Abstract
Background and Aims
The phytohormone abscisic acid (ABA) plays a vital role in various aspects of plant growth and development and in adaptation of plants to various environmental stresses. Cell response to ABA is initiated by ABA perception with a cell receptor. Recently, three distinct ABA receptors have been identified, opening a door to uncover the initial events of ABA signal transduction. The aim of this Botanical Briefing is to present a perspective of the ABA receptors identified.
Scope
This Briefing offers an introduction to the three ABA receptors identified and an analysis of the complexity and multiplicity of ABA receptors, and provides some viewpoints on future research.
Key words: Abscisic acid receptors (ABA receptors), abscisic acid signalling, FCA (flowering time control protein A), ABAR (putative ABA receptor), CHLH (magnesium–protoporphyrin IX chelatase subunit H), GCR2 (G-protein-coupled receptor 2)
INTRODUCTION
The phytohormone abscisic acid (ABA) plays a vital role in various aspects of plant growth and development, including embryo maturation, seed dormancy, germination, post-germinative growth, and the transition from vegetative to reproductive growth. ABA is also a central, hormonal, signal to regulate adaptation of plants to stressful environments, such as drought, cold and salt stresses, by regulating stomatal aperture and the expression of stress-responsive genes (Leung and Giraudat, 1998; Finkelstein et al., 2002; Himmelbach et al., 2003). ABA functions through a complex network of signalling pathways where the cell response is initiated by ABA perception which triggers downstream signalling cascades to induce the final physiological effects. Numerous downstream components involved in ABA signal transduction have been identified by genetic approaches (for reviews, see Leung and Giraudat, 1998; Finkelstein et al., 2002; Himmelbach et al., 2003), leading to considerable progress in understanding ABA signalling pathways. Recently, however, significant progress has been made in the search for receptors that perceive ABA. Three distinct loci have been found to encode ABA receptors: FCA, controlling flowering time (Razem et al., 2006); and ABAR/CHLH and GCR2, regulating the best-characterized ABA-responses including seed germination, seedling growth and stomatal movement (Shen et al., 2006; Liu et al., 2007). This progress in ABA receptor identification will allow us to uncover the initial events of ABA signalling in plant cells and to establish the relationship between the receptors and the currently known regulatory framework or to explore novel pathways of ABA signal transduction.
FCA IS A NUCLEAR RECEPTOR FOR ABA CONTROLLING FLOWERING
ABA has long been known to inhibit flowering, and several components involved in the signalling pathway have been suggested (for a review, see Finkelstein and Rock, 2002), but a flowering-related receptor perceiving ABA had remained elusive until FLOWERING TIME CONTROL PROTEIN A (FCA), an important regulator of floral transition, was identified as an ABA receptor that controls ABA-dependent flowering (Razem et al., 2006).
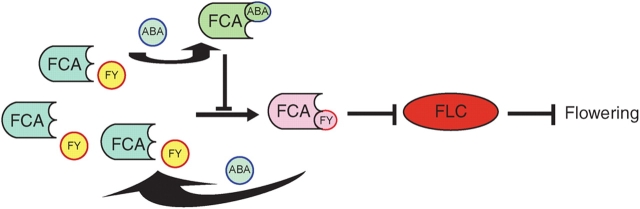
There are four major floral promotion pathways identified in Arabidopsis thaliana for mediating the timing of floral transition (Mouradov et al., 2002; Boss et al., 2004). The photoperiod pathway controls flowering by induction of floral initiation in response to the long days of spring or early summer, while the vernalization pathway promotes flowering in response to extended exposure to low temperatures that mimic winter conditions. The photoperiod and vernalization responses are mediated by pathways that specifically control response to these environmental cues, whereas the third, autonomous pathway, and the fourth, gibberellin (GA) pathway, appear to function independently of these environmental signals (Mouradov et al., 2002; Boss et al., 2004). These multiple pathways, however, converge to regulate the transcription of a set of integrator genes to control flowering. FLOWERING LOCUS C (FLC), a MADS box transcription factor (Michaels and Amasino, 1999), is a central repressor of floral transition for integrating the vernalization and autonomous pathways (Sheldon et al., 2000; Mouradov et al., 2002; Boss et al., 2004; Isabel and Dean, 2006). FCA, an important positive regulator of flowering involved in the autonomous pathway, is a nuclear RNA-binding protein (Macknight et al., 1997) that regulates 3'-end processing and has a tryptophan–tryptophan (WW) protein interaction domain near its C-terminus and two RNA recognition motifs near its N-terminus (Macknight et al., 1997; Quesada et al., 2003). FCA interacts with a second RNA 3′-end-processing protein, FY (for Flowering Locus Y), which binds to the WW domain on FCA. This interaction is required for two possibly linked processes in FCA function (Quesada et al., 2003; Simpson et al., 2003). One process is negative auto-regulation of FCA expression through promoting premature cleavage and polyadenylation in intron 3 of its precursor mRNA (pre-mRNA), thus increasing the premature FCA mRNA encoding the major form of inactive, truncated protein FCAβ and correspondingly decreasing the mature FCA mRNA encoding the active, full-length protein FCAγ. Another process is to down-regulate FLC expression through a direct action on FLC pre-mRNA or indirectly via an FLC regulator. The FCA–FY interaction to control FLC expression is a key process in flowering regulation (Mouradov et al., 2002; Boss at al., 2004; Isabel and Dean, 2006) (Fig. 1). It is in the regulation of this interaction that ABA is involved to control flowering.
Fig. 1.
ABA-triggered FCA signalling delays flowering time. FCA–FY interaction represses FLC expression and thus promotes flowering. ABA disrupts FCA–FY interaction by binding to FCA, up-regulating FLC expression by de-repressing it, and thus delaying flowering. Arrows indicate positive regulation; black bars indicate repression; the grey bar indicates de-repression [diagram modified from Finkelstein (2006) and Schroeder and Kuhn (2006)].
As a matter of fact, to explore ABA receptors, Hill and his colleagues screened a barely complementary DNA expression library with anti-idiotypic antibodies and isolated a protein which binds ABA, called ABAP1 (Razem et al., 2004). They found that ABAP1 is homologous to the Arabidopsis FCA protein, and showed that Arabidopsis FCA also binds ABA with a high affinity (the equilibrium-disassociation constant Kd = 19 nm) (Razem et al., 2006). This binding affinity is high enough to sense physiological concentrations of ABA present in plants. Competition assays revealed that FCA binds to naturally occurring, physiologically active (+)-ABA but not physiologically inactive ABA isomer (–)-ABA, indicating that the ABA binding is stereo-specific to the physiologically active form of ABA. These properties of binding meet the primary criteria of an ABA receptor. The next question was whether FCA regulates ABA-related processes in floral transition, functioning at the primary events as a flowering signal receptor. In-vitro and in-vivo assays demonstrated that the binding of ABA to FCA disrupts FCA–FY interaction. The ABA-binding site of the FCA molecule is near the C-terminus, shielding but not including the FY-binding WW domain. As expected, ABA–FCA binding-induced dissociation of the FCA–FY complex abolishes downstream signalling. The disruption of downstream signalling includes two processes: (1) a loss of function of auto-regulation of FCA expression with a decrease in the premature FCA mRNA encoding the inactive FCAβ as well as a corresponding increase in the mature FCA mRNA encoding the active FCAγ, and (2) an enhancement of expression of the central flowering repressor FLC. Importantly, in addition to these molecular events triggered by ABA binding to FCA, the ABA–FCA binding-induced dissociation of the FCA–FY complex results in the expected physiological consequence – a significant delay in flowering time. Further experiments with the flowering mutants fca-1 and fy-1, as well as ABA biosynthesis mutant aba1 and ABA signalling mutant abi-2, showed that FCA specifically mediates the ABA-regulated floral transition process. The ABA-induced functions also specifically depend on the physiologically active form of ABA. These findings demonstrate that FCA is an ABA receptor controlling ABA-dependent floral transition (Razem et al., 2006). FCA also functions in the response of lateral root growth to ABA, but not in the other major ABA-related responses including seed maturation, germination, post-germinative growth and stomatal movement (Razem et al., 2006). This provides evidence for multiplicity of ABA receptors to mediate ABA signalling in response to different developmental or environmental cues. A model of FCA function to mediate ABA-controlled flowering signalling is presented in Fig. 1. Further studies will be needed, e.g. to explore whether ABA binding to FCA could initiate downstream mRNA processing of other genes than FLC to mediate other responses such as lateral root growth, and to analyse whether RNA processing is a more general mechanism of cell signalling (Finkelstein, 2006; Schroeder and Kuhn, 2006).
ABAR/CHLH IS A PLASTID RECEPTOR FOR ABA CONTROLLING MAJOR ABA RESPONSES
Similarly to the identification of the FCA receptor, the search for ABA receptors began with biochemical approaches to isolate ABA-binding proteins (Zhang et al., 1999, 2001, 2002). An ABA-specific binding protein from broad bean leaves was purified to apparent homogeneity by affinity chromatography, and this protein was shown to be functionally a putative ABA receptor in guard cell protoplasts (Zhang et al., 2002). Thus, the ABA-binding protein was named ABAR (for putative ABA receptor). Based on sequencing information of the purified protein, a cDNA fragment was isolated from broad bean (Vicia faba) leaves and found to encode the C-terminal half of about 770 amino acids of the putative H subunit (CHLH) of the magnesium–protoporphyrin IX chelatase (Mg-chelatase). The purified, yeast-expressed product of the cDNA fragment encoding the broad bean ABAR/CHLH could specifically bind ABA. This suggested that CHLH might be involved in ABA perception as an ABA receptor.
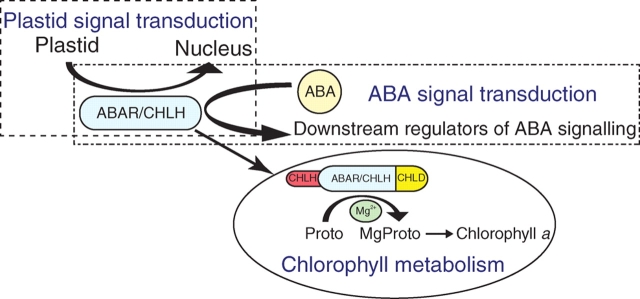
Mg-chelatase is composed of three subunits CHLD, CHLI and CHLH, and catalyses the insertion of Mg2+ into protoporphyrin-IX to form Mg–protoporphyrin-IX, the first step unique to chlorophyll synthesis (Walker and Willows, 1997) (Fig. 2). CHLH plays a central role in Mg–protoporphyrin-IX biosynthesis as a monomeric, protoporphyrin-IX-binding protein (Walker and Willows, 1997; Karger et al., 2001). In relation to its enzymic function, CHLH plays a key role in mediating plastid-to-nucleus retrograde signalling in which, under stressful environments, damaged plastids send signals to the nucleus to control the expression of nuclear genes that encode plastid proteins to co-ordinate gene expression in both organelles (Fig. 2). The Arabidopsis genomes uncoupled 5 (gun5) mutant, resulting in a single amino acid Ala990 → Val mutation in CHLH, revealed that CHLH is involved in plastid-to-nucleus retrograde signalling by controlling metabolism of the tetrapyrrole signal Mg–protoporphyrin-IX or sensing the signal (Mochizuki et al., 2001; Surpin et al., 2002; Strand et al., 2003; Nott et al., 2006).
Fig. 2.
Triple functions of ABAR/CHLH. ABAR/CHLH, interacting with two other subunits of Mg-chelatase, CHLI and CHLD, catalyses the insertion of Mg2+ into protoporphyrin-IX (Proto) to form Mg–protoporphyrin-IX (MgProto), the first step unique to chlorophyll synthesis. ABAR/CHLH regulates also the plastid-to-nucleus retrograde signalling, and mediates ABA signalling as a plastid receptor for ABA.
Testin was done to find out if ABAR/CHLH functions in ABA signalling in the reference plant Arabidopsis thaliana (Shen et al., 2006). Both yeast-expressed and natural Arabidopsis ABAR/CHLH protein specifically bind ABA with high binding-affinities (Kd values 32–35 nm), approximately corresponding to physiological concentrations of ABA. Down-regulation of expression of the ABAR/CHLH gene by RNA interference (RNAi) reduced, but up-regulation of the gene by over-expression enhanced, numbers of ABA-binding sites (maximum binding volume, Bmax), whereas neither of these manipulations modified the ABA-binding affinity (unchanged Kd value). ABA binding to ABAR/CHLH specifically depended on physiologically active ABA, i.e. (+)-ABA. The ABA binding displayed typical ligand–receptor binding characteristics. Additionally, as an ABA and protoporphyrin-IX double-ligand-binding protein, ABAR/CHLH binds ABA independently of protoporphyrin-IX, supporting the suggestion that ABA signal perception by ABAR/CHLH is distinct from protoporphyrin-IX metabolism.
Plants underexpressing ABAR via RNAi and antisense transgenic manipulation showed significant ABA-insensitive phenotypes in seed germination, post-germinative growth, ABA-induced promotion of stomatal closure and inhibition of stomatal opening. Down-regulation of the ABAR expression also reduced the expression of a subset of ABA-positively responsive genes, but enhanced the expression of several ABA-negatively responsive genes. In contrast, the ABAR-overexpressors displayed ABA-supersensitive phenotypes, were more resistant to water loss from their leaves and were more tolerant to drought, but the RNAi- and antisense-plants were more sensitive to dehydration and drought stress. A T-DNA insertion loss-of-function mutant, abar-1, is lethal – probably owing to a distortion in late embryonic development. Plants of a chemically (17β-oestradiol)-regulated inducible ABAR-RNAi line showed, after induction by 17β-oestradiol, ABA-insensitive phenotypes in ABA-induced promotion of stomatal closure and inhibition of stomatal opening. In addition, like the stable ABAR-RNAi lines, the cch mutant, an allele of the gun5 with a single amino acid mutation Pro642 → Leu, showed ABA-insensitive phenotypes in seed germination, post-germinative growth and stomatal movement. The cch mutation decreased ABA-binding activity of ABAR/CHLH, which explains its ABA-insensitive phenotypes, whereas the gun5 has no ABA-related phenotype, probably because the gun5 mutation does not affect ABA-binding to ABAR/CHLH. These findings demonstrate that ABAR/CHLH is a plastid ABA receptor positively controlling major ABA responses. A series of additional, pharmaceutical assays as well as genetic analysis with mutants defective in chlorophyll metabolism or plastid retrograde signalling showed that ABAR/CHLH-mediated ABA signalling is a process distinct from chlorophyll metabolism and Mg–protoporphyrin IX-mediated plastid retrograde signalling (Shen et al., 2006) (see Fig. 2). Recently, however, ABSCISIC ACID INSENSITIVE4 (ABI4), a transcription factor involved in ABA signalling, was shown to be a target of the plastid retrograde signalling that functions downstream of GUN5/ABAR/CHLH (Koussevitzky et al., 2007). This raises the question of whether the hormone signal and retrograde signals cross-talk to control nuclear gene expression. Current studies suggest that hormone signalling shares some components with chloroplast-to-nucleus signalling, but the two pathways do not appear to interact (Shen et al., 2006; Koussevitzky et al., 2007). However, given the important role of abscisic acid in regulating the expression of nuclear genes that encode chloroplast proteins, it is worth further investigating whether the two signalling pathways intersect at some currently unidentified nodes or in some as-yet unknown manner (Zhang, 2007).
The mechanism of ABAR perception of ABA and downstream signalling events are still unknown so it is necessary to explore the ABAR–ABA binding mechanism and its biological significance, to screen the functionally interacting partners of ABAR and to elucidate the signalling mechanism. In this regard, we believe that the mechanisms of the ABAR/CHLH-mediated ABA-signalling pathway may be diversified in different tissues/cells or during developmental stages. It was observed that the transgenic ABAR-RNAi or ABAR-antisense lines or cch mutant plants showed the strongest, stable, ABA-insensitive phenotypes in stomatal movement, whereas for germination, and especially post-germinative growth, the ABA-insensitive phenotypes were weaker, which was more apparent when the ABAR levels were down-regulated to a certain extent (Shen et al., 2006) This suggests that some threshold concentration of this protein may exist to modify the signalling process for seed germination and seedling growth, or that other ABA receptors may function redundantly or distinctly at least in these developmental processes.
GCR2 IS A PLASMA MEMBRANE RECEPTOR FOR ABA, CONTROLLING MAJOR ABA RESPONSES
Ligand signalling via guanine nucleotide (G)-binding protein (G protein)-coupled receptors (GPCRs) is a ubiquitous transmembrane signalling mechanism in a variety of eukaryotic organisms, including plants, fungi and animals (Jones and Assmann, 2004; McCudden et al., 2005). G proteins consist of three different subunits, Gα, Gβ and Gγ, which form a heterotrimeric complex. GPCRs are a class of proteins typically with a seven-transmembrane domain (7TM) structure composed of an extracellular N-terminus, seven hydrophobic stretches of about 20 amino acids linked by alternating intracellular and extracellular loops, and a cytoplasmic C-terminal tail. The heterotrimers of G-protein are membrane-bound by their close association with the intracellular faces of GPCRs. Heterotrimeric G protein complexes are intracellular partners of the GPCRs, linking ligand perception by GPCRs with downstream effectors. GDP-bound Gα subunits bind tightly to the obligate heterodimer of Gβγ, which aids Gα localization to the plasma membrane and is essential for functional coupling to GPCRs. In addition, the binding of Gβγ to Gα inhibits dissociation of GDP from Gα (reviewed in Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004; Assmann, 2005; McCudden et al., 2005). In the classical signalling paradigm, in response to activation by agonist binding to a GPCR, the inactive G-heterotrimeric complex Gα-GDP/Gβγ converts to an active conformation, promoting GDP release from, and GTP binding to, the Gα subunit, which results in both dissociation of the G-protein complex from the GPCR and liberation of Gα from Gβγ. Either the liberated form of Gα (Gα-GTP) or the free Gβγ dimer or both participate in signalling to downstream effector proteins. Signalling is terminated by the intrinsic GTPase activity of Gα, which hydrolyses GTP to GDP, thereby allowing Gα to re-associate with the Gβγ dimer, and thus reforming the inactive heterotrimeric complex associated with the GPCR (reviewed in Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004; Assmann, 2005; McCudden et al., 2005).
The Arabidopsis genome encodes one canonical Gα (GPA1) subunit (Ma et al., 1990; Ma, 1994), one Gβ (AGB1) subunit (Weiss et al., 1994; Mason and Botella, 2001), two Gγ (AGG1 and AGG2) subunits (Mason and Botella, 2000, 2001) and about 25 candidate GPCRs with a seven-transmembrane topology that characterizes this receptor family (Grill and Christmann, 2007). Heterotrimeric G proteins and GPCRs were reported to regulate ABA signal transduction in Arabidopsis. An Arabidopsis GPCR, GCR1, interacting with GPA1, was shown to be a negative regulator of ABA signalling involved in seed germination, post-germinative growth and stomatal response (Pandey and Assmann, 2004; Pandey et al., 2006). GPA1 is involved in ABA signalling as a negative regulator controlling seed germination and post-germinative growth (Ullah et al., 2002; Lapik and Kaufman, 2003; Pandey et al., 2006), but as a positive regulator controlling stomatal opening where GCR1 and GPA1 have opposite effects on ABA signalling (Wang et al., 2001). Consistently to GPA1, the Gβ subunit AGB1 also negatively regulates ABA signalling in seed germination and post-germinative growth (Pandey et al., 2006). However, the GCR1 does not appear to function as an ABA receptor, as ABA was not reported to be a functional ligand of GCR1.
To identify a plasma membrane receptor for ABA in Arabidopsis, Ma and his colleagues found a gene encoding a putative GPCR, GCR2, a typical plasma membrane protein belonging to the 7TM family (Liu et al., 2007). They showed that, like GCR1 (Pandey and Assmann, 2004), GCR2 has the ability to interact with the Gα subunit GPA1, thus forming a possible complex. Unlike GCR1 (Pandey and Assmann, 2004; Pandey et al., 2006), however, GCR2 positively regulates ABA signalling: (a) loss-of-function of the GCR2 gene results in ABA-insensitive phenotypes in major ABA-responses, including ABA-induced inhibition of seed dormancy, germination, post-germinative arrest of seedling growth, inhibition of stomatal opening, promotion of stomatal closure and inhibition of the inward K+ channel in guard cells, and this loss of function also represses some ABA-positively responsive genes; and (b), in contrast, GCR2 overexpression leads to ABA-hypersensitive phenotypes in these ABA-induced physiological processes. These findings reveal that GCR2 is a positive regulator in ABA signalling (Liu et al., 2007). Additionally, to test genetic interactions between GCR2 and GPA1, the assays with gcr2gpa1 double mutants and GCR2- and GPA1-transgenic manipulation were performed. The results showed that GCR2 functions together with GPA1 to transduce ABA signal at least in guard cell regulation (Liu et al., 2007). As GPA1 negatively regulates ABA signalling in seed germination and early seedling growth (Pandey and Assmann, 2004; Pandey et al., 2006), GCR2 should negatively regulate GPA1 function in these developmental processes, which is different from stomatal regulation.
GCR2 was shown to bind specifically the physiologically active (+)-ABA [but not the inactive ABA isomers (–)-ABA or trans-ABA] with a Kd value of 20 nm (Liu et al., 2007), which is consistent with the physiological concentration range of (+)-ABA in plant tissues. Furthermore, (+)-ABA disrupts the GCR2–GPA1 interaction, but (–)-ABA or trans-ABA cannot (Liu et al., 2007). These findings suggest that GCR2 can perceive an ABA signal, and that this ligand-binding event leads to the dissociation of the GCR2–G protein complex to release Gα and the Gβγ dimer, thus activating the downstream signalling events mediated such as by phospholipase Dα1 (Zhao and Wang, 2004) or cupin-domain protein AtPirin1 (Lapik and Kaufman, 2003), which supports the suggestion that GCR2 is an ABA receptor. As both GCR1 and GCR2 have the same interacting partner Gα subunit GPA1, the GPA1 appears to represent a node at which different signalling pathways converge (Grill and Christmann, 2007).
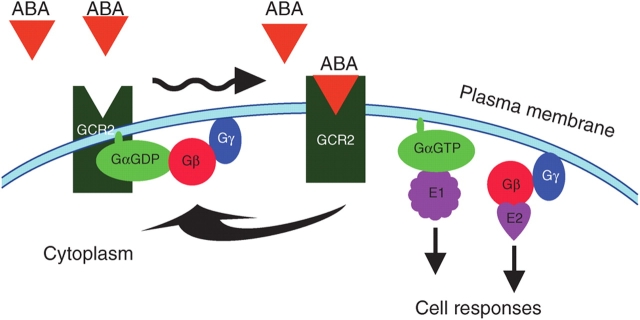
It has been noted that the gcr2 mutants still display ABA responses, which may be because of functional redundancy with other GCR2-related proteins, given that there are two other GCR2 homologues in the Arabidopsis genome (Grill and Christmann, 2007). On the other hand, the relatively weak phenotypes in the gcr2 mutants are consistent with the occurrence of multiple ABA receptors in plant cells, possibly involving at least the intracellular ABA perception site ABAR/CHLH (Shen et al., 2006). A model schematizing how GCR2 works to mediate the ABA signal is presented in Fig. 3. Further work might be focused, for example, on exploring whether GCR2–AGB1/AGG1/AGG2 interactions are required for ABA signalling and on screening the effectors downstream of these interactions to elucidate complex GCR2-mediated ABA-signalling network.
Fig. 3.
ABA-binding to GCR2 activates the plasma membrane receptor. ABA binding to GCR2 disassociates the inactive heterotrimeric G protein (Gα-GDP/Gβγ) from GCR2, facilitating the exchange of GTP for GDP on the Gα subunit, and releasing Gα-GTP from the heterodimer Gβγ. The liberated Gα-GTP and the free heterodimer Gβγ can interact separately with downstream effector proteins (E1, E2) to relay the signal, resulting finally in cell responses. The signalling is terminated by reforming the inactive heterotrimeric G protein associated with GCR2.
CONCLUDING REMARKS: MULTIPLE RECEPTORS PERCEIVE THE ABA SIGNAL
A receptor was traditionally believed to initiate multiple, branching, signalling pathways in response to its ligand binding. However, the events of signal perception and downstream relay may be made more complicated by the possible existence of multiple classes of receptors for a signal. Several lines of evidence support the suggestion that multiple, different, ABA receptors perceive the ABA signal. First, some partly active isomers, such as (–)-ABA and the ABA metabolites 8′-hydroxy-ABA and phaseic acid, activate some responses; in addition, there are different active groups on the ABA molecule, which may interact with different ABA-binding proteins (Walton, 1983; Walker-Simmons et al., 1994, 1997; Nyagulu et al., 2005). These diversities in the structure-defined stereo-specificity suggest the existence of different ABA perception sites. Secondly, ABA perception sites may be different in response to different environmental cues, and they may be organ-, tissue- or even cell-specific and developmental stage-specific Thirdly, plant cells possess the ABA perception sites inside and outside the cells. Previous pharmaceutical studies by manipulating apoplasmic-side or cytosolic-side ABA concentrations mainly in guard-cell and barley aleurone-protoplast systems indicated that there exist both extracellular and intracellular ABA perception sites in cells (reviewed in Finkelstein et al., 2002). The identification of the nuclear receptor FCA, plastid receptor ABAR/CHLH and plasma membrane receptor GCR2 for ABA confirms these early findings provided by physiological approaches. Do other ABA receptors, however, exist to mediate ABA signalling in addition to the three identified ABA receptors? As mentioned above, FCA mediates ABA-dependent flowering control, and also regulates lateral root growth, but not other major ABA responses (Razem et al., 2006). ABAR/CHLH strongly regulates the stomatal response, but relatively weakly affects other responses (Shen et al., 2006). GCR2-mediated responses are also apparently relatively weak (Liu et al., 2007). These studies suggest the possible presence of other additional ABA receptors to mediate ABA signal redundantly or distinctly in response to developmental or environmental cues, or at different cellular compartments.
In looking for additional receptors for ABA, the most obvious candidates would be close homologues of the receptors identified. The Arabidopsis genome harbours a single gene copy for ABAR/CHLH, whereas both FCA and GCR2 have several homologues in the Arabidopsis genome (Finkelstein, 2006; Grill and Christmann, 2007). On the other hand, analysis of ABA-specific-binding domains on FCA, ABAR/CHLH and GCR2 molecules will aid identification of candidate receptors with the same ‘ABA-binding pocket’. Additionally, efforts with biochemical approaches may still be useful in screens for distinct ABA receptor candidates. Isolation of ABA-binding proteins by use of carefully designed affinity probes has been shown to be efficient in the identification of FCA and ABAR/CHLH receptors (Razem et al., 2004, 2006; Zhang et al, 2002; Shen et al., 2006) where the biochemical and genetic approaches converge. Finally, forward genetic screens with possibly improved screening strategies may be auspicious for the isolation of ABA-receptor mutants, as leaky mutations may not necessarily be lethal in ABA receptor genes in which a loss-of-function mutation is lethal. For example, the cch mutant, which is defective in the ABAR/CHLH gene, is ABA-response defective but not lethal (Shen et al., 2006); also, in some receptors with a multigene family, even loss-of-function mutants survive well, such as gcr2 mutants (Liu et al., 2007). The identification of the three ABA receptors also suggests that functional redundancies may be not a big problem to hinder isolating candidate ABA receptors with forward genetic approaches, as clear ABA-related phenotypes have been observed in these ABA-receptor mutants identified (Razem et al., 2006; Shen et al., 2006; Liu et al., 2007). We are waiting for identification of novel ABA receptors through both old and new methods to uncover the complex and diverse ABA signalling mechanisms.
NOTE ADDED IN PROOF
We have noted the debate about whether GCR2 is an ABA receptor or a G-protein-coupled receptor (Gao et al., 2007; Johnston et al., 2007). Given the role of GCR2 in ABA-related stomatal regulation and that the authors have provided additional evidence that GCR2 is a new type of non-classical G-protein-coupled receptor (Liu et al., 2007), we have retained our text about GCR2 as an ABA receptor.
ACKNOWLEDGEMENTS
The work in the authors' laboratory was supported by National Natural Science Foundation of China (grant nos. 30330420, 30421002, 30471193 and 30671444) and by the National Key Basic Research Program of China (grant no. 2003CB114302).
LITERATURE CITED
- Assmann SM. G protein go green: a plant G protein signaling FAQ sheet. Science. 2005;310:71–73. doi: 10.1126/science.1118580. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell. 2004;16(Suppl.):S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. Studies of abscisic acid perception finally flower. The Plant Cell. 2006;18:786–791. doi: 10.1105/tpc.106.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Rock CD. Abscisic acid biosynthesis and response. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis book. Rockville, MD: American Society for Plant Biologists; 2002. pp. 1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;(Suppl.):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Christmann A. A plant receptor with a big family. Science. 2007;315:1676–1677. doi: 10.1126/science.1140761. [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Isabel B, Dean C. The timing of development transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Jones AM, Assmann SM. Plants: the latest model system for G-protein research. EMBO Reports. 2004;5:572–578. doi: 10.1038/sj.embor.7400174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger GA, Reid JD, Hunter CN. Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry. 2001;40:9291–9299. doi: 10.1021/bi010562a. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler T, Hong F, Sachetto-Martins G, Surpin M, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Lapik YR, Kaufman LS. The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α-subunit GPA1 and regulates seed germination and early seedling growth. The Plant Cell. 2003;15:1578–1590. doi: 10.1105/tpc.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, et al. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- McCudden CR, Hains MD, Kimple RJ, Sideroski DP, Willard FS. G-protein signaling: back to the future. Cellular and Molecular Life Science. 2005;62:551–577. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. GTP-binding proteins in plants: new members of an old family. Plant Molecular Biology. 1994;26:1611–1636. doi: 10.1007/BF00016493. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky M, Meyerrowitz EM. Molecular cloning and characterization of GPA1, a G protein α subunit gene from Arabidopsis thaliana. Proceedings of National Academy of Sciences of the USA. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Mason MG, Botella JR. Completing the heterodimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proceedings of National Academy of Sciences USA; 2000. pp. 14784–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR. Isolation of a novel G protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochimica et Biophysica Acta. 2001;1520:147–153. doi: 10.1016/s0167-4781(01)00262-7. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani N, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proceedings of National Academy of Sciences of the USA; 2001. pp. 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. Control of flowering time: interacting pathways as a basis for diversity. The Plant Cell. 2002;14(Suppl.):S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology. 2006;57:730–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Nyangulu J, Galka M, Jadhav A, Gai Y, Graham C, Nelson K, et al. An affinity probe for isolation of abscisic acid-binding proteins. Journal of American Chemical Society. 2005;127:1662–1664. doi: 10.1021/ja0429059. [DOI] [PubMed] [Google Scholar]
- Pandey S, Assmann SM. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. The Plant Cell. 2004;16:1616–1632. doi: 10.1105/tpc.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiology. 2006;141:243–256. doi: 10.1104/pp.106.079038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insight from Arabidopsis and rice mutants. Current Opinion in Plant Biology. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson GG. Auto-regulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO Journal. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem FA, Luo M, Liu JH, Abrams SR, Hill RD. Purification and characterization of a barley aleurone abscisic acid-binding protein. Journal of Biological Chemistry. 2004;279:9922–9929. doi: 10.1074/jbc.M311064200. [DOI] [PubMed] [Google Scholar]
- Razem FA, El-Kereamy A, Abrams SR, Hill RD. The RNA-binding protein FCA is an abscisic acid receptor. Nature. 2006;439:290–294. doi: 10.1038/nature04373. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kuhn JM. Abscisic acid in bloom. Nature. 2006;439:277–278. doi: 10.1038/439277a. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proceedings of National Academy of Sciences of the USA; 2000. pp. 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg–protoporphyrin IX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J. Signal transduction between the chloroplasts and the nucleus. The Plant Cell. 2002;(Suppl.):S327–S338. doi: 10.1105/tpc.010446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang SC, Jones AM. Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiology. 2002;129:897–907. doi: 10.1104/pp.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CJ, Willows RD. Mechanism and regulation of Mg-chelatase. Biochemical Journal. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons M, Rose P, Shaw A, Abrams SR. The 7′-methyl group of abscisic acid is critical for biological activity in wheat embryo germination. Plant Physiology. 1994;106:1279–1284. doi: 10.1104/pp.106.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons M, Holappa LD, Abrams GD, Abrams SR. ABA metabolites induce group 3 LEA mRNA and inhibit germination in wheat. Physiologia Plantarum. 1997;100:474–480. [Google Scholar]
- Walton D. Structure–activity relationships of abscisic acid analogs and metabolites. In: Addicott F, editor. Abscisic acid. New York, NY: Praeger Publishers; 1983. pp. 113–146. [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM. G protein regulation of ion channels and abscisic acid signaling in arabidopsis guard cells. Science. 2001;292:2070–2072. doi: 10.1126/science.1059046. [DOI] [PubMed] [Google Scholar]
- Weiss CA, Garnatt CW, Mukai Y, Hu Y, Ma H. Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proceedings of National Academy of Sciences of the USA; 1994. pp. 9554–9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DP. Signaling to the nucleus with a loaded GUN. Science. 2007;316:700–701. doi: 10.1126/science.1142703. [DOI] [PubMed] [Google Scholar]
- Zhang DP, Zhang ZL, Chen J, Jia WS. Specific abscisic acid-binding sites in mesocarp of grape berry: properties and subcellular localization. Journal of Plant Physiology. 1999;155:324–331. [Google Scholar]
- Zhang DP, Chen SW, Peng YB, Shen YY. Abscisic acid-specific binding sites in the flesh of developing apple fruit. Journal of Experimental Botany. 2001;52:2097–2103. doi: 10.1093/jexbot/52.364.2097. [DOI] [PubMed] [Google Scholar]
- Zhang DP, Wu ZY, Li XY, Zhao ZX. Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiology. 2002;128:714–725. doi: 10.1104/pp.010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang X. Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. Journal of Biological Chemistry. 2004;279:1794–1800. doi: 10.1074/jbc.M309529200. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zeng Q, Guo J, Cheng J, Ellis BE, Jin-Gui C. Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. Plant Journal. 2007;52:1001–1013. doi: 10.1111/j.1365-313X.2007.03291.x. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Temple BR, Jin-Gui C, Gao Y, Moriyama EN, Jones AM, Siderovski DP, Willard FS. Comment on a G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;318:914c. doi: 10.1126/science.1143230. doi:10.1126/science. 1143230. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li W, Ma L. Response to comment on a G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;318:914d. doi: 10.1126/science.1135882. doi:10.1126/science. 1143320. [DOI] [PubMed] [Google Scholar]