Abstract
Background
Phylogenetic and phylogeographic investigations have been previously performed to study the evolution of the olive tree complex (Olea europaea). A particularly high genomic diversity has been found in north-west Africa. However, to date no exhaustive study has been addressed to infer putative polyploidization events and their evolutionary significance in the diversification of the olive tree and its relatives.
Methods
Representatives of the six olive subspecies were investigated using (a) flow cytometry to estimate genome content, and (b) six highly variable nuclear microsatellites to assess the presence of multiple alleles at co-dominant loci. In addition, nine individuals from a controlled cross between two individuals of O. europaea subsp. maroccana were characterized with microsatellites to check for chromosome inheritance.
Key Results
Based on flow cytometry and genetic analyses, strong evidence for polyploidy was obtained in subspp. cerasiformis (tetraploid) and maroccana (hexaploid), whereas the other subspecies appeared to be diploids. Agreement between flow cytometry and genetic analyses gives an alternative approach to chromosome counting to determine ploidy level of trees. Lastly, abnormalities in chromosomes inheritance leading to aneuploid formation were revealed using microsatellite analyses in the offspring from the controlled cross in subsp. maroccana.
Conclusions
This study constitutes the first report for multiple polyploidy in olive tree relatives. Formation of tetraploids and hexaploids may have played a major role in the diversification of the olive complex in north-west Africa. The fact that polyploidy is found in narrow endemic subspecies from Madeira (subsp. cerasiformis) and the Agadir Mountains (subsp. maroccana) suggests that polyploidization has been favoured to overcome inbreeding depression. Lastly, based on previous phylogenetic analyses, we hypothesize that subsp. cerasiformis resulted from hybridization between ancestors of subspp. guanchica and europaea.
Key words: Flow cytometry, hexaploidy, High Atlas, Macaronesia, Olea europaea, olive, SSR, tetraploidy
INTRODUCTION
The cultivated olive (Olea europaea subsp. europaea) is one of the most important crop species in the Mediterranean Basin, and its early cultivation was reported in both the eastern and western Mediterranean regions about 6000 years ago (Zohary and Hopf, 2000; Terral et al., 2004). In addition to the cultivated olive, the O. europaea complex (Green and Wickens, 1989) includes five non-Mediterranean subspecies: subsp. laperrinei distributed in Saharan massifs (Hoggar, Aïr, Jebel Marra); subsp. cuspidata from South Africa to south Egypt and from Arabia to north India and south-west China; subsp. guanchica in the Canary Islands; subsp. maroccana in the Agadir Mountains (Morocco); and subsp. cerasiformis in Madeira (Green, 2002). Phylogeographic studies on the olive complex have been performed recently to investigate the relationships between all subspecies and to determine the origins of the Mediterranean olives (e.g. Rubio de Casas et al., 2006; Besnard et al., 2007b). It was shown that O. europaea subsp. cuspidata has diverged early from North African and Mediterranean taxa. A high diversity was reported in north-west Africa where four subspecies occur (subspp. europaea, maroccana, guanchica and cerasiformis), because of geographic isolation and fragmentation of habitat in this area.
All the species of Olea that have been investigated previously by chromosome counts [i.e. O. europaea (subspp. europaea and cuspidata), O. capensis, O. paniculata, O. dioica and O. salicifolia; for a review see Green and Wickens (1989)] have been reported as 2n = 46, except for an early count of n = approx. 24 in O. europaea (Anderson, 1931). However, artificial triploids and tetraploids have been isolated from cultivars ‘Frantoio’ and ‘Leccino’ mutants (Rugini et al., 1996). Although many genetic and cytogenetic studies have been performed in the last decade on O. europaea subsp. europaea (e.g. Katsiotis et al., 1998; Bitonti et al., 1999; Minelli et al., 2000; Contento et al., 2002), surprisingly no exhaustive studies were performed on the possible occurrence of polyploids in the genus Olea and particularly in some taxa of the O. europaea complex. Additionally, using highly variable multi-allelic loci, polyploidy was not found in a large sample of both wild and cultivated Mediterranean olives (e.g. Khadari et al., 2003; Bandelj et al., 2004; Belaj et al., 2004; Breton et al., 2006). However, polyploidy was recently suspected to occur in O. europaea subspp. cuspidata (from Iran) and maroccana based on microsatellites (Rallo et al., 2003), but no specific investigations were performed to test levels of polyploidy.
Newly developed analytical tools such as flow cytometry and multiallelic molecular markers (microsatellites) can help to determine ploidy levels rapidly in closely related taxa in the absence of meristematic tissues. In the present work, a sample of trees of the six subspecies of the O. europaea complex was characterized using flow cytometry and highly polymorphic microsatellites.
MATERIALS AND METHODS
Plant material
Thirty-one wild trees belonging to the six subspecies of the O. europaea complex were sampled from ten locations (Table 1). Five distant populations across the native range (from Iran, Yemen, Kenya, Reunion and South Africa) were analysed to test polyploidy in subsp. cuspidata (Rallo et al., 2003). Furthermore, three invasive trees (subsp. cuspidata) from Australia, two Spanish cultivars (‘Arbequina’ and ‘Manzanilla’; subsp. europaea) and three controlled hybrids [subspp. laperrinei (Hoggar) × cuspidata (Iran)] were investigated. Genomic DNA from fresh leaves or leaves desiccated in silica gel was extracted using a 2X CTAB method (Besnard et al., 2000) for the 39 individuals.
Table 1.
List of material characterized using flow cytometry and microsatellite analyses
| Taxon | Geographic origin | Collection | N | Ploidy level |
|---|---|---|---|---|
| Olea europaea L. subsp. europaea | ||||
| var. sylvestris (Mill.) Lehr | Trassierra, Spain (wild) | UCM | 3 | 2x |
| var. europaea | ‘Arbequina’, Spain (cultivar) | MA | 1 | 2x |
| ‘Manzanilla’, Spain (cultivar) | MA | 1 | 2x | |
| O. europaea subsp. laperrinei (Batt. & Trab.) Cif. | La Source, Algeria | INRAM | 1 | 2x |
| O. europaea subsp. guanchica P. Vargas et al. | Tenerife, Spain | CIT | 3 | 2x |
| O. europaea subsp. cerasiformis G. Kunkel & Sunding | Funchal, Madeira, Portugal | UNIL | 4 | 4x |
| O. europaea subsp. maroccana (Greuter & Burdet) P. Vargas et al. | Immouzzer, Morocco | UNIL | 3 | 6x |
| O. europaea subsp. Cuspidate (Wall. ex G. Don) Cif. | Kerman, Iran | INRAM | 3 | 2x |
| Almihwit, Yemen | INRAM | 3 | 2x | |
| Rift Valley, Kenya | INRAM | 3 | 2x | |
| La Providence, La Réunion | INRAM | 3 | 2x | |
| Grahamstown, South Africa | UNIL | 5 | 2x | |
| Campbelltown, Australia | UNIL | 3 | 2x | |
| O. europaea subsp. laperrinei × subsp. cuspidata | Hybrids ‘Hoggar × Iran’ | UNIL | 3 | 2x |
The ploidy level of each individual was estimated using 2C DNA content (Fig. 1) and allelic variation at six microsatellite loci (see Supplementary information available online).
N, number of individual(s) per population analysed in the present study; CIT, Cabildo Insular de Tenerife; INRAM, Institut National de Recherche Agronomique de Montpellier; MA, Royal Botanic Garden of Madrid; UCM, Universidad Complutense de Madrid; UNIL, University of Lausanne.
Flow cytometry analyses
The C-value of each olive sample was estimated using flow cytometry. For each individual, intact nuclei were extracted from approx. 0·2 cm2 of leaf tissue in a Petri dish. The plant material was chopped for 30 s with a sharp razor blade in 200 µL of extraction buffer (Partec, Münster, Germany). Three hundred microlitres of extraction buffer were added and the mix was incubated for about 1 min. The nuclei suspension was then filtered in a 30-μm CellTrics® disposable filter (Partec). For each sample, the filtrate was mixed with 1 µL of chicken erythrocyte nuclei (CEN) or CEN Singlets (BioSure, Grass Valley, CA, USA) as an internal standard. Two millilitres of staining buffer (Partec; containing 10 mg mL–1 of RNase) were then added and the mix was incubated for at least 30 min in the dark. The samples were then randomly analysed using a CyFlow (Partec). Data acquisition and analysis were performed in real time with a standard Pentium PC connected to the CyFlow. The software used was FloMax Version 2·0 (Partec). For each individual, the 2C DNA content was estimated by comparing the mean peak intensity of its nuclei with the mean peak intensity of the chicken erythrocyte nuclei (2C DNA = 2·5 pg) following Doležel and Bartoš (2005). Only measurements with coefficients of variation smaller than 8% were accepted. To assess the significance of genome size difference between invasive and native olives of subsp. cuspidata, a Mann–Whitney U-test was performed using the JMP® software version 5·0 (SAS Institute Inc., 2002).
Nuclear microsatellite polymorphism characterization
To characterize the 39 trees, six nuclear microsatellite (SSR) loci were used: ssrOeUA-DCA1, ssrOeUA-DCA3, ssrOeUA-DCA8, ssrOeUA-DCA9, ssrOeUA-DCA14 (Sefc et al., 2000) and EMO03 (de la Rosa et al., 2002). For these loci, absence or low frequency of null alleles was reported in subspp. europaea, laperrinei and cuspidata by Baali-Cherif and Besnard (2005) and Besnard et al. (2007a). Moreover, allele number and gene diversity revealed at the six loci were high in these taxa rendering sufficient variability to distinguish diploids (maximum of two alleles per locus) from polyploids (presence of more than two alleles per locus). PCR conditions and electrophoresis procedures were used as previously described (Baali-Cherif and Besnard, 2005). For each locus, all alleles revealed (based on their size in base pairs) from each individual were scored and a matrix of allelic phenotypes was constructed.
Genetic characterization of a subsp. maroccana offspring
To check for abnormalities in chromosome inheritance leading to aneuploid formation, offspring from a controlled cross between two individuals of subsp. maroccana (S1 and S2, from Immouzzer Ida-Outanane) we analysed. This cross was performed in a greenhouse at INRA, Montpellier. Nine F1 plants were obtained (from 21 seeds) and analysed with the six highly variable microsatellite loci as previously described.
RESULTS
Flow cytometry analyses
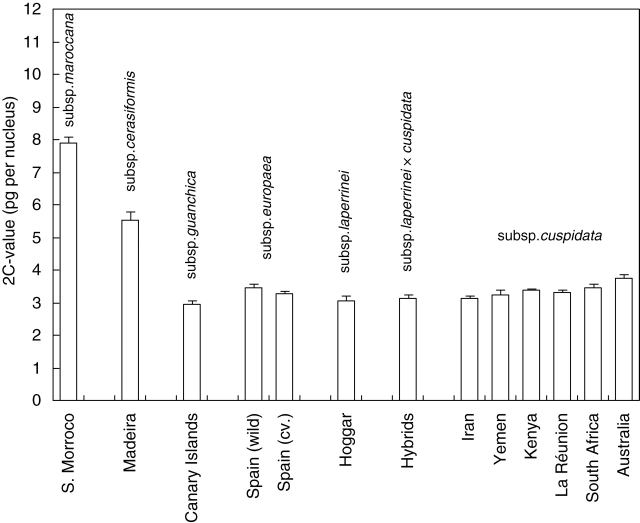
The flow cytometry analyses of 39 olive tree samples showed that the genome size is variable in the O. europaea complex and three discrete DNA values were distinguished among infraspecific taxa (Fig. 1): subsp. maroccana (2C value = 7·88 ± 0·19 pg), subsp. cerasiformis (2C value = 5·52 ± 0·28 pg) and populations of the four remaining subspecies (2C value ranging from 2·93 to 3·75 pg).
Fig. 1.
Flow cytometric analysis of the olive complex: comparative genome content (in pg per nucleus) between different taxa and populations of the olive complex. For each provenance, the mean genome size was estimated from the individuals analysed. Standard deviation based on two to five individuals (see Table 1) is given for each provenance, except for the sample of O. europaea subsp. laperrinei for which the only individual sampled was independently characterized three times.
Twenty genome sizes were estimated for subsp. cuspidata ranging from 3·06 pg to 3·86 pg (Fig. 1). DNA content was highest for the Australian (2C-value = 3·75 ± 0·12 pg) and lowest for the Iranian population (2C-value = 3·11 ± 0·08 pg). Genome size of the introduced Australian trees was significantly larger than those of native trees (z = 2·65, d.f. = 1, P = 0·008, N = 20). The chromosome number of one sample from Australia was, however, determined to be 2x = 46, as for native trees (N. Galland, unpubl. data).
Nuclear microsatellite polymorphism characterization
The SSR allelic phenotypes for all samples analysed is given in the Supplementary Information (available online). A total of 146 alleles was revealed for the six loci (between 23 and 28 per locus). Three or more alleles in most loci were detected in single individuals for subspp. maroccana and cerasiformis (Supplementary information available online). Individuals of subsp. maroccana displayed a maximum of six alleles per locus whereas individuals of subsp. cerasiformis displayed a maximum of four alleles per locus.
Genetic characterization of offspring from a controlled cross in subsp. maroccana
Among the six loci used, ssrOeUA-DCA8 and ssrOeUA-DCA14 displayed a high level of polymorphism with five or six alleles in maroccana trees S1 and S2 (see Supplementary information available online). These two loci were then considered to check for abnormality in chromosome inheritance leading to aneuploid progenies. No abnormality was observed on ssrOeUA-DCA14 since all F1 trees displayed three alleles from each parent. In contrast, some irregularities in the allele inheritance of ssrOeUA-DCA8 were found since individuals m5 and m8 received only two of the six alleles found in parent S1 (Fig. 2).
Fig. 2.
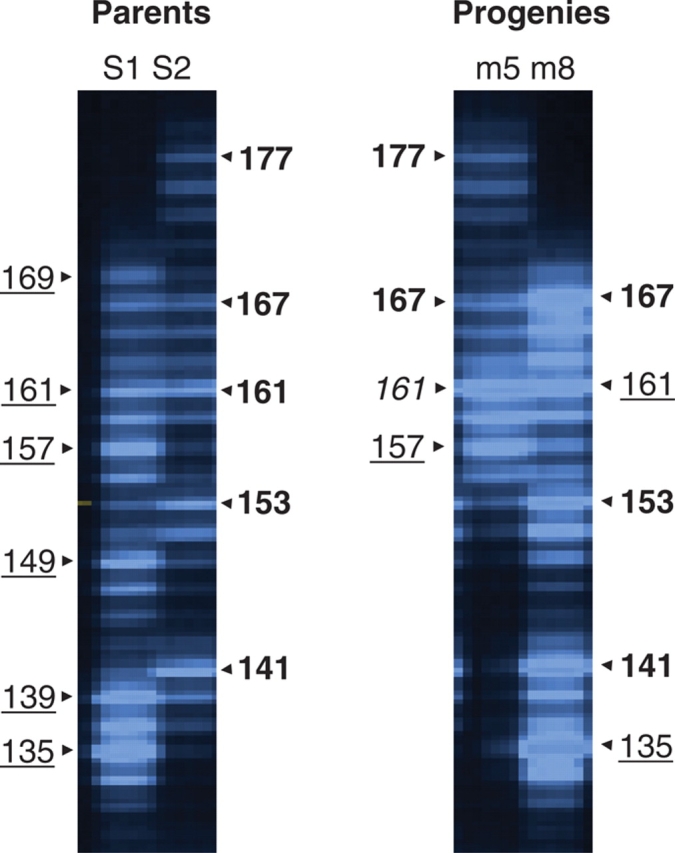
Inheritance of alleles at locus ssrOe-DCA8 in two F1 trees (m5 and m8) of the cross between individuals S1 × S2 of O. europaea subsp. maroccana. Electrophoresis of PCR products was carried out on a denaturing 5% acrylamide gel using an automated sequencer (ABI377; Applied Biosystems). For each individual, the size of each allele is indicated in the margin. The underlined sizes indicate alleles from the female (S1) and the sizes in bold-faced type indicate alleles from the male (S2). In individual m5, allele 161 is in italic because it was probably inherited from both parents. The F1 trees m5 and m8 only received two of the six alleles revealed in parent S1 (m5: alleles 161 and 157; m8: alleles 135 and 161).
DISCUSSION
Genome content in the olive complex
The flow cytometry analyses indicated unambiguously that O. europaea subspp. maroccana and cerasiformis have a larger genome than the other subspecies, which is, given the approximately multiplicative 2C values, likely to be related to polyploidy (Leitch and Bennett, 2004). The DNA content estimations for subsp. cuspidata from Kenya were similar to those previously reported in the literature (Bitonti et al., 1999); the three Kenyan individuals had a 2C-value of 3·38 ± 0·04 pg whereas Bitonti et al. (1999) reported a 2C-value of 3·2 and 3·4 for two trees from the living collections at Kew, also collected in Kenya. The mean 2C-value estimation for Mediterranean samples (subsp. europaea) was 3·39 ± 0·13 for wild individuals and cultivars. This estimation was intermediate between those previously reported for Italian cultivars (2C-value between 3·90 pg and 4·66 pg; Rugini et al., 1996; Bitonti et al., 1999) and those recently reported for Portuguese trees (2C-value between 2·90 and 3·19 pg; Loureiro et al., 2007). Such different results are likely to be due to comparisons of different genotypes and methodological differences in flow cytometry. The internal cytometry reference standards were different in these three investigations: Sorghum bicolor in Bitonti et al. (1999), Pisum sativum in Loureiro et al. (2007) and CEN in the present study. It is therefore not possible to determine if these differences are due to genome size variation between Mediterranean populations or to methodological reasons. We agree that DNA-content variation of small magnitude may be technique-related, as recently reported for several plant species (Greilhuber, 2005).
It was also shown in the present study that genome size of the introduced Australian trees was significantly larger than those of other trees of subsp. cuspidata sampled in the native range. Minor differences of genome size between introduced and native populations could be related to the presence of a variable content of repeated elements (such as tandem repeats or transposable elements; Bennetzen et al., 2005; Piegu et al., 2006). Such a correlation has already been described by Bitonti et al. (1999) in the olive complex but additional investigations are still needed to identify causes behind genome size variation in olive diploids. For more precise genome size comparisons between olive populations and subspecies, it is recommended that a larger sample of individuals is used and genome size estimations are repeated as recently suggested by Loureiro et al. (2007).
Agreement between cytometry and SSR analyses
Individuals of O. europaea subspp. maroccana and cerasiformis displayed three or more alleles at most loci. These SSR characterizations are in agreement with the DNA-ploidy levels measured with flow cytometry and suggest that subsp. maroccana is hexaploid and subsp. cerasiformis tetraploid. This agrees with the conclusions of Rallo et al. (2003) with regard to subsp. maroccana; however, the present results do not confirm polyploidy in Iranian populations of subsp. cuspidata as suggested from SSR variation by Rallo et al. (2003). Except for subspp. maroccana and cerasiformis, the remaining four subspecies of the olive complex showed a maximum of two alleles per locus, supporting a diploid level (see Supplementary information available online). Diploidy has also been suggested from previous studies of SSR variation for subspp. laperrinei (94 genets; Baali-Cherif and Besnard, 2005), europaea (157; Besnard et al., 2007a), and cuspidata (115; Besnard et al., 2007a). An extended sample (181 trees from additional populations of the six subspecies) characterized with the same (ssrOeUA-DCA3, ssrOeUA-DCA8) and additional nuclear SSR loci gave similar results (C. Garcia-Verdugo et al., unpubl. data).
Abnormal chromosome inheritance leading to aneuploids has been reported in several polyploid taxa (Ramsey and Schemske, 2002; Comai, 2005). It is, however, difficult to count chromosomes accurately in polyploid olive trees because of their expected high number (i.e. 4x = 92 or 6x = 138). As an alternative method, highly variable microsatellites can be useful to test failures in regular chromosome inheritance in progenies. For one locus (ssrOeUA-DCA8; Fig. 2), allelic inheritance from parent S1 did not fit our expectations (i.e. inheritance of three parental alleles) in two F1 trees. This observation proves that inheritance of chromosomes in olive polyploids may have irregularities due likely to multivalent formation during meiosis leading to aneuploid formation (Ramsey and Schemske, 2002).
Origin of polyploidy in the olive complex
The present study is the first report of natural polyploids in O. europaea. Diversification of the Macaronesian and south-west Moroccan olives by at least two polyploidization events strongly supports the taxonomic treatment proposed by Vargas et al. (2001) and Green (2002). They distinguished three closely related subspecies in this area (subspp. guanchica, cerasiformis and maroccana), and each of these taxa is characterized by a specific ploidy level in the present study. Polyploidy is a prominent process in plant evolution which has already been reported in other genera of Oleaceae (Fraxinus, Jasminum; Taylor, 1945). In addition, tribe Oleeae (in which genus Olea is included) is derived from an ancient polyploid lineage (Wallander and Albert, 2000). The origins of polyploidy have been discussed by several authors (for reviews, see Soltis et al., 2004; Comai, 2005). A high degree of genomic and biochemical flexibility may offer to polyploids some opportunities to colonize new habitats (Levin, 2002). In contrast to extensive polyploidization in arctic plants (Stebbins, 1984; Brochmann et al., 2004), a relationship between polyploidy and narrow endemic taxa is observed here, since O. europaea subspp. maroccana (hexaploid) and cerasiformis (tetraploid) are only found in restricted areas. These two taxa are endemic to subtropical areas of the Agadir Mountains and Madeira, respectively. Moreover, the Agadir Mountains are considered as an important refugium since numerous endemic plants have been reported in this area (Médail and Quézel, 1999). Strong geographic isolation associated with episodic reduced population size could lead to strong genetic erosion provoking a mutational meltdown. Polyploidization may have thus been favoured to overcome inbreeding depression by providing the possibility of maintaining higher gene diversity than in diploids (Brochmann et al., 2004; Soltis et al., 2004). However, the populations of subspp. cerasiformis and maroccana are endangered (Hess et al., 2000; Médail et al., 2001) and presently occur in sympatry with the Mediterranean olive tree (subsp. europaea). Particularly, O. europaea subsp. maroccana is considered to be one of the ten most threatened trees in the Mediterranean Basin (Barbero et al., 2001; Médail et al., 2001). It is thus suspected that some disadvantages of polyploidy, including larger genome size and difficulties at both mitosis and meiosis (for a review, see Comai, 2005), may also affect the success of these taxa.
The precise origins of polyploids, via autopolyploidization or segmental allopolyploidization, have still to be determined. Based on phylogenetic data (from nuclear ribosomal and plastid DNA), historical processes of reticulation lead to the hypothesis that allopolyploidy occurs (Besnard et al., 2007b). Wild plants of O. europaea subsp. cerasiformis from Madeira display a close genetic relationship with subsp. guanchica (Canary Islands) based on the plastid genome, whereas they appear to be closely related to Western Mediterranean olives (subsp. europaea) based on nuclear DNA (Hess et al., 2000; Besnard et al., 2007b). This could mean that subsp. cerasiformis originated from hybridization between ancestors of subspp. guanchica and europaea. In the case of the hexaploid subsp. maroccana, the available molecular data only support a close phylogenetic relationship with the diploid subsp. guanchica. The origin of a diploid endemic to an oceanic archipelago (subsp. guanchica) from ancestors related to hexaploid populations (subsp. maroccana; Hess et al., 2000) deserves further investigation. Additional phylogenetic evidence using single-copy nuclear genes may also be useful in such a study (e.g. Fortune et al., 2007).
SUPPLEMENTARY INFORMATION
Supplementary information is available online at http://aob.oxfordjournals.org/ and consists of a matrix of allelic phenotypes at the six microsatellite loci for all trees analysed in the present study.
ACKNOWLEDGEMENTS
We thank Anne-Catherine Pasche (Fribourg University) for her help in flow cytometry analyses, Christian Parisod and Pascal-Antoine Christin for helpful discussions, and two anonymous reviewers for their constructive comments. In addition, we are grateful to Nathalie Moutier, José Augusto Carvalho and Monique Besnard for providing fresh olive samples and/or management of wild olive collections.
LITERATURE CITED
- Anderson A. Studien über die embryologie der familien Celastraceae, Oleaceae und Apocynaceae. Lunds Universitets Årsskrift, N. F. Avd. 1931 2, bd. 27 (no. 7) [Google Scholar]
- Baali-Cherif D, Besnard G. High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Annals of Botany. 2005;96:823–830. doi: 10.1093/aob/mci232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelj D, Jakše J, Javornik B. Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica. 2004;136:93–102. [Google Scholar]
- Barbero M, Loisel R, Médail F, Quézel P. Signification biogéographique et biodiversité des forêts du bassin méditérranéen. Bocconea. 2001;13:11–25. [Google Scholar]
- Belaj A, Cipriani G, Testolin R, Rallo L, Trujillo I. Characterization and identification of the main Spanish and Italian olive cultivars by simple-sequence-repeat markers. HortScience. 2004;39:1557–1561. [Google Scholar]
- Bennetzen JL, Ma JX, Devos K. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Khadari B, Villemur P, Bervillé A. Cytoplasmic male sterility in the olive (Olea europaea L.) Theoretical and Applied Genetics. 2000;100:1018–1024. [Google Scholar]
- Besnard G, Henry P, Wille L, Cooke D, Chapuis E. On the origin of the invasive olives (Olea europaea L., Oleaceae) Heredity. 2007;a 99:608–619. doi: 10.1038/sj.hdy.6801037. [DOI] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. Plastid and nuclear DNA polymorphism reveals historical processes of isolation and reticulation in the olive tree complex (Olea europaea L.) Journal of Biogeography. 2007;b 34:736–752. [Google Scholar]
- Bitonti MB, Cozza R, Chiappetta A, Contento A, Minelli S, Ceccarelli M, et al. Amount and organization of the heterochromatin in Olea europaea and related species. Heredity. 1999;83:188–195. doi: 10.1046/j.1365-2540.1999.00564.x. [DOI] [PubMed] [Google Scholar]
- Breton C, Tersac M, Bervillé A. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. Journal of Biogeography. 2006;33:1916–1928. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen AC, Elven R. Polyploidy in artic plants. Biological Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nature Reviews Genetics. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Contento A, Ceccarelli M, Gelati MT, Maggini F, Baldoni L, Cionini PG. Diversity of Olea genotypes and the origin of cultivated olives. Theoretical and Applied Genetics. 2002;104:1229–1238. doi: 10.1007/s00122-001-0799-7. [DOI] [PubMed] [Google Scholar]
- Doležel J, Bartoš J. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune PM, Schierenbeck KA, Ainouche AK, Jacquemin J, Wendel JF, Ainouche ML. Evolutionary dynamics of waxy and the origin of hexaploid Spartina species (Poaceae) Molecular Phylogenetics and Evolution. 2007;43:1040–1055. doi: 10.1016/j.ympev.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Green PS. A revision of Olea L. (Oleaceae) Kew Bulletin. 2002;57:91–140. [Google Scholar]
- Green PS, Wickens GE. The Olea europaea complex. In: Tan K, Mill RR, Elias TS, editors. The Davis and Hedge Festschrift. Edinburgh: Edinburgh University Press; 1989. pp. 287–299. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Kadereit JW, Vargas P. The colonization history of Olea europaea L. in Macaronesia based on internal transcribed spacer 1 (ITS-1) sequences, randomly amplified polymorphic DNAs (RAPD), and inter simple sequence repeats (ISSR) Molecular Ecology. 2000;9:857–868. doi: 10.1046/j.1365-294x.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- Katsiotis A, Hagidimitriou M, Douka A, Hatzopoulos P. Genomic organization, sequence interrelationships, and physical localization using in situ hybridization of two tandemly repeated DNA sequences in the genus. Olea. Genome. 1998;41:527–534. [PubMed] [Google Scholar]
- Khadari B, Breton C, Moutier N, Roger JP, Besnard G, Bervillé A, et al. The use of molecular markers for germplasm management in a French olive collection. Theoretical and Applied Genetics. 2003;106:521–529. doi: 10.1007/s00122-002-1079-x. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. New York, NY: Oxford University Press; 2002. [Google Scholar]
- Loureiro J, Rodriguez E, Costa A, Santos C. Nuclear DNA content estimations in wild olive (Olea europaea L. ssp. europaea var. sylvestris Brot.) and Portuguese cultivars of O. europaea using flow cytometry. Genetic Resources and Crop Evolution. 2007;54:21–25. [Google Scholar]
- Médail F, Quézel P. The phytogeographical significance of south-west Morocco compared to the Canary Islands. Plant Ecology. 1999;140:221–244. [Google Scholar]
- Médail F, Quézel P, Besnard G, Khadari B. Systematics, ecology and phylogeographic significance of Olea europaea L. subsp. maroccana (Greuter & Burdet) P. Vargas et al., a relictual olive tree in south-west Morocco. Botanical Journal of the Linnean Society. 2001;137:249–266. [Google Scholar]
- Minelli S, Maggini F, Gelati MT, Angiolillo A, Cionini PG. The chromosome complement of Olea europaea L.: characterization by differential staining of the chromatin and in-situ hybridization of highly repeated DNA sequences. Chromosome Research. 2000;8:615–619. doi: 10.1023/a:1009286008467. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallo P, Tenzer I, Gessler C, Baldoni L, Dorado G, Martín A. Transferability of olive microsatellite loci across the genus. Olea. Theoretical and Applied Genetics. 2003;107:940–946. doi: 10.1007/s00122-003-1332-y. [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics. 2002;33:589–639. [Google Scholar]
- de la Rosa R, James CM, Tobutt KR. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Molecular Ecology Notes. 2002;2:265–267. [Google Scholar]
- Rubio de Casas R, Besnard G, Schoenswetter P, Balaguer L, Vargas P. Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L. Theoretical and Applied Genetics. 2006;113:575–583. doi: 10.1007/s00122-006-0306-2. [DOI] [PubMed] [Google Scholar]
- Rugini E, Pannelli G, Ceccarelli M, Muganu M. Isolation of triploid and tetraploid olive (Olea europaea L.) plants from mixoploid cv. ‘Frantoio’ and ‘Leccino’ mutants by in vivo and in vitro selection. Plant Breeding. 1996;115:23–27. [Google Scholar]
- SAS Institute Inc. JMP® 5 Administrator's guide to annually licensed Windows and Macintosh versions. Cary, NC: 2002. Version 5. [Google Scholar]
- Sefc KM, Lopes MS, Mendonca D, Rodrigues dos Santos M, Laimer da Câmara Machado M, da Câmara Machado A. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Molecular Ecology. 2000;9:1171–1173. doi: 10.1046/j.1365-294x.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytologist. 2004;161:173–191. [Google Scholar]
- Stebbins GL. Polyploidy and the distribution of the arctic-alpine flora: new evidence and a new approach. Botanica Helvetica. 1984;94:1–13. [Google Scholar]
- Taylor H. Cyto-taxonomy and phylogeny of the Oleaceae. Brittonia. 1945;5:337–367. [Google Scholar]
- Terral JF, Alonso N, Capdevila RBI, Chatti N, Fabre L, Fiorentino G, et al. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. Journal of Biogeography. 2004;31:63–77. [Google Scholar]
- Vargas P, Muñoz Garmendia F, Hess J, Kadereit JW. Olea europaea subsp. guanchica and subsp. maroccana (Oleaceae), two new names for olive tree relatives. Anales del Jardin Botanico de Madrid. 2001;58:360–361. [Google Scholar]
- Wallander E, Albert VA. Phylogeny and classification of Oleaceae based on rps16 and trnL-F sequence data. American Journal of Botany. 2000;87:1827–1841. [PubMed] [Google Scholar]
- Zohary D, Hopf M. Domestication of plants in the Old World. 3rd edn. Oxford: Clarendon Press; 2000. [Google Scholar]