Abstract
Background and Aims
Highly variable, yet possibly convergent, morphology and lack of sequence variation have severely hindered production of a robust phylogenetic framework for the genus Ophrys. The aim of this study is to produce this framework as a basis for more rigorous species delimitation and conservation recommendations.
Methods
Nuclear and plastid DNA sequencing and amplified fragment length polymorphism (AFLP) were performed on 85 accessions of Ophrys, spanning the full range of species aggregates currently recognized. Data were analysed using a combination of parsimony and Bayesian tree-building techniques and by principal co-ordinates analysis.
Key Results
Complementary phylogenetic analyses and ordinations using nuclear, plastid and AFLP datasets identify ten genetically distinct groups (six robust) within the genus that may in turn be grouped into three sections (treated as subgenera by some authors). Additionally, genetic evidence is provided for a close relationship between the O. tenthredinifera, O. bombyliflora and O. speculum groups. The combination of these analytical techniques provides new insights into Ophrys systematics, notably recognition of the novel O. umbilicata group.
Conclusions
Heterogeneous copies of the nuclear ITS region show that some putative Ophrys species arose through hybridization rather than divergent speciation. The supposedly highly specific pseudocopulatory pollination syndrome of Ophrys is demonstrably ‘leaky’, suggesting that the genus has been substantially over-divided at the species level.
Key words: AFLP, DNA sequencing, hybridization, introgression, Ophrys, pseudocopulation, species delimitation, systematics
INTRODUCTION
The genus Ophrys will be familiar to many botanists, conservationists and orchid enthusiasts, having become widely recognized as a model system for exploring floral evolution. Both molecularly and morphologically distinct (Bateman et al., 2003), the genus occurs within a clade of genera delimited by a chromosome number of 2n, 36 (records of tetraploids in Ophrys are rare: Bernardos et al., 2003). Like Serapias, but unlike most other related genera, Ophrys has a relatively narrow distribution that encompasses much of Europe but does not extend east of Asia Minor.
The genus is most remarkable for the complex morphology of the flower in general, and of the insect-like labellum in particular (Fig. 1). This is thought to have evolved primarily through pollinator mimicry. Pollination occurs by sexual deception through a process termed pseudocopulation; the flower (the mimic) imitates a female of one or more pollinator species (the model) in order to attract males of the same species (the operator). Pollinators are usually sexually inexperienced male bees of Andrenidae, Anthophoridae, Megachilidae, Apidae and Colletidae, wasps of Sphecidae and Scolidae and, in two cases, beetles of Scarabidae (Paulus, 1997). Males of the pollinator species are attracted to the flowers through a combination of olfactory, visual and tactile stimuli. Several active compounds in the scent trigger behavioural responses in the pollinator when present in specific ratios (Schiestl et al., 1999). The visual signals are based on shape and chromatic variation in the flower (including a highly reflective region termed the speculum), whereas the main tactile component is variation in pilosity across the labellum. Once attracted, males attempt to mate with the flower, and pollinia are transferred to the pollinator through the adhesion of viscid discs. Further attempts to mate with other Ophrys flowers result in the transfer of pollinia, thereby effecting cross-pollination.
Fig. 1.

(A–J) Single representative species of each of the ten clades of Ophrys delimited in the ITS tree, presented in alphabetical order (i.e. image A represents clade A) and at a constant magnification (original images were slides taken at 1:1 scale; thus, the long axis of each image represents 35 mm): (A) O. insectifera, Hampshire, UK; (B) O. tenthredinifera, Sicily; (C) O. speculum, Sicily; (D) O. bombyliflora, Sicily; (E) O. bilunulata, Crete; (F) O. apifera, Kent, UK; (G) O. sphegodes, Dorset, UK; (H) O. apulica, Gargano, Italy; (I) O. heldreichii, Crete; (J) O. attica, Peloponnese. (K–L) Two examples from Crete of natural hybrids between highly divergent clades within Ophrys: (K) O. bombyliflora (Group D) × O. cretensis (Group G); (L) O. iricolor (Group E) × O. spruneri grigoriana (Group G). All photographs by R. M. Bateman.
This remarkable pollination syndrome may help explain the wide range of often subtle morphological differentiation evident among putative species of Ophrys. Centuries of predominantly morphological study have resulted in monographs of the genus that recognize from as few as 16 species plus 34 subspecies (Sundermann, 1980) or 19 species plus 46 subspecies (Faurholdt and Pedersen, 207), through 150 species forming 29 complexes (Devillers and Devillers-Terschuren, 1994), to as many as 252 species forming 32 complexes (Delforge, 201). Species-rich classifications have become dominant in recent years, reflecting in part the increasing influence of a species concept in Ophrys systematics that even subtly distinct morphological variants each have a specific, dedicated pollinator. It is further assumed that this supposed precise and intimate relationship forms a reliable pre-zygotic mating barrier around each putative species (Delforge, 205). However, other lines of evidence have challenged these finely drawn species boundaries. For example, breeding experiments performed by S. Malmgren (pers. comm., 2006) regularly recovered multiple morphological ‘species’ from single selfed Ophrys flowers. Also, Soliva and Widmer (2003) clearly demonstrated recent and apparently ubiquitous introgression among at least some members of the O. sphegodes s.l. group, while Gulyás et al. (2005) reported similar gene flow among certain members of the O. fuciflora s.l. group.
The higher-level classification of Ophrys also reflects inferences about pollination biology. The genus was previously divided into two sections (treated as subgenera by some authors) based on suites of morphological characters later inferred to dictate the position of the pollinator during pseudocopulation. A ‘head-up’ (cephalic) position, with the abdomen pressed against the labellum, characterized section Ophrys (often referred to as section Euophrys). In contrast, a ‘head-down’ (abdominal) position, in which pollinia are removed via the abdomen, characterized section Pseudophrys. However, field observations, in the form of close-up video footage (F. Schiestl, pers. comm., 2005), clearly show that the widely accepted and allegedly mutually exclusive concepts of abdominal and cephalic pseudocopulation are apocryphal, an observation reinforced by the natural occurrence of ‘wide crosses’ between members of the two supposed sections (e.g. Bateman and Devey, 2006; Fig. 1K, L).
Controversies over species delimitation in the genus are not simply theoretical debates between evolutionary biologists. Being both numerous and charismatic, Ophrys species figure prominently in local and national conservation strategies across Europe, and thus attract a great deal of conservation attention. Once a species has been named it must then be considered seriously by conservationists, potentially diverting limited resources from arguably more deserving cases. It is therefore highly desirable that each of these species be distinct and stable, rather than a subjective and poorly tested taxonomic entity.
Here, a suite of DNA-based techniques, spanning the boundary between systematics and population genetics, is applied in order to clarify species delimitations. Multiple accessions have been obtained from many of the species groups recognized by Delforge (2005), and are subjected to DNA-based analyses. First, phylogenetic trees have been generated from sequence data obtained from fast-evolving regions of both the nuclear and plastid genomes. Secondly, a set of multi-locus markers were generated and scored for each accession using the AFLP technique (Vos et al., 1995) and subjected to multivariate ordination. The AFLP technique uses restriction enzymes to cut genomic DNA. Complementary double-stranded adaptors are ligated to the ends of the restricted fragments. A subset of these restriction fragments is then further amplified using a pair of primers complementary to the adaptor and restriction site fragments. Fragments are visualized on denaturing polyacrylamide gels where polymorphisms are apparent as presence or absence of peaks. AFLP markers sample restriction endonuclease sites widely across the nuclear genome by selective amplification (Remington et al., 1999). AFLP markers were initially developed for population and species delimitation studies (Mueller and Wolfenbarger, 1999), but they have also been used to resolve relationships among closely related species (e.g. Richardson et al., 2003).
Then the possibilities of integrating these techniques are reviewed, along with appropriate morphological analyses, into a metapopulation-based approach to species delimitation. The objectives include:
circumscribing minimum resolvable genetically distinct entities within the genus;
determining the degree of morphological convergence evident within the genus;
inferring whether some putative Ophrys species may have arisen through hybridization rather than through divergent speciation;
assessing the likelihood of gene flow between genetically distinct entities, both sympatric and allopatric.
MATERIALS AND METHODS
Generating sequences
Accessions of Ophrys species and outgroup taxa used in this study are listed in Appendix 1. Genomic DNA was extracted from both fresh and silica-dried material. Extractions followed the 2× CTAB protocol (Doyle and Doyle, 1987), but used a CsCl2/ethidium bromide density gradient (1·55 g mL−1) for purification (Creeth and Denborough, 1970).
Table 1.
PCoA summary statistics
| Axis | Eigenvalues | % of variance | Cumulative (%) |
|---|---|---|---|
| 1 | 6·104 | 24·8 | 28·4 |
| 2 | 1·907 | 7·9 | 32·6 |
| 3 | 1·414 | 5·9 | 38·5 |
Amplification of the trnH–psbA intergenic spacer was carried out in 50 µL reactions, containing 45 µL of 2·5 mm Mg PCR master mix (Abgene Ltd, Epsom, UK), 1·5 µL bovine serum albumin (0·04 %), 0·6 µl H20, 60 ng of each primer, trnH F (Tate and Simpson, 203) and psbA R (Sang et al., 1997) with approx. 40 ng DNA template. The PCR profile was as follows: initial denaturation of 94 °C for 2 min, followed by 28 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min, extension at 72 °C for 3 min, followed by a final extension of 7 min at 72 °C.
Amplification of the trnD–trnT intergenic spacer was carried out in 50-μL reactions, containing 45 µL Abgene PCR mastermix (2·5 mm Mg), 1·5 µl bovine serum albumin (0·04 %), 0·8 µL H20, 50 ng of each primer trnD F and trnT R (Demesure et al., 1995) and approx. 40 ng DNA template. The PCR profile was as follows: initial denaturation of 94 °C for 2 min, followed by 28 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min, extension at 72 °C for 3 min, followed by a final extension of 7 min at 72 °C.
Amplification of internal transcribed spacers 1 and 2 (ITS) and the 5·8S gene was carried out in 50-μL reactions, containing 45 µL PCR Abgene mastermix (1·5 mm Mg), 1·5 µL bovine serum albumin (0·04 %), 60 ng H20, 0·6 µL of each ITS primer, 17SE and 25SE (Sun et al., 1994) and approx. 40 ng DNA template. The PCR profile was as follows: initial denaturation of 94 °C for 2 min, followed by 28 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min, extension at 72 °C for 3 min, followed by a final extension of 7 min at 72 °C.
Any ITS trace files showing evidence of introgression in the form of heterogeneous ITS copies were cloned into a vector (pGem-T Easy Vector, Cat. No. A1360; Promega Ltd, Madison, WI, USA) to isolate single sequences. The ITS region was then re-amplified from the transformed bacterial colonies using the M13 primers contained in the kit and a small portion of the colony as the DNA template. Cloning was undertaken for the following taxa: O. aegirtica, O. apifera, O. apulica, O. araneola, O. bombyliflora, O. bornmuelleri, O. bremifera, O. dyris, O. garganica, O. lacaitae, O. mammosa, O. murbeckii, O. pallida, O. phillipei and O. rhodia. Ten ITS clones were obtained for each of the above taxa, though only the example of each clone type with the best sequence was included.
All PCR products were purified using DNA purification columns according to the manufacturers' protocols (QIAquick; Qiagen Ltd, Crawley, UK.). Dideoxy cycle sequencing was then performed using the chain termination method and ABI Prism Big Dye version 3·1 reaction kit, following the manufacturers' protocols (Applied Biosystems Inc., Warrington, UK). The products were run on an ABI 3700 Genetic Analyser, also according to the manufacturers' protocols. Sequence editing and assembly of contigs were performed using Sequence Navigator and AutoAssembler software programs (ABI). All sequences were aligned by eye, following the guidelines of Kelchner (2000).
For the AFLP analysis, a primer trial was conducted using 14 primer combinations to identify pairs of selective primers that would be appropriate to the study. The standard primer Mse1–AGG and a modified (by a fourth selective base) EcoRI–CTAT primer (both 5 mm) were used, following the manufacturers' instructions (MWG Biotech Ltd. UK), to produce AFLP profiles for all Ophrys species, since this combination yielded a suitable number of bands and variation among loci. The addition of an extra base to reduce the number of peaks produced was described by Vos et al. (1995), Fay and Krauss (2003) and Fay et al. (2005) for use in cases where the genome of the plant in question is substantially larger than those of plants for which the AFLP kits are optimized.
Data analysis strategy
The ITS data were analysed using the Fitch parsimony model (equal weight, unordered (Fitch, 1971) and a bootstrapping (Felsenstein, 1985) approach using the software program PAUP 4·0b2A (Swofford, 2001). One thousand replicates were performed using the sub-tree pruning and re-grafting (SPR) algorithm, with MulTrees on but holding only five trees at each step, to reduce the time spent in swapping on large numbers of potentially suboptimal trees. Support for branches was evaluated by bootstrapping using 1000 random addition replicates with simple sequence addition, SPR swapping and holding five trees at each stage.
A Bayesian analysis was also conducted using Mr Bayes version 2·01 (Ronquist and Huelsenbeck, 2003) for the ITS matrix. Prior to the Bayesian analysis, a model of DNA evolution was obtained using the Model Test software version 3·0 (Posada and Crandall, 1998) following the authors' protocols. Model Test implements three different model selection frameworks: hierarchical likelihood ratio tests, Akaike information criterion, and Bayesian information criterion, and the model selected was HKY85. For the analysis itself, 5 000 000 cycles were performed, sampling one tree every 20 generations. A graph of generation number versus log likelihood values was plotted and any trees preceding the plateau phase (burn in) were discarded. A 50 % majority rule consensus was constructed in PAUP 4·0b2A from the remainder of the trees with the ‘include compatible grouping’ and ‘show frequency of all observed bipartitions’ options selected. To test whether taxon order could affect tree topology and nodal support, five replicates of 500 000 cycles with randomized taxon order were also performed, sampling a tree every 40 generations and comparing the results. No incongruence was detected between these topologies and those resulting from the 5 000 000-generation analysis.
Analyses of the plastid datasets, both individually and in combination, were carried out using a maximum parsimony approach incorporating a Fitch parsimony model (equal weight, unordered) and a bootstrapping approach using the software program PAUP 4·0b2A. Analysis settings followed those detailed for the ITS analysis.
AFLP analysis was performed using primers selected for use across the genus. Fragment data were analysed using Genescan (version 2·02) and Genotyper (version 1·1) analysis software (ABI). The AFLP traces were carefully compared by eye to ensure homology of bands. Markers with evidence of ‘false negative’ peaks (small, unscorable peaks in a size range where other samples have larger, scorable peaks) were discarded from all samples. This screening strategy prevented the potential introduction of artefacts into the data due to uneven amplification among samples. Bands ranging in size from 50 bp to 500 bp were scored as present or absent. Principal co-ordinates analysis (PCoA), using the Jaccard similarity coefficient (Jaccard, 1908) to exclude shared zeros, was performed using the program ‘R’ Version 4·0 (Casgrain and Legendre, 2001).
RESULTS
DNA sequencing
Figure 2 shows one of 6287 equally most-parsimonious trees obtained with maximum parsimony. The trees produced by both parsimony and Bayesian inferences were congruent with respect to the groups recovered. The analyses were undertaken with outgroups included but, in order to facilitate the display of the resulting tree as a phylogram, these taxa were reduced to a single aggregate terminal in the tree presented.
Fig. 2.
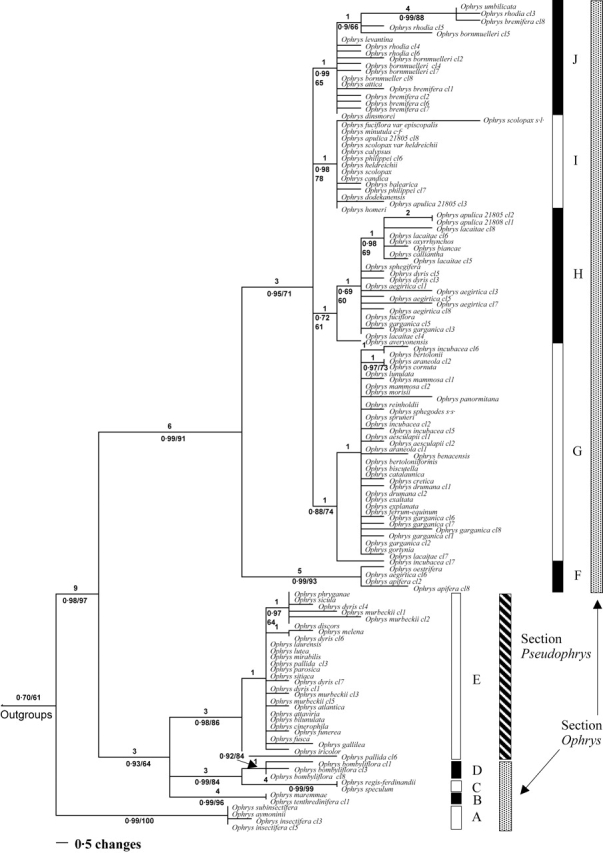
One of 6287 equally parsimonious trees for the transcribed spacers 1 and 2 (ITS) and the 5·8S gene dataset. Numbers above lines represent branch lengths. Of the numbers below the branches, the first is the Bayesian posterior probability (pp) and the second the bootstrap percentage (bp). Values <50 % not shown.
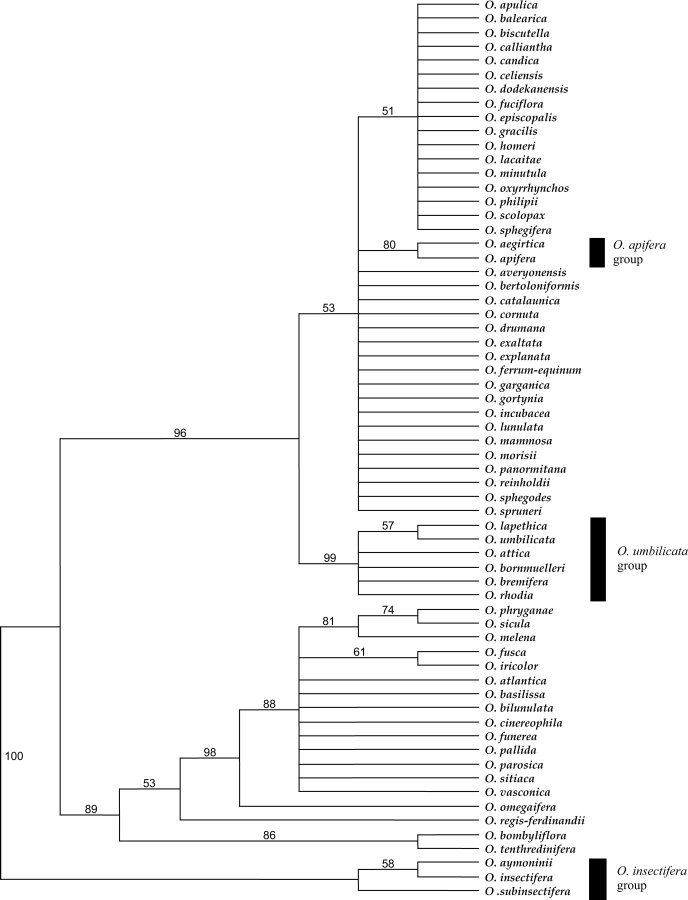
The analyses confirm that Ophrys is monophyletic [posterior probability (pp) 0·98, bootstrap percentage (bp) 97], as shown in previous studies (Cozzolino et al., 2001; Bateman et al., 2003). Outgroups are placed in the following order: Neotinea maculata, Orchis italica, Steveniella satyrioides as successive sister clades to a combined ((Anacamptis laxiflora + Serapias lingua) Ophrys) clade. Ophrys can be divided into ten clades. These clades (labelled A–J in Fig. 2) are, with one exception, subsets of section Ophrys, namely the groups corresponding to O. insectifera (A), O. tenthredinifera (B), O. speculum (C), O. bombyliflora (D), O. apifera (F), O. sphegodes s.l. (G), O. fuciflora s.l. (H), O. scolopax s.l. (I) and O. umbilicata (J). Section Pseudophrys (E) forms a monophyletic group nested within a paraphyletic section Ophrys (sensu Delforge).
The O. insectifera clade (A) is strongly supported (pp 0·99, bp 100) and is placed with weak support as sister to the rest of Ophrys (pp 0·70, bp 61). Within the clade containing groups A–E, the O. tenthredinifera group (B) is strongly supported as a monophyletic entity with a posterior probability of 0·99 (bp 96). The O. speculum group (C) is strongly supported (pp 0·99, bp 99) and is sister to the morphologically contrasting O. bombyliflora group (D) (pp 0·99, bp 84). Group E (Section Pseudophrys) is well supported with a posterior probability of 0·98 (bp 86). Although the precise relationships between group B, groups (C and D) and group E remain uncertain, they form a clade in all the most parsimonious trees. This clade receives a posterior probability of 0·93 (bp 64) indicating that these groups are more closely related to each other than to any other Ophrys species sampled.
Cloning of the ITS amplification products from O. dyris produced heterogeneous ITS copies, and the respective placements of these copies challenge the monophyly of section Pseudophrys. Three copy types were, as expected, placed in Group E (i.e. section Pseudophrys), but the other two were located in the O. fuciflora clade (H), part of section Ophrys.
There is strong support (pp 0·99, bp 91) for a clade containing several of the groups assigned to section Ophrys by Delforge (groups F, G, H, I and J), but this excludes other groups traditionally assigned to section Ophrys, specifically the O. insectifera, O. speculum, O. bombyliflora and O. tenthredinifera groups. Within this clade the O. apifera group (F) (pp 0·99, bp 93) is strongly supported as sister to a clade containing the O. sphegodes s.l., O. fuciflora s.l., O. scolopax s.l. and O. umbilicata s.l. groups (G, H, I and J, respectively) with a posterior probability of 0·95 (bp 71). In addition to O. apifera, this group contains O. oestrifera, which is treated by Delforge and many others as a synonym of O. apifera. Additionally, this group contains one ITS copy type from O. aegirtica.
There is good support for an O. sphegodes clade (G) with a posterior probability of 0·88 (bp 74), although relationships within the group are poorly resolved. Some taxa previously identified as members of this group also appear in other clades due to their possession of heterogeneous ITS copies. For example, O. garganica has ITS copies that are also placed in the O. fuciflora s.l. clade, suggesting either this species is of relatively recent hybrid origin or introgression has occurred in the genealogy of the sampled specimen.
There is weak support (pp 0·72, bp 61) for the O. fuciflora s.l. clade (H), which also contains taxa with ITS copies appearing elsewhere in section Ophrys. As previously stated, O. aegirtica has one copy type that is strongly supported as belonging to the O. apifera clade. Some copy types from O. dyris are also located in the O. fuciflora s.l. clade. In addition, O. lacaitae possesses an ITS copy type that is placed in the O. sphegodes clade (G), and O. apulica has ITS copies that appear in the O. scolopax clade (I).
Clade I contains taxa that generally correspond to the O. scolopax s.l. group and is well supported (pp 0·98, bp 78). Clade J (pp 0·99, bp 65) consists of taxa that had previously been placed in the O. bornmuelleri, O. scolopax and O. umbilicata groups sensu Delforge; consequently the groups corresponding to O. bornmuelleri and O. scolopax appear paraphyletic, as members of both of these groups are distributed between more than one clade.
No well-supported examples of incongruence were found between phylogenetic reconstructions produced by maximum parsimony and Bayesian analysis.
Figure 3 shows one of the 2053 most-parsimonious cladograms for plastid data; specifically, the combined trnH–psbA intergenic spacer and trnD–trnT intergenic spacer data matrices. The topology is largely congruent with that of the nuclear ITS tree described from Figure 2. The main differences are the position of the O. insectifera group, which in this analysis is placed as sister to all other Ophrys, and the placement of the O. umbilicata group rather than the O. apifera group as sister to the non-Pseudophrys clade. Both of these incongruencies have little bootstrap support in the plastid tree.
Fig. 3.
One of the 2053 most parsimonious trees for the combined trnH–psbA intergenic spacer and trnD–trnT intergenic spacer data matrices. Numbers represent the bootstrap percentage (bp).
AFLP
With 79 accessions spanning 74 putative species, the AFLP dataset contains fewer taxa than the sequencing analysis, due to the greater sensitivity to DNA quality of AFLP in comparison to DNA sequencing. All ingroup taxa (listed in Appendix 1) were originally included in the AFLP study, but some AFLP traces from the analysis were subsequently discarded due to weak signal strength.
The primer combination selected resulted in the generation of 165 AFLP markers, each accession possessing between 22 and 41 peaks. The sample of O. dyris had the most peaks (41), the next highest having only 35. This may reflect hybrid additivity, as this accession is the only sample that fell outside sections Ophrys and Pseudophrys as depicted in the PCoA. The PCoA analysis derived from AFLP data (Fig. 4A) shows partitioning of Ophrys species into four discrete clusters that correspond to amalgamations of groups recovered in the ITS analysis. Letters in parentheses refer to clades recovered in the ITS analysis. Clusters correspond to the O. insectifera group (A), the O. tenthredinifera group (B–D), section Pseudophrys (E) and the O. apifera group (F–J). The O. apifera cluster itself contains two groups: the O. apifera group (F) and the O. fuciflora group (G–J). Subgroup fuciflora contains two clusters that correspond to the O. umbilicata aggregate (J) and an aggregate that is a combination of subgroup O. sphegodes s.l., subgroup O. fuciflora s.l. and subgroup O. scolopax s.l. (G, H and I, respectively). Details of taxa contained within each of the above-mentioned clusters are given in Appendix 2.
Fig. 4.
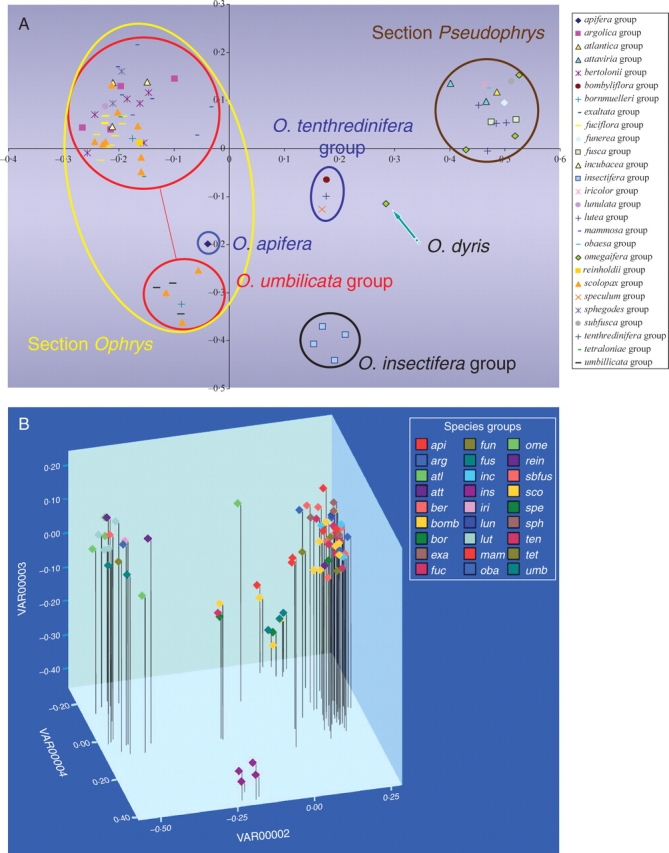
(A) PCoA plot of AFLP results. (b) Three-dimensional PCoA plot of AFLP results. Species groups: api, apifera; arg, argolica; atl, atlantica; att, attaviria; ber, bertolonii; bomb, bombyliflora; bor, bornmuelleri; exa, exaltata; fuc, fuciflora; fun, funerea; fus, fusca; inc, incubacea; ins, insectifera; iri, iricolor; lun, lunulata; lut, lutea; mam, mammosa; oba, obaesa; ome, omegaifera; rein, reinholdii; sbfus, subfusca; sco, scolopax; spe, speculum; sph, sphegodes; ten, tenthredinifera; tet, tetraloniae; umb, umbilicata.
Ophrys dyris, of the O. omegaifera group, is placed between the section Pseudophrys cluster and the cluster representing the larger subset of section Ophrys. It is apparent from the combination of sequencing and AFLP analysis that either this particular accession represents an inter-sectional hybrid individual or the species per se is of hybrid origin. Since only one accession labelled O. dyris was included in this particular analysis, it is impossible at present to determine which of these scenarios is correct. Ophrys aegirtica, which showed evidence of hybridization with O. apifera in the ITS analysis, is positioned on the periphery of the larger section Ophrys cluster and, when viewed in the three-dimensional PCoA space (Fig. 4B), it is the closest member of this cluster to O. apifera.
The groups corresponding to O. bornmuelleri and O. scolopax (sensu Delforge) are split between different clusters in the PCoA. Some members are found in the O. umbilicata cluster, whereas others are positioned in the larger section Ophrys subset Table 1.
DISCUSSION
Species concepts
One of the most crucial decisions to be made when dealing with the systematics of any group, but in particular a group of closely related taxa like Ophrys distributed in a biodiversity hotspot such as the Mediterranean, is selecting the appropriate species concept and species delimitation method. Since all subsequent descriptions and implications depend on these initial assumptions, these need to be specified for any investigation of this genus.
Many of the previous attempts to classify Ophrys species have adopted a loosely defined and intuitive approach to taxonomy, based largely on field observations while incorporating analysis of some micro-morphological characters (e.g. Devillers and Devillers-Terschuren, 1994; Delforge, 2001, 2005). In effect, the problems posed by homoplasy are ignored. These are especially important in a pseudocopulatory pollination scenario, in which sympatric taxa are supposedly reliant on differential pollinator attraction and pseudocopulatory positioning for reproductive success. Convergence in morphology is likely if plants from separate lineages share pollinators. It was due to the problems associated with potential homoplasy that the phenetic species concept (at least, when applied to only morphological characters) was discarded as inappropriate for this study.
The data presented in this study and elsewhere (e.g. Soliva and Widmer, 2003) clearly show evidence of introgression in many of the taxa sampled. Since intersectional hybrids are evident (Bournérias and Prat, 2005; Bateman and Devey, 2006), adoption of the biological species concept (Mayr, 1963), which recognizes actually or potentially inbreeding individuals as members of the same species, is impractical. Nevertheless, some consideration of isolation through pollinator behaviour, as inferred by patterns of introgression, seems desirable for species delimitation in this group. In older species groups, where sufficient time has passed for fixed genetic variation to accumulate to a level detectable by conventional sequencing, the adoption of a pattern-based approach is relatively straightforward. However, within a genus such as Ophrys, where there is an absence of complete differentiation and lineage sorting due to the suspected recentness of the radiation and the likelihood of continuing introgression, an approach incorporating a combination of the two concepts is more appropriate. Specifically, for this study a fundamentally phylogenetic pattern cladistic approachadopted, but it still incorporated a mechanistic component in which putative species are viewed as participants in an ongoing evolutionary process, rather than the fixed end-products of the process. It is this ongoing introgression, both within and between members of different species groups, which gives rise to the ‘fuzzy margins’ of the species clusters described here.
We believe that this combined ‘pattern–process’ model is a pragmatic compromise that best reflects the morphology and evolutionary biology of the group. This approach is broadly congruent with that of Sundermann (1980) and Bateman (2001), but differs radically from that of Devillers and Devillers-Terschuren (1994) and Delforge (1995 et seq.), in which many minor variants and morphological oddities are categorized as full species.
With this in mind, two interpretations are possible for the patterns of variation shown in Ophrys: an incipient speciation scenario and a reticulate evolution scenario.
(1) Incipient speciation, in which several species clusters are recognized. The self-imposed criteria for the species name chosen to represent these clusters are prioritized as follows: (a) they must fall wholly within one of the lettered groups described in the Results section, (b) they must be geographically widespread, (c) they must be central to a cluster of genetically distinct entities, and (d) they must be central to a morphologically distinct suite of characters. These species clusters are equivalent to the clades recovered in the ITS and AFLP analyses and it is assumed that they actually attract a specific spectrum of pollinators. In addition to these species, there also exist reproductively suboptimal species, which are still sufficiently successful in terms of pollinator attraction to maintain populations and reproduce to a level where it is not uncommon to find their offspring. This is demonstrated by the presence of heterogeneous peaks in ITS electropherograms for some accessions prior to cloning. This variation demonstrates evidence of relatively recent gene flow, where insufficient time has passed for complete copy conversion. The contribution of each progenitor lineage can be estimated as the ratio of copy types that would be proportional to the ratio of peak heights (Y. Pillon, pers comm, 2005). These species may be present due to the existence of odour-bouquet variability within populations produced by negative frequency-dependent selection in response to pollinator learning (Moya and Ackerman, 1993), which would lead to mistakes by pollinators and hence to introgression between species.
In terms of lowered fitness through lowered reproductive viability, members of Orchidaceae pose an additional problem. Due to the abundance of dust-like seed produced by Ophrys (typically 5000–10 000 per fertilized capsule), even a dramatically reduced fruiting success can result in hundreds or thousands of viable seeds. Thus, infrequent aberrant pollination events can permit the dispersal of the offspring of these slightly suboptimal ‘species’ and thereby induce ‘fuzzy margins’ to species clusters. This situation may occur because the group is too young for divergence to have taken place and for reliable genetic or behavioural barriers to reproduction to have become established. There may, in addition, be an anthropogenic component to Ophrys speciation, as many species favour ‘man-made’ (anthropogenic) and ‘man-maintained’ (anthropostatic) environments such as grazed but unimproved scrubland and grassland. In this case, there may not have been any time in the past when reproductive isolation was complete (such as glacial refugia). Phenotypic variation in floral morphology, and probably more importantly pheromone mimicry among sympatric individuals, would attract different suites of pollinators and lead to the formation of putative ‘prospecies’ by the erection of initially weak reproductive barriers. Different ‘species’ may coalesce if they share pollinators and introgress to a level where no pure individuals of the parent species remain (hybrid speciation). An example of speciation based on geographical vicariance could be the O. umbilicata group (as presently circumscribed), in which the members are located exclusively in the eastern Mediterranean.
(2) Reticulate evolution by the mechanism of incipient genetic homogenization. At some point in the past, there was sufficient isolation of Ophrys populations, perhaps in glacial refugia, to lead to speciation. Due to the subsequent increase in areas habitable by Ophrys, reflecting post-glacial recolonization and/or anthropogenic effects on European biogeography, the ranges now overlap, allowing increased levels of hybridization and introgression between species that still lack effective mechanisms of reproductive isolation. If this scenario is valid, homogenization of the genomes of formerly distinct Ophrys species is likely to increase through time, potentially as a result of anthropogenic disturbance and possibly climate change.
The two scenarios outlined above need not be mutually exclusive. However, it is important to determine how much gene exchange between diverging populations or groups of genetically distinct entities is possible without arresting, or even reversing, the divergence. Introgression among populations tends to make gene pools progressively more similar. Nevertheless, populations or meta-populations may continue to diverge despite some introgression, if it is subordinate to divergent selection or stochastic processes such as genetic drift. Net genetic convergence or divergence, at least in sympatry, will be determined by the balance of these opposing forces. Exactly how much isolation is required (be it contrasting geographical distributions, habitat preferences or pollinators) to permit prospecies formation and divergence in sympatry depends upon the intensity of the differentiating selection. It seems probable that, within Ophrys, the average level of genetic divergence slightly exceeds that of genetic reticulation. There may be a breakout threshold within Ophrys in terms of levels of genetic differentiation, which has been exceeded by each of the groups A–J in Fig. 2. This threshold would represent a point at which phenotypic or genotypic differences between groups or populations have a negative effect on the likelihood of successful reproduction given the available pollinators in those locations. Once this threshold has been passed and additional divergence takes place, lineages may undergo an evolutionarily positive feedback loop as fixed changes accrue more rapidly due to increased isolation. It is interesting to speculate, though almost impossible to determine, how many times this threshold has been approached, only for introgression to eradicate the divergence.
Phylogeny reconstruction
Variation observed within the sampled plastid loci (Fig. 2) among Ophrys species was, as expected, low. A total of 2211 nucleotide characters was sequenced from two non-coding regions. Analysing data for each plastid locus independently provided insufficient potentially parsimony-informative characters for well-resolved phylogenetic reconstructions. Even when the regions were combined, few of the groups were resolved with high levels of bootstrap support. Combining the plastid datasets into a single matrix provided increased resolution, but nevertheless only 4 % of characters proved potentially parsimony-informative. Improved resolution was obtained from the ITS analysis, in which 21 % of characters proved potentially parsimony-informative. Even then, clade resolution was concentrated along the spine of the tree, with little or no resolution near the tips. High consistency and retention indices indicated that most of the variable sites were congruent. Low bootstrap support was expected for many clades, as re-sampling within a matrix is likely to remove groups defined by only one or two character states. No hard incongruence was found among the different partitions. The O. insectifera group proved the most labile, albeit with only weak bootstrap support for any particular position. The O. apifera group was also somewhat labile, typically appearing as sister to the remainder of section Ophrys but occasionally as sister to the O. umbilicata group.
Unfortunately, due to the rarity and conservation protection offered to many Ophrys species, in some cases it was only possible to collect very small amounts of plant tissue to avoid excessive damage to the plant. This lack of material and therefore the limited amount of extracted DNA available meant that it was not possible to amplify all the DNA-based markers for all of the sampled taxa. It was the combination of this limitation and the presence of polymorphic ITS sequences that prevented production of a combined plastid and nuclear DNA phylogenetic reconstruction.
These results demonstrate that, within a potentially actively evolving group such as Ophrys, species-level trees can be difficult to reconstruct when based on DNA sequence data alone. Sequences from loci routinely exploited for phylogenetic purposes may not contain sufficient signal to make confident statements about relationships, even when the group has been well sampled for a large number of nucleotides in rapidly evolving regions of the various genomes. In an attempt to overcome this limitation, AFLP techniques were incorporated. The use of AFLP data for tree-building purposes can present problems due to the assumption of homology of all co-migrating fragments and the difficulties associated with assigning homology to alleles of differing sizes. However, this approach may be justified as the use of modern automated genetic analysers greatly increases the accuracy of fragment size measurement, and therefore resolution of the technique with regard to the co-migrating fragments. More pragmatically, within this project at least, results are consistent with those produced by the sequence analyses, and this mirrors the congruence between datasets described by other authors using the same combination of techniques (e.g. Richardson et al., 2003; Goldman et al., 2004; Koopman, 2005).
As putative species within groups in the genus Ophrys are so closely related, it is almost impossible to rule out chimaeric recombination within those groups, though even if it had taken place the groups are so similar in terms of ITS sequences that any conclusions reached would not be affected. Sequences were visually inspected for evidence of chimeras; however, wherever heterogeneous sequences indicating hybridization or introgression between groups (notably O. dyris) were found, cloned sequences fell in either one or the other putative parental groups and not in intermediate positions in the phylogenetic analysis, suggesting chimaeric recombination had not taken place. There is the possibility that chimeric sequences could be generated from combining fragments of ITS copy types inherited from different species groups but had these sequences been amplified, the analyses of nuclear and plastid sequencing and those of AFLP data would have displayed incongruencies that were not evident.
The best-supported estimate of relationships appeared to be from combined analysis of AFLP and sequence data: the retention index remained high and, although the consistency index was lowered, it remained sufficiently high for continued statistical confidence in tree topology. Additionally, analysis of AFLP, plastid DNA and nuclear DNA all independently recovered the same groups, including the novel, geographically constrained O. umbilicata group, indicating that the combined data provide relatively stable and reliable estimates of relationships at the level of species groups.
The results produced in this molecular study show some incongruence with those of previous studies based solely on morphological characters. For example, we refute the suggestion of Devillers & Devillers-Terschuren (1994) that section Ophrys is best divided into three assemblages organized around the O. insectifera, O. speculum and O. bombyliflora–fuciflora–sphegodes groups. Their study also used shape of the labellum, details of the stigmatic cavity and organization of labellum pilosity as morphological synapomorphies to suggest that O. speculum is sister to the rest of the section, but the present molecular data strongly reject this inference.
Analysis of the O. aegirtica sample used in this study demonstrates conclusively that introgression between members of different groups has taken place. As this accession was collected from the type locality of the species, we can be confident that no misidentification has taken place. Because the O. aegirtica site was within 200 m of a population of O. apifera s.s., we propose that O. aegirtica is of recent hybrid origin and should be considered a nothospecies.
The Bayesian posterior probabilities and levels of bootstrap support for some nodes within the phylogram (Fig. 2), particularly those that would assign sister relationships between the larger, better differentiated groups, are relatively low. Only once posterior probabilities exceed >0·9 and bootstrap percentages exceed 85 % can we be reasonably confident of support for a particular tree topology. Hence, we are confident that these species groups are meaningful entities, but the relationships among them must still be considered cryptic. Although the fine detail observed within phylograms has only moderate support and collapses into several large polytomies, the fact that the taxa are sufficiently closely related to allow reliable scoring of AFLP bands across the entire genus provides an indication of the closeness of the relationships among these species.
We believe the congruence between the ITS and AFLP datasets, which respectively sample a very small section of the nuclear genome and many loci distributed across the genome, lend credibility to our conclusions. Taken together, these data refute claims of Paulus and Gack (1990), that the supposedly species-specific pseudocopulatory pollination syndrome of Ophrys provides reliable reproductive isolation among species.
Two explanations are feasible for the poorer differentiation within the fuciflora–scolopax–sphegodes clade using AFLP analysis relative to the ITS analysis. The first is evolutionary and the second technical. Firstly, in species of recent origin, few changes will have become fixed and even then only in a few functional genes directly involved in the speciation event, which form a small proportion of the whole genome sampled by AFLP. Less constrained regions of the genome, such as the ITS spacers, are not likely to have participated in the speciation event but will subsequently accumulate changes more rapidly than most other regions (Bateman, 1999). There may not have been sufficient time for the accumulation of changes in plastid regions (which in general accrue sequence changes more slowly) to track those occurring in more rapidly mutating regions. As AFLP techniques sample restriction sites from across the genome there will be evolutionary rate heterogeneity between the sites sampled. Consequently, phylogenetic patterns recovered by the quickly evolving markers may be swamped by the addition of regions that evolve more slowly.
The second, technical factor reflects the rationale behind AFLP primer selection. The initial focus of the present investigation was based on a need to obtain primers for use across the genus. Consequently, primers that could potentially have differentiated between closely related members of section Ophrys were not selected. Further studies (D. S. Devey et al., unpubl. res.) will therefore be needed to address Ophrys systematics within the major species groups recognized here.
Taxonomic implications
Delimitation of the O. scolopax s.l. group is based mainly on the occurrence and size of the ‘horns’ on the lateral lobes of the labellum. The present data suggest that the O. scolopax group, as currently delimited (Figs 2 and 3), is polyphyletic; several lineages have independently converged on this visually distinct morphology. We assert that the presence of long ‘horns’ cannot be used as a synapomorphy delimiting the group.
Some sampled taxa show evidence of introgression, as ITS copy types appear in multiple, well-supported clades. It is therefore proposed that they be treated as taxa of hybrid origin and excluded from the aforementioned groupings. The taxa are as follows: O. aegirtica, O. apulica, O. araneola, O. dyris, O. garganica, O. lacaitae, O. mammosa, O. murbeckii, O. pallida and O. phillipei. Should these patterns be repeated in a wider range of accessions of these taxa, we suggest that for systematic and conservation purposes, they be treated as hybrids.
We attribute these patterns to introgression rather than any contamination of samples, as in many cases evidence of hybridization was expected from field observations of the taxa concerned. Additionally, careful examination of samples at every step of the extraction and PCR process, and robustness of data produced in other laboratory studies using the same extraction techniques indicates that the chances of cross-contamination are remote.
Several species groups (sensu Delforge, 2005) are polyphyletic with respect to ITS sequences, including the O. bornmuelleri group, the O. tetraloniae group, the O. scolopax group and the O. tenthredinifera group.
Biogeography
This study clearly demonstrates biogeographic partitioning within section Ophrys. Members of the O. umbilicata group (as redelimited using genetic evidence), which are genetically distinct and therefore presumably reproductively isolated from the remainder of the section, occur exclusively to the south-east of the Balkans. Within the group, Ophrys umbilicata s.s. has the widest reported distribution, stretching from Greece in the north-west to Israel and Jordan in the south and Asiatic Turkey in the east. As the accession used in this study was collected on Cyprus, any future study should analyse O. umbilicata from the northern and western extremes of its distribution. The distribution of the O. umbilicata group (as recircumscribed here) is centred on Cyprus, but two members are located in southern Greece. One species from section Ophrys (O. levantina) was sequenced from Cyprus and did not appear in this clade, thus ruling out the possibility that all the relevant species from this island are undergoing extensive introgression.
General conclusions
Despite the interesting results currently being produced by researchers in the field of pheromone-mediated pollinator attraction, until the odour bouquets of many more putative species (and many individuals sampled from a wide geographical spectrum within species) have been examined and overlaid onto DNA-based frameworks, it will be impossible to determine with any degree of certainty which component(s) of pollinator attraction selection is acting upon. In particular, pollinator data need to be obtained from the full range of target species and their relatives, ideally sampled throughout the flowering season. Only then can gene flow be estimated directly, rather than indirectly though genetic fingerprinting of the orchids themselves as reported here.
As a general approach to Ophrys systematics, we suggest caution with regard to segregation of the genus into trivial partitions. The partitions must be demonstrably real and should ideally be based on a combination of discrete characters: genetic, chemical and/or morphological. Such characters should be prioritized over arbitrary divisions of continuous morphological spectra that are hypothesized to coincide with species-specific pollinator relationships. This is particularly important, as the identification of many putative pollinators is itself contentious, and frequently based on largely anecdotal – and, most crucially, geographically restricted – evidence of pollinator visitation. Although, for both systematics and conservation purposes, it would be remiss to ignore a biologically significant species or species group, at the same time it is a severe hindrance to conservation bodies to have to factor into their plans poorly substantiated taxa or taxonomic groupings. This is particularly true with relation to supposed island endemics, which would receive a high conservation priority as they stand but would be considerably less significant if proved to be trivially distinct local morphological variants of the same species or comparatively transient hybrid swarms.
ACKNOWLEDGEMENTS
The authors thank the John Spedan Lewis Foundation (Howard Frank Layzell Scholarship) for funding this research. We also acknowledge Mark Chase, Paula Rudall, Salvatore Cozzolino and Laure Civeyrel for their constructive insights and help with fieldwork. Additionally, Barry Tattersall and other members of the Hardy Orchid Society, and Giuseppe Tosi in Italy provided invaluable plant samples and local knowledge. Finally, thanks also go to staff and colleagues in the Jodrell Laboratory for their continuing support and technical expertise.
APPENDIX 1
Table A1.
List of all Ophrys taxa, Genbank ID numbers and voucher information
| Species | Voucher | Source | ITS Genbank ID | trnD–trnT Genbank ID | trnH–psbA Genbank ID |
|---|---|---|---|---|---|
| Anacamptis laxiflora (Lam.) Bateman, Pridgeon and Chase | Bateman 4 | Crete, marsh imm. W Frangocastello Castle, E Hora Sf.,12 April 1996 | AM711747 | AM711971 | AM711707 |
| Neotinea maculata (Desf.) Stearn | Bateman 35 | 1·2 km SE Karines, Karines–Spili road, Crete; 19 April 1996 | AM711744 | AM711970 | AM711706 |
| Ophrys aegirtica P.Delforge | Fay 555 | AM711759 AM711760 | AM711905 | AM711641 | |
| AM711761 | |||||
| AM711762 | |||||
| Ophrys aesculapii Renz | Chase O- 901 K | Oncidiinae, Kew 1984-8117 | AM711787 AM711788 | N/A | N/A |
| Ophrys apifera Huds. | Chase 13839 | England, Avon, off A432 between Bristol and Chipping Sodbury, Cuckoo Lane, 16 June 2002 | AM711789 AM711790 | AM711906 | AM711642 |
| Ophrys apulica | Bateman 705 | Bosco di Pianelle, Martina Franca, Italy | AM711794 AM711795 | AM711907 | AM711643 |
| Ophrys apulica (Danesch) O.Danesch & E.Danesch ex Gölz & H.R.Reinhard | Bateman 696 | Massafra-Martina Franca, Italy | AM711791 | N/A | N/A |
| AM711792 | |||||
| AM711793 | |||||
| Ophrys araneola Rchb. | Chase O- 701 K | Orchidinae, Kew 1983-8174 | AM711796 AM711797 | N/A | N/A |
| Ophrys atlantica Munby | Bateman 208 | Alhaurin el Grande, Malaga, Spain | AM711798 | AM711908 | AM711644 |
| Ophrys attaviria D.Rückbr., U.Rückbr., Wenker & S.Wenker | Bateman 209 | Alhaurin el Grande, Malaga, Spain | AM711799 | N/A | N/A |
| Ophrys attica Soó | Bateman 1158 | AM711800 | AM711909 | AM711645 | |
| Ophrys aveyronensis (J.J.Wood) H.Baumann & Künkele | Bateman 1236 | AM711801 | AM711910 | AM711646 | |
| Ophrys aymoninii (Breistr.) Buttler | Bateman 1235 | AM711802 | AM711911 | AM711647 | |
| Ophrys balearica P.Delforge | Bateman 257 | Cabo Gros, Alcudia, Mallorca | AM711803 | AM711912 | AM711648 |
| Ophrys basilissa | DSD14 | N/A | AM711913 | AM711649 | |
| Ophrys benacensis (Reisigl) O.Danesch & E.Danesch | Bateman 571 | Picnic site NW Gardola, N Lake Garda, NW Verona, Italy | AM711804 | N/A | N/A |
| Ophrys bertoloniformis O.Danesch & E.Danesch | AM711806 | AM711914 | AM711650 | ||
| Ophrys bertolonii Moretti | Devey DSD 60A | Sicily, Forest of Ficuzza April 2004 | AM711805 | N/A | N/A |
| Ophrys biancae Macch. | Bateman 527 | 4·1 km Pantalica-Ferla, SW Sortino, Syracusa, Sicily | AM711807 | N/A | N/A |
| Ophrys bilunulata Risso | Chase 16363 | Crete, SW Bridge, Kanevos-Kali Sikea rd, NW Spili, c. 14 April 1996 | AM711808 | AM711915 | AM711651 |
| Ophrys biscutella O.Danesch & E.Danesch | Bateman 653 | Monte Sacro, Gargano, Italy | AM711809 | AM711916 | AM711652 |
| Ophrys bombyliflora Link | Bateman 22 | SW Bridge, Kanevos–Kali Sikea road, NW Spili, Crete; 14 April 1996 | AM711810 AM711811 | AM711917 | AM711653 |
| Ophrys bornmuelleri (s.s.) Schulze | Bateman 396 | SE Cyprus | AM711766 | AM711918 | AM711654 |
| AM711767 | |||||
| AM711768 | |||||
| AM711769 | |||||
| AM711770 | |||||
| Ophrys bremifera (c.f.) Stev. ex M.Bieb. | Bateman 297 | Skopelos, Greece | AM711812 | AM711919 | AM711655 |
| AM711813 | |||||
| AM711771 | |||||
| AM711772 | |||||
| Ophrys calliantha Bartolo & Pulv. | Devey DSD43 | Sicily E Pantalica. April 2004 | AM711815 | AM711920 | AM711656 |
| Ophrys calypsus M.Hirth & H.Spaeth | Bateman 445 | Agiasos, Lesvos | AM711816 | N/A | N/A |
| Ophrys candica (E.Nelson ex Soó) H.Baumann & Künkele | R. Bateman 037 | Crete 29a/96 | AM711814 | AM711921 | AM711657 |
| Ophrys catalaunica O.Danesch & E.Danesch. | Bateman 321 | Barcelona, Spain. | AM711817 | AM711922 | AM711658 |
| Ophrys celiensis | DSD11 | N/A | N/A | AM711923 | |
| Ophrys cinerophila Paulus & C.Gack | Bateman 225 | Tekke, Cyprus | AM711818 | AM711924 | AM711660 |
| Ophrys cornuta Stev. ex M.Bieb. | Bateman 287 | Kokkonicasto, Greece | AM711819 | AM711925 | AM711661 |
| Ophrys cretica (Vierh.) Erich Nelson | Chase O- 709 K | Orchidinae, Kew 1983-5694 | AM711820 | N/A | N/A |
| Ophrys dinsmorei | AM711822 | N/A | N/A | ||
| Ophrys discors Bianca | Cozzolino 1827 | AM711821 | N/A | N/A | |
| Ophrys dodekanensis H.Kretzschmar & Kreutz | Bateman 1148 | AM711823 | AM711926 | AM711662 | |
| Ophrys drumana (c.f.) P.Delforge | Civegiel et al. 502 | France, nr Murs. similar to Delforge ed.1lower photo page 439. | AM711824 | AM711927 | AM711663 |
| AM711825 | |||||
| Ophrys dyris Maire | Bateman 93 | Faunia, Portugal | AM711749 | N/A | N/A |
| AM711750 | |||||
| AM711751 | |||||
| AM711752 | |||||
| AM711753 | |||||
| AM711754 | |||||
| Ophrys episcopalis Poir. | Bateman 17 | AM711826 | AM711932 | AM711668 | |
| Ophrys exaltata Ten. | Bateman 365 | C sicily | AM711827 | AM711928 | AM711664 |
| Ophrys explanata (Lojac.) P.Delforge | Devey DDA | Sortino, Sicily 2004 | AM711828 | AM711929 | AM711665 |
| Ophrys ferrum-equinum Desf. | Chase O- 707 K | Orchidinae, Kew 1984 2540 | AM711829 | AM711930 | AM711666 |
| Ophrys fuciflora (F.W.Schmidt) Moench | Devey DSD29 | Sicily, E Pantalica 2004 | AM711830 | AM711931 | AM711667 |
| Ophrys funerea Viv. | Chase 16364 | Crete, SW Bridge, Kanevos-Kali Sikea rd, NW Spili, c. 14 April 1996 | AM711831 | AM711933 | AM711669 |
| Ophrys fusca subsp. fusca Link | Chase O- 711 K | Orchidinae, Kew 1983-5696 | AM711832 | AM711934 | AM711670 |
| Ophrys galilaea H.Fleischm. & Bornm. | Cozzolino 865 | AM711836 | N/A | N/A | |
| Ophrys garganica E.Nelson ex O.Danesch & E.Danesch | Bateman 671 | Monte San Angelo, Gargano, Italy | AM711833 | AM711935 | AM711671 |
| AM711834 | |||||
| AM711835 | |||||
| AM711764 | |||||
| AM711765 | |||||
| Ophrys gortynia (Baumann & Künkele) Paulus | Bateman 24 | Prov.: Hillock imm. N Agia Triada ruins, Timbaki, Crete; 16 April 1996 | AM711837 | AM711936 | AM711672 |
| Ophrys gracilis | N/A | AM711937 | AM711673 | ||
| Ophrys heldreichii Schltr. | Bateman 13 | Orchidinae | AM711734 | N/A | N/A |
| AM711838 | |||||
| Ophrys homeri M.Hirth & H.Spaeth | Bateman 1104 | AM711839 | AM711938 | AM711674 | |
| Ophrys incubacea Bianca ex Tod. | Devey DSD62A | Sicily, Forest of Ficuzza; coll. April 2004 | AM711783 | AM711939 | AM711675 |
| AM711784 | |||||
| AM711785 | |||||
| Ophrys insectifera L. | Fay 570 | Italy,W Verona, E L Garda, Bizzano-St Zeno | AM711840 | AM711940 | AM711676 |
| Ophrys iricolor Desf. | Bateman 33 | Crete 24/96 | AM711841 | AM711941 | AM711677 |
| Ophrys lacaitae Lojac. | Devey DSD36 | Sicily, E Pantalica 2004 | AM711708 | AM711942 | AM711678 |
| AM711709 | |||||
| AM711842 | |||||
| AM711763 | |||||
| AM711777 | |||||
| Ophrys lapethica | AM711943 | AM711679 | |||
| Ophrys laurensis Geniez & Melki | Cozzolino 1417 | AM711710 | N/A | N/A | |
| Ophrys levantina Gölz & H.R.Reinhard | Bateman 230 | Limasol, Cyprus | AM711711 | N/A | N/A |
| Ophrys lunulata Parl. | Devey DSD1 | Sicily, Pantalica Road. Limestone bank across gorge from Sortino 2004 | AM711712 | AM711944 | AM711680 |
| Ophrys lutea Biv. | Fay 543 | AM711713 | N/A | N/A | |
| Ophrys mammosa Desf. | Bateman 12 | Orchidinae | AM711714 | AM711945 | AM711681 |
| AM711715 | |||||
| Ophrys maremmae O.Danesch & E.Danesch. | Cozzolino 974 | AM711716 | N/A | N/A | |
| Ophrys melena (Renz) Paulus & Gack | Devey DSD66A | Sicily, Forest of Ficuzza; April 2004 | AM711717 | AM711946 | AM711682 |
| Ophrys minutula c.f. Gölz & H.R.Reinhard | Bateman 1107 | AM711718 | AM711947 | AM711683 | |
| Ophrys mirabilis P.Geniez & F.Melki | Cozzolino 1408 | AM711719 | N/A | N/A | |
| Ophrys morisii (Martelli) G.Keller & Soó | Bateman 1232 | AM711720 | AM711948 | AM711684 | |
| Ophrys murbeckii H.Fleischm. | Cozzolino 1329 | AM711773 | N/A | N/A | |
| AM711774 | |||||
| AM711775 | |||||
| AM711776 | |||||
| Ophrys oestrifera M.Bieb. | Bateman 682 | Vieste-peschicchi, Gargano, Italy | AM711721 | N/A | N/A |
| Ophrys omegaifera H.Fleischm. | Bateman 364 | Purchased | N/A | AM711949 | AM711685 |
| Ophrys oxyrrhynchos Tod. | Devey DSD50 | Sicily, Forest of Ficuzza; April 2004 | AM711722 | AM711950 | AM711686 |
| Ophrys pallida Raf. | Devey DSD59A | Sicily, Forest of Ficuzza; coll. April 2004 | AM711723 | AM711951 | AM711687 |
| AM711724 | |||||
| AM711725 | |||||
| AM711726 | |||||
| Ophrys panormitana (Tod.) Soó | Bateman 541 | Piazza Amerina, Sicily | AM711727 | AM711952 | AM711688 |
| Ophrys parosica P.Delforge | Bateman 1125 | AM711728 | AM711953 | AM711689 | |
| Ophrys philippei Gren. | Bateman 1234 | AM711779 | AM711954 | AM711690 | |
| AM711780 | |||||
| Ophrys phryganae J.Devillers-Terschuren & P.Devillers | Chase 16355 | Crete, zig-zag above Gouverneto Gorge, Akrotiri, c. 11 April 1996 | AM711729 | AM711955 | AM711691 |
| Ophrys regis-ferdinandii (Renz) Buttler | Chase O- 905 K | Orchidinae, Kew 1984-2613 | AM711730 | AM711956 | AM711692 |
| Ophrys reinholdii Sprun. ex Boiss. | Bateman 1146 | AM711731 | AM711957 | AM711693 | |
| Ophrys rhodia (H.Baumann & Künkele) P.Delforge | Bateman 229 | Tekke, Cyprus | AM711756 | AM711958 | AM711694 |
| AM711755 | |||||
| AM711757 | |||||
| AM711758 | |||||
| Ophrys scolopax Bory & Chaub. | Chase O- 703 K | Orchidinae, Kew 1984-2590 | AM711732 AM711733 | AM711959 | AM711695 |
| Ophrys sicula Tineo | Bateman 36 | 5·1 km E Spili, Spili–Gerakari road, Crete; 20 April 1996 | AM711735 | AM711960 | AM711696 |
| Ophrys sitiaca Paulus, C.Alibertis & A.Alibertis | Bateman 39 | SW T-junction, Spili–Agia Galini/Melanbes, Crete; 21 April 1996 | AM711736 | AM711961 | AM711697 |
| Ophrys speculum Link | AM711737 | N/A | N/A | ||
| Ophrys sphegifera Willd. | Bateman 211 | Alhaurin el Grande, Malaga, Spain | AM711738 | AM711962 | AM711698 |
| Ophrys sphegodes | DSD17 | AM711739 | AM711963 | AM711699 | |
| Ophrys splendida | DSD21 | N/A | AM711964 | AM711700 | |
| Ophrys spruneri Nyman | Bateman 001 | Crete 4/96 | AM711740 | AM711965 | AM711701 |
| Ophrys subinsectifera C.E.Hermos. & J.Sabando | Bateman 318 | Navarra, Spain | AM711741 | AM711966 | AM711702 |
| Ophrys tenthredinifera Willd. | Chase 15846 | Source: RBG-Kew, LivColl.2002-745 [Provenance: Italy] | AM711743 | AM711967 | AM711703 |
| Ophrys umbilicata Desf. | Bateman 397 | SE Cyprus | AM711742 | AM711968 | AM711704 |
| Ophrys vasconica | DSD19 | N/A | AM711969 | AM711705 | |
| Orchis italica | Cotrim, Pinto, Chase & Fay 456 A | Casais Robustos, nr Vila Moreira Alcanena, Portugal, 24 March 2001 | AM711745 | N/A | N/A |
| Serapias lingua L. | Bateman 8 | Crete, marsh imm. W Frangocastello Castle, E Hora Sf.,12 April 1996 | AM711748 | N/A | N/A |
| Steveniella satyrioides Schlechter | Güner 12838 | Turkey, A4 Bolu, 5 km W of Mengen, open scrub and meadows, 590 m a.s.l., 7 May 2000 | AM711746 | N/A | N/A |
APPENDIX 2
Table A2.
The four clusters of Ophrys species as indicated by PCoA analysis derived from AFLP data (Fig. 4A)
| Ophrys species | PCoA cluster | Ophrys species | PCoA cluster |
|---|---|---|---|
| Ophrys aegirtica | Section Ophrys large subset (SOLS) | Ophrys panormitana | SOLS |
| Ophrys aesculapii | SOLS | Ophrys philippeii | SOLS |
| Ophrys apulica 2 | SOLS | Ophrys reinholdii | SOLS |
| Ophrys apulica 5 | SOLS | Ophrys scolopax | SOLS |
| Ophrys apulica 7 | SOLS | Ophrys scolopax | SOLS |
| Ophrys apulica 8 | SOLS | Ophrys sphegifera | SOLS |
| Ophrys araneola | SOLS | Ophrys sphegodes | SOLS |
| Ophrys argolica | SOLS | Ophrys spruneri | SOLS |
| Ophrys averyonensis | SOLS | Ophrys atlantica | Section Pseudophrys |
| Ophrys balearica | SOLS | Ophrys attaviria | Section Pseudophrys |
| Ophrys bertolonii | SOLS | Ophrys basilissa | Section Pseudophrys |
| Ophrys bertoloniiformis | SOLS | Ophrys bilunulata | Section Pseudophrys |
| Ophrys biscutella | SOLS | Ophrys cinereophila | Section Pseudophrys |
| Ophrys calliantha | SOLS | Ophrys funerea | Section Pseudophrys |
| Ophrys calypsus | SOLS | Ophrys fusca s.s. | Section Pseudophrys |
| Ophrys candica | SOLS | Ophrys iricolor | Section Pseudophrys |
| Ophrys catalaunica | SOLS | Ophrys lutea | Section Pseudophrys |
| Ophrys celiensis | SOLS | Ophrys melena | Section Pseudophrys |
| Ophrys cornuta | SOLS | Ophrys pallida | Section Pseudophrys |
| Ophrys dodecanensis | SOLS | Ophrys parosica | Section Pseudophrys |
| Ophrys drumana | SOLS | Ophrys phryganae | Section Pseudophrys |
| Ophrys episcopalis | SOLS | Ophrys sicula | Section Pseudophrys |
| Ophrys exaltata | SOLS | Ophrys sitiaca | Section Pseudophrys |
| Ophrys explanata | SOLS | Ophrys vasconica | Section Pseudophrys |
| Ophrys ferrum-equinum | SOLS | Ophrys aymoninii | insectifera |
| Ophrys fuciflora | SOLS | Ophrys insectifera | insectifera |
| Ophrys garganica | SOLS | Ophrys insectifera | insectifera |
| Ophrys gortynia | SOLS | Ophrys subinsectifera | insectifera |
| Ophrys gracilis | SOLS | Ophrys bombyliflora | bombyliflora/speculum/tenthredinifera (BST) |
| Ophrys homeri | SOLS | Ophrys speculum | BST |
| Ophrys heldreichii | SOLS | Ophrys tenthredinifera | BST |
| Ophrys incubacea | SOLS | Ophrys apifera | apifera |
| Ophrys lacaitae | SOLS | Ophrys oestrifera | apifera |
| Ophrys lucis | SOLS | Ophrys attica | umbilicata |
| Ophrys lunulata | SOLS | Ophrys bornmuelleri | umbilicata |
| Ophrys mammosa | SOLS | Ophrys bremifera | umbilicata |
| Ophrys minutula | SOLS | Ophrys lapethica | umbilicata |
| Ophrys morisii | SOLS | Ophrys rhodia | umbilicata |
| Ophrys oxyrrhynchos | SOLS |
LITERATURE CITED
- Bernardos S, Amich F, Gallego F. Karyological and taxonomic notes on Ophrys (Orchidoideae, Orchidaceae) from the Iberian Peninsula. Botanical Journal of the Linnean Society London. 2003;142:395–406. [Google Scholar]
- Bateman RM. Integrating molecular and morphological evidence of evolutionary radiations. In: Hollingsworth PM, Bateman RM, Gornall RJ, editors. Molecular systematics and plant evolution. London: Taylor and Francis; 1999. [Google Scholar]
- Bateman RM. Evolution and classification of European orchids: insights from molecular and morphological characters. Journal Europäischer Orchideen. 2001;33:33–119. [Google Scholar]
- Bateman RM, Devey DS. When orchids challenge an island race. Orchid Review. 2006;114:98–102. [Google Scholar]
- Bateman RM, Hollingsworth PM, Preston J, Yi-Bo L, Pridgeon AM, Chase MW. Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae) Botanical Journal of the Linnean Society. 2003;142:1–40. [Google Scholar]
- Bournérias M, Prat D. Les orchidées de France, Belgique et Luxembourg. 2nd edn. Paris: Collection Pathénope (Biotope). Société Francaise d'Orchidophilie, Mèze; 2005. [Google Scholar]
- Casgrain P, Legendre P. The R Package for multivariate analysis. Version 4·0d6. User's manual. Departement de sciences biologiques, Universite de Montreal; 2001. [Google Scholar]
- Cozzolino S, Aceto S, Caputo P, Widmer A, Dafni A. Speciation processes in eastern Mediterranean Orchis s.l. species: molecular evidence and the role of pollination biology. Israeli Journal of Plant Science. 2001;49:91–103. [Google Scholar]
- Creeth JM, Denborough MA. The use of equilibrium-density-gradient methods for the preparation and characterization of blood-group specific glycoproteins. Biochemical Journal. 1970;117:879–891. doi: 10.1042/bj1170879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delforge P. Orchids of Britain and Europe Harper Collins. London: 1995. ed. 1. [Google Scholar]
- Delforge P. Guide des orchidées d'Europe, d'Afrique du Nord et du Proche-Orient Harper Collins. London: 2001. ed. 2. [Google Scholar]
- Delforge P. Guide des orchidées d'Europe, d'Afrique du Nord et du Proche-Orient Harper Collins. London: 2005. ed. 3. [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Devillers P, Devillers-Terschuren J. Essai d'analyse systématique du genre Ophrys. Les Naturalistes Belges. 1994;75:273–400. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Fay MF, Cowan RS, Leitch IJ. The effects of nuclear DNA content (C-value) on the quality and utility of AFLP fingerprints. Annals of Botany. 2005;95:237–246. doi: 10.1093/aob/mci017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MF, Krauss SL. Orchid conservation genetics in the molecular age. In: Dixon K. W., Kell S. P., Barrett R. L., Cribb P. L., editors. Orchid conservation. Natural History Publications, Kota Kinabalu; 2003. pp. 91–112. [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies – an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416. [Google Scholar]
- Goldman DH, Jansen RK, van den Berg C, Leitch IJ, Fay MF, Chase MW. Molecular and cytological examination of Calopogon (Orchidaceae, Epidendroideae): circumscription, phylogeny, polyploidy, and possible hybrid speciation. American Journal of Botany. 2004;91:707–723. doi: 10.3732/ajb.91.5.707. [DOI] [PubMed] [Google Scholar]
- Gulyás G, Sramkó G, Molnár AV, Rudnóy S, Illyés Z, Balázs T, et al. Nuclear ribosomal DNA ITS paralogs as evidence of recent interspecific hybridization in the genus Ophrys (Orchidaceae) Acta Biologica Cracoviensia Series Botanica. 2005;47:61–67. [Google Scholar]
- Jaccard P. Nouvelles recherches sur la distribution florale. Bulletin, Société Vaudoise des Sciences Naturelles. 1908 [Google Scholar]
- Kelchner SA. The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden. 2000;87:482–498. [Google Scholar]
- Koopman WJM. Phylogenetic signal in AFLP data sets. Systematic Biology. 2005;54:197–217. doi: 10.1080/10635150590924181. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge, MA: The Belknap Press; 1963. [Google Scholar]
- Moya SJ, Ackerman D. Variation in the floral fragrance of Epidendrum ciliare (Orchidaceae) Nordic Journal of Botany. 1993;13:41–47. [Google Scholar]
- Mueller UG, Wolfenbarger LL. AFLP genotyping and fingerprinting. Trends in Ecology and Evolution. 1999;14:389–394. doi: 10.1016/s0169-5347(99)01659-6. [DOI] [PubMed] [Google Scholar]
- Paulus AF. Signale in der Bestauberanlockung: Weibchenimitation als Bestaubungsprinzip bei der mediterranen Orchideengattung Ophrys. Verhaltens Zoolozische–Botanische Gesellschoft Oesterreichs. 1997;134:133–176. [Google Scholar]
- Paulus HF, Gack C. Pollinators as prepollinating isolation factors: evolution and speciation in Ophrys (Orchidaceae) Israel Journal of Botany. 1990;39:43–79. [Google Scholar]
- Pedersen HA, Faurholdt N. Ophrys: a guide to the bee orchids of Europe. London: Kew Publishing; 2007. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Remington DL, Whetten RW, Liu B-H, O'Malley DM. Construction of an AFLP genetic map with nearly complete genome coverage in Pinus taeda. Theoretical and Applied Genetics. 1999;98:1279–1292. doi: 10.1007/s001220051194. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Fay MF, Cronk QCB, Chase MW. Species delimitation and the origin of populations in island representatives of Phylica (Rhamnaceae) Evolution. 2003;57:816–827. doi: 10.1111/j.0014-3820.2003.tb00293.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) American Journal of Botany. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Schiestl FP, Ayasse M, Paulus HF, Lofstedt C, Hansson BS, Ibarra F, et al. Orchid pollination by sexual swindle. Nature. 1999;399:421–422. [Google Scholar]
- Soliva M, Widmer A. Gene flow across species boundaries in sympatric, sexually deceptive Ophrys (Orchidaceae) species. Evolution. 2003;57:2252–2261. doi: 10.1111/j.0014-3820.2003.tb00237.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- Sundermann H. Europäische und Mediterrane Orchideen. 3rd edn. Hildesheim: Brücke-Verlag Kurt Schmersow; 1980. [Google Scholar]
- Swofford DL. PAUP* 4·0: phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Systematic Botany. 2003;28:723–737. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Rijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]