that degraded and originally homoeologous 35 rDNA arrays were never present in the PNA-E clade. Such arrays could have been largely lost in modern PNA-E taxa as concerted evolution among closely related but homoeologous nrDNA arrays may act rapidly (e.g. Arnheim et al., 1980; Grimm et al., 2006). Also, the degree of pseudogeny probably is connected to differential transcription and intragenomic competition that have been documented in artificial polyploids containing several homoeologous but potentially functional rDNA arrays (e.g. Chen and Pikaard, 1997; Komarova et al., 2004). Therefore, scenario 2 is possible as well, although not represented in ITS data: As in the natural allopolyploid Nicotiana tabacum (Volkov et al., 1999; Lim et al., 2000), it could be possible that highly degraded 35S rDNA remains of an ANA clade ancestor are still part of the nuclear genome of modern PNA-E clade taxa, but have not been amplified. The three pseudorecombinants of two P. mexicana var. mexicana individuals, from which no ITS pseudogenes were obtained, demonstrate potential data bias. A selected amplification and mass detection of specific pseudogenes (via in situ hybridization, PCR-RFLP, etc.) is difficult due to their sequence heterogeneity, the generally high amount of 35S rDNA repeats (Hemleben et al., 1988), and the general set-up of such experiments (determination of ploidy at the population level, characterization of NORs, cultivation, etc.).
Under a third scenario, abundant hybridization and introgression that was not linked to the evolution of P. racemosa species aggregate and P. orientalis could have disguised the systematic relationships of modern taxa in the ANA clade (Fig. 7). The pseudogene lineage would then represent the original 35S rDNA arrays of a central North American species (proto-occidentalis) that underwent intensive hybridization with the ancestor of the modern P. mexicana and introgression of ANA clade ITS variants. The original rDNA arrays (of PNA-E origin) started to degrade, whereas the introgressive rDNA arrays (of ANA origin) retained functionality. This would explain why the pseudogenes obtained are still amplifiable using a proofreading polymerase (Vent®) and recognizable as distinct ITS variants (pseudogenes A to C). One could expect that the original rDNA arrays occur(red) in great numbers and continued to be repaired for some time after hybridization. The few pseudogenes and pseudorecombinants in P. mexicana and the potentially permeable species boundaries in the ANA clade (see ‘Recent hybrids’; Fig. 6) are in agreement with the assumption of former and ongoing bidirectional reticulation among Central and North American Platanus, and with evidence from additional nuclear markers (5S intergenic spacer, second intron of LEAFY gene; data will be published elsewhere).
Concluding remarks
A major problem with phylogenetic studies at the genus level is that species in woody angiosperms are often closely related. In view of the high potential for gene flow in Platanus, the boundaries of modern species may be young or permeable. The gene pools of extinct species and lineages were probably affected in a similar way. This is expressed in generally low sequence differentiation among ‘sister’ species of Platanus, partly high intraspecific divergence, and an ITS signal that is to a certain degree incompatible with dichotomizing phylogenetic reconstructions (trees). Network methods provide an effective means of representing such situations (cf. similar results in Hörandl et al., 2005; Winkworth et al., 2005; Whitfield and Lockhart, 2007).
Low genetic differentiation among ancestors of modern species as well as different levels of derivedness can result in weak clade support as has been found for basal relationships in Acer (Grimm et al., 2006; Renner et al., 2007). Evidence from ITS data accumulating over the last years has shown that significant intraspecific divergence is the rule rather than the exception in plants (independent of the ploidy level; Grimm et al., 2006), no matter whether linked to pseudogeny or not (Buckler et al., 1997; Álvarez and Wendel, 2003, among many others). Consequently, similar conditions have to be assumed for extinct or ancestral lineages [e.g. Denk et al. (2005) for Fagus]. ITS pseudogenes and homoeologues (this study) as well as incongruent topologies inferred from plastid and nuclear data (Feng et al., 2005) reflect hybridization and lateral gene flow in Platanus. Such conditions are likely to invoke incoherent phylogenetic models when using bifurcating trees (Lockhart and Cameron, 2001). The resultant patterns of intrageneric evolution and sequence divergence are complex. This is not surprising in view of the fossil history of the genus and family that may have undergone repeated radiations and extinction during the Late Cretaceous and (Early) Tertiary (Friis et al., 1988; Kvaček et al., 2001; Kvaček and Manchester, 2004) involving the extinction of one subgenus (Kvaček et al., 2001). In Platanus, this history is mirrored by the ITS variability encountered in modern species.
Incomplete homogenization among nrDNA arrays and their biparental heritage make the spacers of the multicopy nrDNA, such as ITS1 and ITS2, a powerful tool to trace the formation of species within a temporal and spatial framework. Multigene analyses including low-copy nuclear markers and (uniparentally inherited) plastid sequences may be hampered by generally lower divergence and the loss of reticulate signals and homoeologues (e.g. Razafimandimbison et al., 2004; Feng et al., 2005; Draper et al., 2007 for mosses; but see Hidalgo et al., 2004). In contrast, the comparably high fixation rates in ITS and the possibility of detecting homoeologues and pseudogenes in allopolyploids and (ancient) hybrids provide the basis for analysing a key element of evolution, the formation of new species. The underlying mechanisms often are inconsistent with fundamental requirements of cladistic analyses, such as discrete and discontinuous biological species as terminals of a tree and the assumption that evolution follows a strictly dichotomous pattern, resulting in fully compatible data. This drawback can be overcome by using phylogenetic networks and sequence motif analysis.
SUPPLEMENTARY INFORMATION
Supplementary information is available online at http://aob.oxfordjournals.org/ and consists of four files as follows.
Supplementary Information 1. Voucher information and gene bank accession numbers.
Supplementary Information 2. Pairwise genetic distances between clones of Platanus calculated for completely sequenced clones based on the optimal substitution model HKY + Γ (DT-ModSel) with parameters set according to AIC (MrModeltest).
Supplementary Information 3. Compilation of phylogenetically informative sites and regions in the ITS summarized in ten sequence motives (cf. Fig. 4). Pseudogenous data and site variabilities restricted to a single taxon (e.g. site variability diagnostic for P. orientalis) are not included.
Supplementary Information 4. Compilation of pseudogenous data; the list includes all sites where (partially) pseudogenous ITS accessions differ from their non-pseudogenous counterparts obtained from the same individuals and taxa.
ACKNOWLEDGEMENTS
Anja Fichtner and Gerwin Gruber accompanied T.D. on his first American collecting trip in 2003. Steven Manchester and Herb Meyer provided general information on Platanus in northern North America. Susana Magallón, Gerardo Salazar and Ruperto Velazquez facilitated the work at MEXU and fieldwork of T.D. in Mexico in 2004. Evangelos Velitzelos collected plant material from Rhodes and Lesbos. Alexandros Stamatakis performed the RAxML analyses on the CIPRES cluster. Technical assistance of Karin Stögerer is gratefully acknowledged. The Swedish Research Council funded fieldwork of T.D. in USA and Mexico. G.W.G. and K. Stögerer were funded by the German Science Foundation grant Gr3124/1-1,2 to G.W.G.
This paper is dedicated to Anja Fichtner.
LITERATURE CITED
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proceedings of the National Academy of Sciences of the USA. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CD, Carr TG, Harris SA, Hughes CE. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution. 2003;29:435–455. doi: 10.1016/j.ympev.2003.08.021. [DOI] [PubMed] [Google Scholar]
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence of angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- Barker NP, Weston PH, Rourke JP, Reeves G. The relationships of the southern African Proteaceae as elucidated by internal transcribed spacer (ITS) DNA sequence data. Kew Bulletin. 2002;57:867–883. [Google Scholar]
- Besnard G, Tagmount A, Baradat P, Vigouroux A, Bervillé A. Molecular approach of genetic affinity between wild and ornamental Platanus. Euphytica. 2002;126:401–412. [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. Plastid and nuclear DNA polymorphism reveals historical processes of isolation and reticulation in the olive tree complex (Olea europaea) Journal of Biogeography. 2007;34:736–752. [Google Scholar]
- Bryant D, Moulton V. NeighborNet: an agglomerative method for the construction of planar phylogenetic networks. In: Guigó R, Gusfield D, editors. Algorithms in bioinformatics, 2nd International Workshop, WABI. Berlin: Springer Verlag; 2002. pp. 375–391. Lecture Notes in Computer Science 2452. Rome, Italy. [Google Scholar]
- Bryant D, Moulton V. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Buckler ES, IV, Ippolito A, Holtsford TP. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145:821–832. doi: 10.1093/genetics/145.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, et al. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. 15 (Erratum) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proceedings of the National Academy of Sciences of the USA; 1997. pp. 3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PR, Pedersen KR, Friis EM, Drinnan AN. Early Cretaceous (Early to Middle Albian) platanoid inflorescences associated with Sapindopsis leaves from the Potomac Group of Eastern North America. Systematic Botany. 1993;18:328–342. [Google Scholar]
- Cronn P, Cedroni M, Haselkorn T, Grover C, Wendel JF. PCR-mediated recombination in amplification products dervied from polyploid cotton. Theoretical and Applied Genetics. 2002;104:482–489. doi: 10.1007/s001220100741. [DOI] [PubMed] [Google Scholar]
- Cronquist A. An integrated system of classification of flowering plants. New York, NY: Columbia Press; 1981. [Google Scholar]
- Denduangboripant J, Cronk QCB. Evolution and alignment of the hypervariable arm 1 of Aeschynanthus (Gesneriaceae) ITS 2 nuclear ribosomal DNA. Molecular Phylogenetics and Evolution. 2001;20:163–172. doi: 10.1006/mpev.2001.0968. [DOI] [PubMed] [Google Scholar]
- Denk T, Grimm GW. Phylogeny and biogeography of Zelkova (Ulmaceae sensu stricto) as inferred from leaf morphology, ITS sequence data and the fossil record. Botanical Journal of the Linnéan Society. 2005;147:129–157. [Google Scholar]
- Denk T, Tekleva M. Comparative pollen morphology and ultrastructure of Platanus. Implications for phylogeny and evaluation of the fossil record. Grana. 2006;45:195–221. [Google Scholar]
- Denk T, Grimm G, Stögerer K, Langer M, Hemleben V. The evolutionary history of Fagus in western Eurasia: evidence from genes, morphology and the fossil record. Plant Systematics and Evolution. 2002;232:213–236. [Google Scholar]
- Denk T, Grimm GW, Hemleben V. Patterns of molecular and morphological differentiation in Fagus: implications for phylogeny. American Journal of Botany. 2005;92:1006–1016. doi: 10.3732/ajb.92.6.1006. [DOI] [PubMed] [Google Scholar]
- Doyle JA, Endress PK. Morphological phylogenetic analysis of basal angiosperms: comparison and combination with molecular data. International Journal for Plant Science. 2000;161:S121–S153. [Google Scholar]
- Draper I, Hedenäs L, Grimm GW. Molecular and morphological incongruence in European species of Isothecium (Bryophyta) Molecular Phylogenetics and Evolution. 2007;42:700–716. doi: 10.1016/j.ympev.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Drinnan I, Crane PR, Hoot SB. Patterns of floral evolution in the early diversification of non-magnoliid dicotyledons (eudicots) Plant Systematics and Evolution. 1994;8:93–122. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Oh S-H, Manos PS. Phylogeny and historical biogeography of the genus Platanus as inferred from nuclear and chloroplast DNA. Systematic Botany. 2005;30:786–799. [Google Scholar]
- Friis EM, Crane PR, Pedersen KR. Reproductive structures of Cretaceous Platanaceae. Biologiske Skrifter. 1988;31:1–55. [Google Scholar]
- Gebhardt C, Ritter E, Debener T, Schachtschabel U, Walkemeier B, Uhrig H, et al. RFLP analysis and linkage mapping in Solanum tuberosum. Theoretical and Applied Genetics. 1989;78:65–75. doi: 10.1007/BF00299755. [DOI] [PubMed] [Google Scholar]
- Grebenstein B, Röser M, Sauer W, Hemleben V. Molecular pyhlogenetic relationships in Aveneae (Poaceae) species and other grasses as inferred from ITS1 and ITS2 sequences. Plant Systematics and Evolution. 1998;213:233–250. [Google Scholar]
- Grimm GW. Tracing the mode and speed of molecular evolution – a case study of genus Acer L. and Fagus L. Tübingen: Eberhard-Karls University; 2003. D.Sc. Thesis (urn:nbn:de:bsz:21-opus-15744) [Google Scholar]
- Grimm GW, Renner SS, Stamatakis A, Hemleben V. A nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analyses of 606 sequences. Evolutionary Bioinformatics. 2006;2:279–294. [PMC free article] [PubMed] [Google Scholar]
- Grimm GW, Denk T, Hemleben V. The evolutionary history and systematics of Acer section Acer – a case study of low-level phylogenetics. Plant Systematics and Evolution. 2007;267:215–253. [Google Scholar]
- Hemleben V, Ganal M, Gerstner J, Schiebel K, Torres RA. Organization and length heterogeneity of plant ribosomal RNA genes. In: Kahl G eds. Architecture of eukaryotic genes; Weinheim: VCH Verlagsgesellschaft mbH; 1988. pp. 371–383. [Google Scholar]
- Hershkovitz MA, Lewis LA. Deep-level diagnostic value of the rDNA-ITS region. Molecular Biology and Evolution. 1996;13:1276–1295. doi: 10.1093/oxfordjournals.molbev.a025693. [DOI] [PubMed] [Google Scholar]
- Hershkovitz MA, Zimmer EA. Conservation patterns in angiosperm rDNA ITS2 sequences. Nucleic Acids Research. 1996;24:2857–2867. doi: 10.1093/nar/24.15.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo O, Garnatje T, Susanna A, Mathez J. Phylogeny of Valerianaceae based on matK and ITS markers, with reference to matK individual polymorphism. Annals of Botany. 2004;93:283–293. doi: 10.1093/aob/mch042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland B, Moulton V. Consensus networks: a method for visualising incompatibilities in collections of trees. In: Benson G, Page R, editors. Algorithms in bioinformatics: 3rd International Workshop, WABI, Budapest, Hungary. Proceedings. Berlin: Springer Verlag; 2003. pp. 165–176. Lecture Notes in Bioinformatics (LNBI) 2812. [Google Scholar]
- Hörandl E, Paun O, Johansson JT, Lehnebach C, Armstrong T, Lockhart P. Phylogenetic relationships and evolutionary traits in Ranunculus s. l. (Ranunculaceae) inferred from ITS sequence analysis. Molecular Phylogenetics and Evolution. 2005;36:305–327. doi: 10.1016/j.ympev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Hsiao J-Y. A numerical taxonomic study of the genus Platanus based on morphological and phenolic characters. American Journal of Botany. 1973;60:678–684. [Google Scholar]
- Hsiao J-Y, Li H-L. A study of the leaf chromatograms of the London plane and its putative parent. American Midland Naturalist. 1975;93:234–239. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Evolution – Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jobst J, King K, Hemleben V. Molecular evolution of the internal transcribed spacers (ITS1 and ITS2) and phylogenetic relationships among species of Cucurbitaceae. Molecular Phylogenetics and Evolution. 1998;9:204–219. doi: 10.1006/mpev.1997.0465. [DOI] [PubMed] [Google Scholar]
- Keller I, Bensasson D, Nichols RA. Transition-transversion bias is not universal: A counter example from grasshopper pseudogenes. PloS Genetics. 2007;3:e22. doi: 10.1371/journal.pgen.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K, Torres RA, Zentgraf U, Hemleben V. Molecular evolution of the intergenic spacer in the nuclear ribosomal-RNA genes of Cucurbitaceae. Journal of Molecular Evolution. 1993;36:144–152. doi: 10.1007/BF00166250. [DOI] [PubMed] [Google Scholar]
- Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA. Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Molecular Biology. 2004;56:439–463. doi: 10.1007/s11103-004-4678-x. [DOI] [PubMed] [Google Scholar]
- Kvaček Z, Manchester SR. Vegetative and reproductive structure of the extinct Platanus neptunii from the Tertiary of Europe and relationships within the Platanaceae. Plant Systematics and Evolution. 2004;244:1–29. [Google Scholar]
- Kvaček Z, Manchester SR, Guo Z-H. Trifoliolate leaves of Platanus bella (Heer) comb. n. from the Palaeocene of North America, Greenland, and Asia and their relationships among extinct and extant Platanaceae. International Journal for Plant Science. 2001;162:441–458. [Google Scholar]
- Leitch I, Bennett M. Polyploidy in angiosperms. Trends in Plant Science. 1997;2:470–476. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Bezdek M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;109:161–172. doi: 10.1007/s004120050424. [DOI] [PubMed] [Google Scholar]
- Lockhart PJ, Cameron SA. Trees for bees. Trends in Ecology and Evolution. 2001;16:84–88. doi: 10.1016/s0169-5347(00)02054-1. [DOI] [PubMed] [Google Scholar]
- Manen J-F. Are both sympatric species Ilex perado and Ilex canariensis secretly hybridizing? Indication from nuclear markers collected in Tenerife. BMC Evolutionary Biology. 2004;4 doi: 10.1186/1471-2148-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslova NP. Extinct and extant Platanaceae and Hamamelidaceae: morphology, systematics, and phylogeny. Paleontological Journal. 2003;37(Suppl.):S467–S590. [Google Scholar]
- Mayol M, Rosselló JA. Why nuclear ribosomal DNA spacers (ITS) tell different stories in Quercus. Molecular Phylogenetics and Evolution. 2001;19:167–176. doi: 10.1006/mpev.2001.0934. [DOI] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. Performance-based selection of likelihood models for phylogeny estimation. Systematic Biology. 2003;52:674–683. doi: 10.1080/10635150390235494. [DOI] [PubMed] [Google Scholar]
- Müller KF. The efficiency of different search strategies for estimating parsimony, jackknife, bootstrap, and Bremer support. BMC Evolutionary Biology. 2005;5:58. doi: 10.1186/1471-2148-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon KC, Poole JM. Revision of the Mexican and Guatemalan species of Platanus (Platanaceae) Lundellia. 2003;6:103–137. [Google Scholar]
- Nylander JAA. MrModeltest. Evolutionary Biology Centre, Uppsala University; 2004. Uppsala, Program distributed by the author. [Google Scholar]
- Pikaard CS. Genomic change and gene silencing in polyploids. Trends in Genetics. 2001;17:675–677. doi: 10.1016/s0168-9525(01)02545-8. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- Razafimandimbison SG, Kellogg EA, Bremer B. Recent origin and phylogenetic utility of divergent ITS putative pseudogenes: a case study from Naucleeae (Rubiaceae) Systematic Biology. 2004;53:177–192. doi: 10.1080/10635150490423278. [DOI] [PubMed] [Google Scholar]
- Renner SS, Beenken L, Grimm GW, Kocyan A, Rickleffs RE. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution. 2007;61:2701–2719. doi: 10.1111/j.1558-5646.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samuel R, Bachmair A, Jobst J, Ehrendorfer F. ITS sequences from nuclear rDNA suggest unexpected phylogenetic relationships between Euro-Mediterranean, East Asiatic and North American taxa of Quercus (Fagaceae) Plant Systematics and Evolution. 1998;211:129–139. [Google Scholar]
- Sanderson MJ, Wojciechowski MF, Hu J-M, Sher Khan T, Brady SG. Error, bias, and long-branch attraction in data of two chloroplast photosystem genes in seed plants. Molecular Biology and Evolution. 2000;17:782–797. doi: 10.1093/oxfordjournals.molbev.a026357. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Senters AE, Zanis MJ, Sangtae K, Thompson JD, Soltis PS, et al. Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. American Journal of Botany. 2003;90:461–470. doi: 10.3732/ajb.90.3.461. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML–VI-HPC: maximum-likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) Champaign, IL, USA: National Illinois History Survey; 2002. [Google Scholar]
- Takhtajan A. Systemat Magnoliophytorum. Leningrad: Nauka; 1987. [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk, Schweizer D, Hemleben V. Elimination and rearrangement of parental rDNA in the allotetraploid. Nicotiana tabacum. Molecular Biology and Evolution. 1999;16:311–320. doi: 10.1093/oxfordjournals.molbev.a026112. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Komarova NY, Hemleben V. Ribosomal DNA and plant hybrids: Inheritance, rearrangement, expression. Systematics and Biodiversity. 2007;5:261–276. [Google Scholar]
- Winkworth RC, Bryant D, Lockhart PJ, Havell D, Moulton V. Biogeographic interpretation of splits graphs: least squares optimization of branch lengths. Systematic Biology. 2005;54:56–65. doi: 10.1080/10635150590906046. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Lockhart PJ. Deciphering ancient rapid radiations. Trends in Ecology and Evolution. 2007;22:258–265. doi: 10.1016/j.tree.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Won H, Renner SS. The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Molecular Phylogenetics and Evolution. 2005;36:581–597. doi: 10.1016/j.ympev.2005.03.011. [DOI] [PubMed] [Google Scholar]