Abstract
Paraneoplastic neurologic disorders (PNDs) offer an uncommon opportunity to study human tumor immunity and autoimmunity. In small cell lung cancer (SCLC), expression of the HuD neuronal antigen is thought to lead to immune recognition, suppression of tumor growth, and, in a subset of patients, triggering of the Hu paraneoplastic neurologic syndrome. Antigen-specific CTLs believed to contribute to disease pathophysiology were described 10 years ago in paraneoplastic cerebellar degeneration. Despite parallel efforts, similar cells have not been defined in Hu patients. Here, we have identified HuD-specific T cells in Hu patients and provided an explanation for why their detection has been elusive. Different Hu patients harbored 1 of 2 kinds of HuD-specific CD8+ T cells: classical IFN-γ–producing CTLs or unusual T cells that produced type 2 cytokines, most prominently IL-13 and IL-5, and lacked cytolytic activity. Further, we found evidence that SCLC tumor cells produced type 2 cytokines and that these cytokines trigger naive CD8+ T cells to adopt the atypical type 2 phenotype. These observations demonstrate the presence of an unusual noncytotoxic CD8+ T cell in patients with the Hu paraneoplastic syndrome and suggest that SCLC may evade tumor immune surveillance by skewing tumor antigen–specific T cells to this unusual noncytolytic phenotype.
Introduction
The paraneoplastic neurologic disorders (PNDs) offer a rare opportunity to study tumor immunity in humans. The subacute sensory neuropathy/encephalomyelopathy syndrome (the Hu syndrome) is characterized by a high-titer antibody response to the HuD protein (also known as ELAVL4), which is normally expressed exclusively in neurons and also in small cell lung cancers (SCLCs) and has been termed an “onconeural antigen” (1, 2). It is believed that HuD expression by tumor cells exposes the antigen to the immune system, generating an HuD-specific immune response. This results in appropriate and partially effective tumor immunity (3, 4). Hu patients typically first present to clinicians with neurological symptoms triggered when this tumor immune response, by unknown means, becomes competent to attack the nervous system.
High-titer antibody to neuronal antigens such as HuD in the serum and cerebrospinal fluid of PND patients provided the initial evidence for an immunologic basis for these disorders (5, 6). Patient antiserum also allowed for the identification and cloning of the HuD antigen (7), which is a member of a multigene family of RNA-binding proteins involved in mRNA stability and translational regulation. The Hu protein family includes HuA (also known as HuR and ELAVL1), a ubiquitously expressed protein not recognized in native form by Hu PND antisera (8, 9), and HuB (also known as ELAVL2), HuC (also known as ELAVL3), and HuD neuron-specific proteins (10).
Since several of the PND antigens, including HuD, are intracellular proteins, it has been hypothesized that CD8+ T cells, which monitor the intracellular compartment, can recognize these antigens and are the effectors of the clinical antitumor and autoimmune neuronal responses (1, 3, 6). This has been shown explicitly in patients with paraneoplastic cerebellar degeneration (PCD) (11, 12), who have gynecologic malignancies expressing the cerebellar degeneration-related protein 2, 62 kDa (CDR2) antigen, an intracellular protein normally expressed in cerebellar Purkinje neurons (13). CDR2-specific CD8+ CTLs are present in the peripheral blood of PCD patients, and they can kill target cells in an HLA-restricted manner (11, 14).
Curiously, parallel efforts to identify HuD-specific T cells over the past decade have not yielded clear results. There have not been consistent or reproducible descriptions of Hu-specific T cells in the peripheral blood of Hu patients (15–18) (R.B. Darnell, unpublished observations), despite significant interest in characterizing the nature of the immune response. In addition to interest in understanding PND pathophysiology, interest in the HuD immune response is heightened because all SCLCs express the HuD antigen, making it an especially attractive tumor immune target. In addition, a large number of the general population of SCLC patients (~15%–20%) have low-titer antibodies to HuD without autoimmunity (19), and this response correlates with improved prognosis, providing an example of a common, naturally occurring tumor immune response (20).
To pursue the hypothesis that Hu patients harbor HuD antigen-specific CD8+ CTLs, we have searched for HLA-restricted HuD peptides that could be recognized by such cells, using an exhaustive in vitro peptide screen. These experiments allowed us to develop HLA-peptide tetramers able to detect HuD-specific T cells in patient blood. Unexpectedly, functional studies of tetramer-reactive T cells, including ELISPOT and cytotoxicity assays, revealed both typical and atypical antigen-specific CD8+ T cells. The latter were inefficient killers and secreted only low levels of IFN-γ but were able to produce large amounts of IL-13 and other Th2-like cytokines in response to specific antigen, and we refer to these cells as type 2 CD8+ T cells. We also found evidence that SCLCs produce Th2 cytokines and that these may contribute to generation of the unusual class of CD8+ T cells identified here. Thus, both classical and what we believe to be a novel type of human noncytotoxic CD8+ T cells may play roles in pathogenesis of the Hu syndrome and more generally in tumor immune interactions.
Results
Screen for candidate HuD peptide epitopes.
To identify HuD peptides able to bind HLA class I molecules, we generated a library consisting of all 386 possible nonamer human HuD peptides, including previously described splice variants (10). We screened these peptides using an approach successfully applied to identify CD8+ T cell epitopes for survivin (21), CDR2 (14), and mycobacterium tuberculosis (22). Peptides were screened for the ability to bind recombinant HLA molecules and further analyzed by determining off-rate and affinity, yielding a net score for each peptide. We initially focused our effort on the class I alleles most commonly found in US populations of mixed European descent. HLA-A0201 and HLA-A0301 yielded a total of 48 peptides with significant net scores (Supplemental Table 1). To refine this list, we reasoned that pathophysiologically relevant human HuD-specific T cells should fail to react with the HuA protein for several reasons: while HuD patients have high-titer antibody responses to the neuronal Hu family members, they have no significant antibody reactivity with mouse tissues expressing HuA (using Western blot or immunofluorescence microscopy), and PND patients do not develop peripheral autoimmunity to non-neuronal tissues that express robust amounts of HuA (10). Eighteen HuD HLA-A0201 and five HLA-A0301 binding peptides with different sequences than HuA (Supplemental Figure 1 and Supplemental Table 1) were identified for further evaluation.
Identification of classical HuD-specific CD8+ T cells in an Hu patient.
Cells from Hu patient 1, an acutely ill HLA-A0301 individual, were used to further screen the HLA-A0301 peptides. CD8+ T cells were cultured for 8 days with autologous peptide-pulsed DCs to expand antigen-specific T cells. CD8+ T cells recognized peptide Hu133 but none of the other candidate peptides, as demonstrated by binding to specific HLA-peptide tetramers (Figure 1A and Supplemental Figure 2). We found that these Hu133-specific CD8+ T cells were functional using chromium release cytotoxicity and IFN-γ elispot assays (Figure 1, B and C). To determine whether Hu133-specific T cells could be detected in neurologically normal control patients, T cells were tested for tetramer staining after 8 days of in vitro expansion. We also tested 3 chronically ill HLA-A0301 Hu patients and did not find significant levels of Hu133-specific T cells (Supplemental Figure 3). We note, however, that 2 of these 3 patients were also HLA-A0201 and did have A0201-Hu–specific T cells (see Figure 2C and Supplemental Table 2), suggesting that their dominant HuD response may be HLA-A0201 restricted. Zero of two A0301 control patients had detectable Hu133-specific T cells in this assay, while all patients and controls showed T cell expansion of influenza nucleoprotein-specific T cells (Supplemental Figure 3).
Figure 1. Demonstration of Hu133-specific T cells that have lytic activity and secrete IFN-γ.
(A) Tetramer staining of cells from patient 1 (HLA-0301) after 1 round of in vitro expansion with A0301-predicted peptides. The percentages of tetramer-positive cells are noted above the boxed populations. (B) The expanded T cells were also tested for functional activity in a CTL assay. Irrelevant peptide condition is designated by red squares, and relevant peptide is designated by blue circles. (C) Similar results were obtained with IFN-γ ELISPOT. Gray bars indicate irrelevant peptide condition, and black bars indicate relevant peptide. Error bars represent SD of the mean of triplicate wells (B and C).
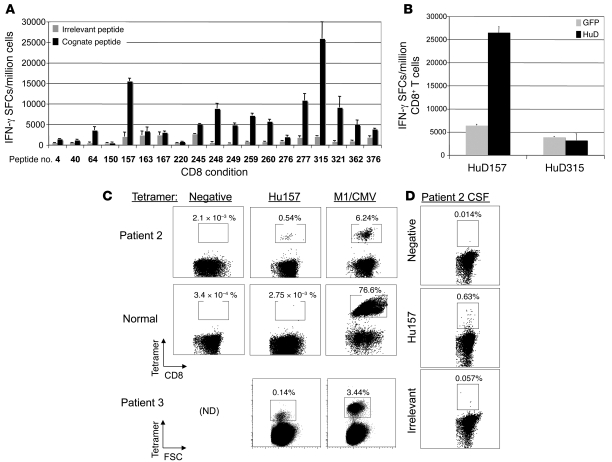
Figure 2. Identification of the Hu157 epitope and Hu157-specific T cells in patients.
(A) Screening of Hu peptides in HLA-A0201 transgenic mice. CD8+ T cells purified from AAD mouse spleen cultures were cultured with stimulator cells pulsed with cognate or irrelevant peptide. A positive response was considered at least a 2-fold increase in signal over an irrelevant A0201 binding control peptide. (B) CD8+ T cells were tested for their ability to recognize whole HuD protein processed and presented by primary HHD kidney cells. Wild-type kidney cells were not recognized (data not shown). Peptide 315 was positive in the first screen (A) but was not processed on HLA-A0201 in the kidney cell experiment (B), indicating possible in vitro priming of T cells to this epitope or a low-affinity T cell response, and was not selected for further experiments with patient T cells. (C) Tetramer staining of patient 2, a normal donor, and patient 3 peripheral blood T cells after 1 round of in vitro expansion with HuD157 peptide. Positive control expansions and tetramer staining were with either M1 or CMV, depending on the response of the donor to these viruses. FSC, forward scatter; ND, not determined. (D) Tetramer staining for Hu157 T cells in CSF of Hu patient 2. The percentages of tetramer-positive cells are noted above the boxed populations in C and D. Error bars represent SD of the mean of triplicate wells (A and B).
Identification of atypical HuD-specific CD8+ T cells in Hu patients.
Given the large number of candidate HuD HLA-A0201 peptides, we first screened the 19 high-scoring HuD HLA-A0201 peptides for their ability to generate immune responses in vivo, using transgenic mice expressing human HLA-A0201 (14, 23). These mice were immunized with replication-defective adenovirus expressing full-length mouse HuD protein (AdV-HuD). CD8+ splenocytes purified from immunized mice and restimulated with HuD peptides showed reactivity to 11 of the 19 candidate A0201-restricted HuD peptide-pulsed targets using IFN-γ elispot (Figure 2A).
As a more stringent follow-up screen, we assayed reactivity of CD8+ T cells to more physiologic amounts of HuD peptides. Primary kidney epithelial cells (KECs) from HLA-A0201 transgenic or wild-type control mice were infected with AdV-HuD and used as targets in an IFN-γ elispot. Of the 11 peptides tested in this secondary screen, only HuD157 T cells recognized KECs expressing the full HuD sequence (Figure 2B). This result indicated that peptide HuD157 could be naturally processed and presented on HLA-A0201.
We were able to detect Hu157-specific CD8+ T cells in the peripheral blood from 2 of 2 chronically ill A0201 Hu patients after restimulation (Hu patients 2 and 3; Figure 2C). In contrast, none of 4 control patients harbored Hu157-specific CD8+ T cells (Figure 2C and Supplemental Figures 4 and 5), demonstrating specificity of the tetramer and indicating that the patient Hu157 cell populations were not due to in vitro priming. Each control patient harbored T cells specific for at least 1 of 2 positive control tetramers specific to influenza matrix protein (M1) or CMV (Figure 2C and Supplemental Figure 4). We also detected a high percentage (~0.5%) of Hu157 tetramer–positive cells directly from cerebrospinal fluid available from Hu patient 2 (Figure 2D). Taken together, these results demonstrated that Hu patients have T cells capable of recognizing Hu157 as a naturally processed HLA-A0201 epitope.
Function of atypical Hu-specific CD8+ T cells.
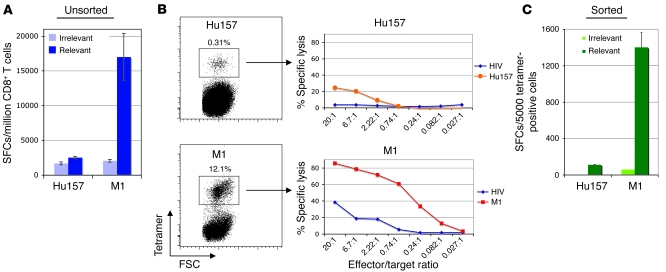
We initially attempted to assess the function of the Hu157-specific CD8+ T cells in our 2 HLA-A0201 Hu patients using standard assays. We consistently found extremely low T cell responses to Hu157-pulsed targets in CTL (data not shown) and IFN-γ elispot (Figure 3A) assays in HuD patient 2, despite positive T cell responses to influenza or CMV peptides. To address the possibility that this might have resulted from a low number of Hu157-responding T cells yielding responses at or below our limits of detection and based on our finding that 1 round of stimulation with Hu157 peptide led to expansion of HuD-specific T cells in patients but not control individuals by tetramer staining (Figure 2C and Supplemental Figure 4), we stimulated cells to obtain sufficient numbers for tetramer FACS sorting. The percentage of Hu157 tetramer–positive cells in the presorted population was at least 1 log lower than M1 tetramer–specific cells, consistent with a low precursor number of Hu157-specific CD8+ T cells present in the unstimulated T cell population (Figure 3B). Sorting cells from both M1- and Hu157-specific T cell populations allowed us to add equivalent numbers of each cell type to the assays.
Figure 3. Weak lytic activity and IFN-γ secretion by Hu157 patient T cells.
Hu157- and M1 tetramer–positive T cells from an Hu patient were expanded 1 time in vitro with peptide, followed by FACS sorting of the tetramer-positive population. Tetramer-positive cells were allowed to recover with irradiated, peptide-pulsed, autologous PBMCs and IL-2. After recovery, the function of the T cells was tested in a CTL assay. (A) ELISPOT with bulk (presort) CD8+ T cell cultures. (B) Tetramer staining showing sorting gate and CTL assay using sorted T cells. The percentages of tetramer-positive cells are noted above the boxed populations. (C) IFN-γ ELISPOT assay with sorted T cells. Error bars in A and C indicate SD of the mean of triplicate wells.
CTL assays using sorted Hu157-specific T cells from Hu patient 2 showed a low but detectable amount of specific lysis when compared with M1-specific T cells (a maximum of ~20% specific lysis seen with Hu157 over that seen with irrelevant peptide, compared with 45% of that seen with influenza, using equivalent numbers of sorted antigen-specific T cells; Figure 3B). In addition, only relatively small numbers of IFN-γ–producing cells were seen in elispot assays using Hu157-specific T cells (100 spot-forming cells [SFCs] for Hu157 compared with 1,400 SFCs for influenza; Figure 3C). Similarly, CTL assays with Hu157 T cells from patient 3 showed 40% specific lysis over that seen with irrelevant peptide, compared with 85% seen with influenza T cells, and low numbers of IFN-γ–producing cells using ELISPOT assay (40 SFCs for Hu157 compared with 540 SFCs for influenza; Supplemental Figure 6). Taken together, these experiments demonstrated that Hu157-specific CD8+ T cells are indeed present in our HLA-A0201 patients but were detectable in traditional assays for CD8+ CTL function only if the population was concentrated by sorting prior to the assay. The low signals in both assays, even with sorted Hu157 T cells, suggested that this population is either relatively unresponsive or harbors cells with functions other than the typical CD8+ IFN-γ secretion and robust lytic activities, for which different assays are needed.
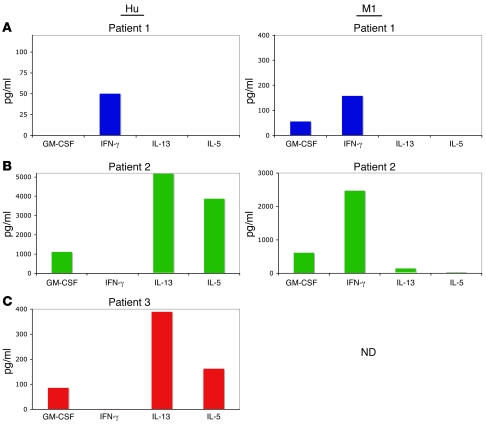
Accordingly, we searched for differences between Hu157-specific T cells and M1-specific T cells. The 2 CD8+ populations were nearly identical with respect to 8 standard cell-surface markers (Supplemental Figure 7). Since cytokine production profiles have been used to discriminate T cell subtypes (24), we measured 25 cytokines in the supernatants of cultures of CD8+ T cells incubated with cognate peptide-pulsed target cells. Multiplex analysis of culture supernatants of Hu-specific CD8+ T cells from Hu patient 1 revealed a classic CTL profile, with the secretion of IFN-γ in response to HuD peptide and influenza peptide (Figure 4A). In stark contrast, the atypical Hu-specific T cells from Hu patients 2 and 3 exhibited robust production of IL-13 (5,130 pg/ml and 389 pg/ml) and IL-5 (3,857 pg/ml and 162 pg/ml) in response to Hu157 peptide but no detectable IFN-γ production (Figure 4, B and C). Similar IL-13 production in response to Hu peptide was obtained with unsorted bulk T cell cultures, indicating that tetramer binding and sorting itself did not result in the unusual T cell phenotype (Supplemental Figure 8). We also found that Hu patient 2 harbored typical T cells specific for influenza, as the CD8+ T cells produced IFN-γ in response to M1-specific peptide (2,471 pg/ml; Figure 4B).
Figure 4. Screening for cytokines secreted by Hu-specific T cells reveals a Th2-like phenotype in some patients.
Supernatants from tetramer-sorted T cells cultured with target cells, pulsed with irrelevant or relevant peptide, were analyzed for the presence of 25 cytokines using a Multiplex kit. (A) Supernatants from patient 1, (B) from patient 2, and (C) from patient 3.
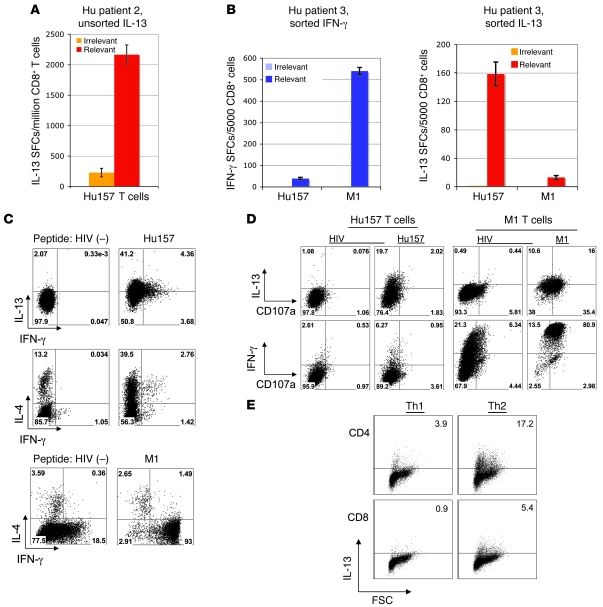
We extended these observations by developing an ELISPOT assay to enumerate T cells producing IL-13 in response to antigen. We cocultured stimulated CD8+ T cells from Hu patient 2 and Hu157 tetramer–sorted T cells from Hu patient 3 with peptide-pulsed T2 cells for 20 hours and counted IL-13–producing cells. Cells from Hu patient 2 showed robust antigen-specific production of IL-13 and in contrast demonstrated weak production of IFN-γ (Figure 5A; 1,937 IL-13 SFCs above background [irrelevant peptide] SFCs/million T cells; 833 IFN-γ SFCs above background [irrelevant peptide] SFCs/million T cells; total number of possible SFCs is 3,100 based on 0.31% tetramer-positive cells in Figure 2B). Similarly, cells from Hu patient 3 showed an IL-13 response to antigen (159 SFCs/5,000 sorted cells; Figure 5B). At the same time, both patients 2 and 3 had an IFN-γ response to M1 peptide (Figure 3A and Figure 5B).
Figure 5. Hu patient T cells are skewed toward production of IL-13 and IL-4 but not IFN-γ.
(A) ELISPOT assay demonstrating the number of IL-13–producing, Hu-specific CD8+ cells in a bulk, unsorted Hu157 culture from patient 2 (same cells as in Figure 3A). (B) ELISPOT results using sorted cells from patient 3, comparing IFN-γ and IL-13 responses. Error bars in A and B indicate SD of the mean of triplicate wells. (C) Intracellular staining for IFN-γ, IL-13, and IL-4, using sorted T cells from Hu patient 2. The top 2 rows show Hu157 cells after irrelevant (HIV) and Hu157 peptide stimulation. The bottom row shows staining of M1-specific cells after irrelevant and M1 peptide stimulation. (D) Staining for Hu157- and M1-specific CD8+ T cells with lytic activity, using CD107a antibody combined with intracellular cytokine staining. The percentages of cells in each quadrant are denoted in the corners of the plots in C and D. (E) Intracellular staining of in vitro–skewed T cells showing production of IL-13 by CD8+ T cells and CD4+ T cells (positive controls) cultured in the presence of Th2 conditions (IL-4) but not Th1 conditions (IL-12, anti–IL-4). Numbers in corners represent the percentage of IL-13+ cells.
We next confirmed that IL-13 was produced from Hu-specific T cells in a cell autonomous manner by analysis of Hu157 tetramer–positive T cells, using intracellular FACS analysis for IL-13. Over 40% of the Hu157-specific T cells from patient 2 were producing IL-13 when assayed at a single 18-hour time point (Figure 5C, top panels). These results demonstrated that IL-13 was specifically being produced by an atypical HuD-specific CD8+ T cell.
Characterization of Hu157-specific type 2 CD8+ T cells.
We characterized the responses of sorted CD8+ T cells to antigen stimulation by measuring production of type 1 and type 2 cytokines, using intracellular FACS. Approximately 30% of the Hu157-specific T cells produced IL-4 specifically in response to antigen (Figure 5C). A small subset (~10%) of either the IL-13– or IL-4–producing cells made IFN-γ in response to antigen (Figure 5C). In contrast, M1-specific T cells stimulated in parallel from the same patient produced almost exclusively IFN-γ and very little IL-4 (Figure 5C). Taken together, our results indicated that Hu157-specific T cells were skewed toward the production of Th2 cytokines, including IL-13, IL-5, and IL-4, and should be considered to be type 2 CD8+ T cells.
Given the modest lytic activity seen in Figure 3, we asked whether the poor ability of IL-13–positive T cells to kill is a function of weak activity of the entire population or whether it derives from a smaller subpopulation of cells. We assayed sorted Hu157 T cells by staining them for intracellular cytokines along with the lysosomal marker CD107a, a marker of lytic effectors (25). Only a small number of these Hu157 T cells were CD107a positive (~4%; Figure 5D, left panels). Ten percent of the IL-13+ and twelve percent of the IFN-γ+ population stained for CD107a. In contrast, M1-specific T cells from the same patient produced almost exclusively IFN-γ (93% positive), with very little IL-4 and IL-13, and approximately 75% displayed lytic function (Figure 5D, right panels). These results indicated that a small population of Hu157-specific T cells, perhaps IL-13+/IFN-γ+ double-positive cells, can act as lytic effectors.
We further characterized the Hu157-specific T cell phenotype by contrasting its gene expression profile with that of M1-specific T cells. Expanded T cells were sorted with Hu157- or M1-specific tetramers, activated for 18 hours, and profiled to ascertain RNA expression using Affymetrix microarray. Given limited clinical material available, we were restricted to analysis of a single patient but were able to repeat experiments using different means of stimulation (autologous DCs pulsed with peptide or with anti-CD28 antibody). We found that the genes for IL-13 and IL-5 were expressed in Hu157 T cells and those for IFN-γ and granzyme B were upregulated in M1-specific T cells in response to specific peptide (Table 1 and Supplemental Table 3). This suggests that the microarray data provided an accurate reflection of the different proteins present in each T cell type (Figures 4 and 5). We also searched for robust changes in gene expression (≥5 fold) that were present in both peptide-stimulated and CD28 antibody-stimulated Hu157 T cells but not in M1-specific T cells (Table 1). Taken together, the data provided evidence that activated Hu157-specific type 2 T cells may specifically induce gene expression of IL13 (18-fold), IL5 (12-fold), and other immune genes (such as CD25, CXCL10, CD30, and GITR; Table 1 and Supplemental Table 3). These data illustrated that there may be a unique gene profile of Hu157-specific T cells and support their classification as type 2 CD8+ T cells.
Table 1 .
Microarray analysis of Hu157-specific type 2 T cells
In order to address whether SCLC tumor cells might contribute to the skewing of HuD-specific T cells toward a type 2 phenotype, we analyzed published microarray data (26) of SCLC and normal lung samples. These data indicated that the Th2 cytokines IL-4 and IL-5 were expressed at 4- and 7-fold higher, respectively, in tumors than in normal lung. Moreover, although IL-13 expression was detected in both normal and tumor samples, the mean IL-13 levels were 2-fold higher in tumors (Table 2).
Table 2 .
Expression of cytokine genes induced in SCLC compared with normal lung expression, determined by analysis of microarray data (26)
We next tested whether these cytokines were able to skew naive CD8+ T cells to a type 2 phenotype. We cultured primary CD8+ T cells in the presence of different cytokine mixtures and measured their cytokine production using intracellular FACS. As a positive control, we cultured CD4+ T cells in the presence of IL-4, which skewed them toward production of Th2-type cytokines as expected (Figure 5E). When we cultured CD8+ T cells in the presence of IL-4, we also found that they produced type 2 cytokines (IL-4 and IL-13; Figure 5E and data not shown). Taken together, these results suggested that in some patients, Th2 cytokine production by SCLC may lead to skewing of the HuD immune response toward production of type 2 CD8+ T cells.
Discussion
The paraneoplastic Hu syndrome is characterized by an immune response identified with a high-titer antibody. However, the target antigen is an intracellular protein expressed in SCLC and neurons, which raised the possibility that there may be a cellular component to the disorder. Moreover, these patients, as well as a significant percentage of the general population of SCLC patients, show evidence of tumor immunity (19, 27), including spontaneous tumor regression (3), further supporting the possibility of a cell-mediated component to the disorder. Yet Hu-specific CTLs have not previously been described in the disorder. Here, we provide an explanation for these observations. Different Hu patients harbor different kinds of HuD-specific T cell responses at low precursor frequencies: either typical CTL responses or atypical, noncytotoxic CD8+ T cells that secrete type 2 cytokines, a characteristic that is missed using standard functional assays to look for antigen-specific CD8+ T cells.
Previous studies have demonstrated type 2 cytokine-producing CD8+ T cells in vitro and have suggested the possibility that such cells might exist in humans. Several groups have shown that in vitro stimulation and skewing of mouse T cells can produce CD8+ T cells that make IL-4 and other type 2 cytokines (25, 28). In humans, cell clones grown from patients with lepromatous leprosy (29) and a subset of HIV patients with Job’s-like syndrome (30) have also been shown to produce IL-4, although follow-up studies have not reported the presence of such cells in the absence of repeated in vitro stimulations. Finally, one report of high IL-13 levels in patients with amyotrophic lateral sclerosis could be interpreted as consistent with the presence of such cells in vivo (31). Here, we detect type 2 CD8+ T cells in Hu patients after one in vitro peptide stimulation, reflecting the low precursor frequency of these cells as indicated by tetramer staining. We do not believe that this stimulation accounts for the type 2 phenotype, since M1-specific T cells from the same patients, treated in parallel, maintain a type 1 phenotype. Moreover, tetramer sorting was not responsible for the type 2 phenotype, since it was also detected when Hu157 peptide was used to stimulate unsorted bulk T cell cultures. Finally, HuD-specific T cells were not detected in the peripheral blood of 6 neurologically normal patients, indicating that the general population does not harbor such T cells, even after in vitro stimulation.
The spectrum of tumor immune response in Hu patients may be a reflection of the spectrum of different antigen-specific T cell types. Patients with more robust tumor immunity (3) may harbor typical IFN-γ–producing CTLs. Others, despite harboring tumors expressing the HuD tumor antigen, may have a more aggressive course with their tumors (4) as a consequence of harboring type 2 CD8+ T cells. This hypothesis is consistent with the observation that type 2 CD8+ T cells have very little cytolytic activity. One explanation for the presence of such cells may be that the tumor itself acts to reshape the immune response, as in the phenomenon of tumor immunoediting demonstrated in animal models by Schreiber and colleagues (32). Cytokines have been noted to contribute to tumor immunoediting, either through direct killing of tumor cells or through the induction of immunosuppressive IL-13–producing NKT cells or CD4+CD25+ regulatory T cells (33). Our data extend these observations by providing evidence that SCLCs express type 2 cytokines and that one of these, IL-4, is sufficient to skew tumor antigen–specific CD8+ T cells to the type 2 phenotype.
It is of interest to consider what role type 2 CD8+ T cells may be playing in the PND. Prior studies have suggested that such cells may play a helper role for eosinophils and antibody production (30, 34, 35). While our patients did not have an elevated eosinophil count, it is clear that a hallmark of the PNDs is a very high-antibody titer, and this was evident in our Hu patients harboring type 2 CD8+ T cells (data not shown). Cytotoxicity of mouse (25, 36) and human type 2 CD8+ T cells has been reported, with relatively weak killing activity in the latter (30). Similarly, our data suggested weak but detectable cytotoxic activity of Hu-specific type 2 CD8+ T cells (Figure 3) and that this may be due to lytic activity of a minor subset of IL-13+/IFN-γ+ cells (Figure 5).
The cytokine profile of Hu-specific type 2 CD8+ T cells is closest to classical CD4+ Th2 cells, in that they produce especially large amounts of IL-13 and IL-5 (by cytokine and gene array) as well as IL-4 (by intracellular FACS); notably, these cells do not produce IL-17, IL-6, or TNF-α, the cytokine hallmarks of autoimmune Th17 cells (24). Cells producing classical type 2 cytokines are often thought of as part of a compensatory response to autoimmune disease, involved in the resolution of an immune response, for example by downregulating type 1 T cells (24, 37). We note that the 2 Hu patients with type 2 CD8+ T cells were in a chronic phase of their illness, suggesting the possibility that acutely ill patients (e.g., Hu Patient 1) harbor a classical IFN-γ CD8+ T cell response and that this can segue into a chronic type 2 T cell response over time. Importantly, Hu157-specific T cells were detected in the cerebrospinal fluid (CSF) of 1 patient (Figure 2), which implicated them as playing a role in brain degeneration.
Our studies defined a clear cell-mediated component to Hu paraneoplastic syndrome. This disorder can be associated with a spectrum of clinical outcomes — from profound tumor immunity (3) to tumor outgrowth and death from cancer (4) and from restricted peripheral nervous system disease, most commonly subacute pan-sensory neuropathy, to isolated central nervous system disease, such as limbic encephalopathy, to a multifocal encephalomyelopathy, with rapid death from neurologic disease. Our data underscore that models of PND pathogenesis should be refined to take into account the observation that different kinds of T cells may be present in different PND patients (6) and that these variants may explain the different responses to tumor and brain seen clinically. Identification of qualitatively different kinds of Hu-specific T cells in different Hu patients provides a new focus for trying to understand the clinical complexity of the syndrome and for developing new therapies that target human immune responses.
Methods
Patients.
All patients were seen at The Rockefeller University Hospital (RUH) and gave informed consent, under protocols approved by The Rockefeller University Institutional Review Board. The Hu antibody was diagnosed in all patients by Western blot against recombinant HuD fusion protein. High resolution HLA typing was performed at the Immunogenetics Laboratory of the University of Maryland Medical Center.
Characteristics of HuD patient 1.
A 59-year-old male smoker was first seen at RUH in November 2006, 5 months after the onset of short-term memory loss, numbness and tingling in the fingertips of both hands, and worsening gait. Workup revealed an asymptomatic lung nodule, which was biopsied and found to be SCLC. When examined at RUH, the patient exhibited profound short-term memory loss, with no recall of events occurring 5 minutes prior, and pronounced emotional lability. Lumbar puncture revealed a CSF pleocytosis of 6 wbc/mm3. One week later, the patient went into cardiac arrest on his way to RUH for treatment and expired. At time of patient’s death, he was in the midst of completing chemotherapy (cisplatin and etoposide) for his cancer and had completed radiation therapy the week prior.
Characteristics of HuD patient 2.
A 74-year-old woman smoker was well until 2005, at which time she presented with progressive numbness in the feet and hands as well as fatigue and weight loss. Workup revealed the anti-Hu antibody in October 2005 and SCLC in June 2006, for which she underwent chemotherapy and radiation therapy. On examination at that time the patient had a moderate pan-sensory neuropathy, with dysesthesia in the distal hands and feet, and moderate gait imbalance requiring a cane. In subsequent follow-up visits, her cancer has remained in remission; however, her neurologic symptoms have progressively worsened to the point that she requires a walker to ambulate and has chronic painful neuropathy.
Characteristics of HuD patient 3.
A 65-year-old woman developed numbness in her feet in spring of 2001. In April 2002, workup revealed SCLC, for which she received chemotherapy and prophylactic brain irradiation. In March 2003, she developed difficulty walking and coordination problems, becoming wheelchair bound over the ensuing months. She was diagnosed with anti-Hu antibody in July of that year. The patient was admitted to RUH in April of 2004, when she had worsening dysarthria and increased tremors of left hand and shoulder. She exhibited a CSF pleocytosis of 13 wbc/mm3 at that time. To date, the patient remains relatively stable with her symptoms, and her cancer remains in remission.
Cell lines and antibodies.
Antibodies for FACS staining were obtained from Becton Dickinson, except FITC-conjugated CD8 (Beckman Coulter). The EL4 cell line expressing HLA-0201 was obtained from the laboratory of Alan Houghton (Memorial Sloan-Kettering Cancer Center). T2 cells were obtained from ATCC.
Peptides.
The peptide library for iTopia screening was purchased from Jerini Peptide Technologies. Peptides for mouse experiments and influenza matrix peptide GILGFVFTL were purchased from American Peptide Company. The CEF peptide mixture was purchased from MabTech. Influenza nucleoprotein peptide for A0301 (ILRGSVAHK) was purchased from AnaSpec. Negative control peptides were HIVgag for A0201 (SLYNTVATL) and cdr2331 for A0301 (CIRRAKAVK).
iTopia screening.
Peptides were dissolved in DMSO (10 mM) and stored in aliquots at –20°C. All screening assays were performed according to the manufacturer’s instructions using the provided reagents (Beckman Coulter Immunomics). For the initial binding assay, each peptide (0.1 mM) was added to plates with wells coated with recombinant HLA molecules, B2m and FITC-labeled anti-HLA class I. Incubation was performed for 18 hours at 21°C, and plates were then washed and read in a fluorescence plate reader (excitation 490 nm, emission 520 nm) (Molecular Devices). For affinity measurements, 8 serial dilutions of the peptides were made and assayed as in the binding assay. For off-rate measurements, peptides were added as in the binding assay, incubated for 18 hours at 21°C, washed, and then shifted to 37°C for the indicated time points. Data were analyzed with the iTopia software using Prism (GraphPad), and iScores were generated using a proprietary formula, which takes into account all 3 parameters measured.
Mice and immunizations.
Wild-type C57BL/6 mice were purchased from The Jackson Laboratory. AAD mice are transgenic for the human HLA-A0201 molecule, with a mouse α3 domain (23) on a C57BL/6 background, and were purchased from The Jackson Laboratory. HHD mice are transgenic for the human HLA-A0201 molecule, with a mouse α3 domain on a B2m–/– background, and were obtained from F. Lemmonier (Institut Pasteur, Paris, France) (14, 38). Transgenic mice were monitored for expression of HLA-A0201 on peripheral blood cells by flow cytometry using the BB7.2 antibody (BD Biosciences — Pharmingen). Mice were immunized intradermally with 100 μl purified virus. On day 0 and day 2, 400 μg of pertussis toxin (Sigma-Aldrich) was administered intraperitoneally. Mice were used for experiments 13 days after adenovirus immunization. All mice were housed in a specific pathogen–free facility and were used according to IACUC regulations under an animal protocol approved by The Rockefeller University.
Adenovirus production.
Control adenovirus expressing GFP and adenovirus expressing GFP and the HuD protein were made by H.J. Okano (Keio University, Tokyo, Japan) using the pAdenoTrack vector and recombination in bacteria as previously described (39). Adenovirus was produced for experiments by transduction of HEK293 cells (ATCC), and virus in the supernatant was purified after 72 hours using the Adenopure kit (Puresyn Inc.) according to kit instructions. Viral titers were determined by serial dilution in 293 cell cultures and counting GFP-positive cells after 24 hours. Virus was used at 1:10,000 titer.
Mouse kidney cell culture.
Primary kidney cell cultures were made by mashing kidneys with the back of a syringe, pipetting until a single-cell suspension was obtained, and passing the suspension over a 70 μm cell strainer. After washing, cells were cultured in D10 medium (DMEM) supplemented with gentamicin, l-glutamine (Cellgro; Mediatech), and 10% FBS (HyClone). Cells were fed by replacing medium on days 4 and 7. On day 7, 10 U/ml recombinant mouse IFN-γ (R&D Systems) was added to the cells. On day 8, 10 μl of purified adenovirus was added to each plate. Cells were harvested the next day for use in the ELISPOT assay.
Peripheral blood isolation.
Blood cells were collected by leukapheresis, under The Rockefeller University Institutional Review Board–approved protocol with informed consent. PBMCs were isolated by density gradient centrifugation over Ficoll-Hypaque (Pharmacia) and separated into T cell–enriched (ER+) and T cell–depleted (ER–) fractions by rosetting with neuraminidase-treated (Calbiochem) sheep red blood cells (Colorado Serum Company). These cell fractions were cryopreserved by freezing in 10% human serum albumin/10% DMSO/RPMI.
DC culture.
Human DCs were generated from PBMCs. Briefly, ER– cells were cultured in the presence of 100 U/ml IL-4 (R&D Systems) and 100 U/ml GM-CSF (Immunex) for 6 days to generate immature DCs. DCs were harvested on day 6 and plated with PGE2 and TNF-α to induce maturation. After 2 days, mature DCs (mDCs) were harvested and used for T cell stimulation.
CD8+ T cell isolation.
ER+ PBMC fractions were thawed and incubated with anti-CD8 MACS beads according to manufacturer’s instructions (Miltenyi Biotec). For experiments with adenovirus-immunized mice, spleens were harvested on day 13, and CD8+ T cells were purified as in the human protocol using anti-mouse CD8a beads (Miltenyi Biotec).
In vitro stimulation of human peripheral blood T cells.
Purified CD8+ T cells were incubated with autologous mDCs at a 30:1 ratio in 24-well plates, with 10 μg/ml peptide in AIM-V medium (Invitrogen) supplemented with 5% FBS. After 1 day of culture, recombinant human IL-2 (Chiron) was added at 10 IU/ml.
In vitro stimulation of mouse spleen cells.
Spleen cells from adenovirus-immunized mice were used for restimulation with indicated peptides. Cells were stimulated at 2 × 106 per well of a 24-well plate, with 0.5 μM peptide and 10 IU/ml recombinant human IL-2 (Chiron) in RPMI with 10% FBS. After 10 days of culture, cells were harvested and CD8+ T cells were isolated by magnetic separation (Miltenyi Biotec). Subsequent stimulations were performed similarly with syngeneic irradiated, peptide-pulsed splenocytes.
Tetramer staining.
All HLA-A0201 tetramers were obtained from Beckman Coulter Immunomics. Each lot of tetramer was tested on a restimulated Hu patient T cell line of known haplotype and known to be reactive by tetramer and ELISPOT. Tetramers were titrated and compared with previous results before use in experiments. For 20 minutes, 1.0 × 106 patient PBMCs or 0.3 × 106 cultured CD8+ cells were incubated with a 1:20 dilution of tetramer at room temperature. Antibody to CD8 was then added to the cells for an additional 10 minutes, and cells were washed and analyzed immediately using a BD FACSCaliber. Dead cells were excluded by gating on Topro-3–negative (Molecular Probes) cells and on the CD8+ population.
Tetramer-positive T cell sorting.
CD8+ T cells were purified by positive selection (Miltenyi Biotec) from the frozen T cell fractions of patient leukapheresates. After 8–12 days of culture with mDCs, the cells were harvested and incubated at 20 × 106/ml with a 1:20 dilution of tetramer. The tetramer-positive population was sorted on a FACSAria instrument (BD Biosciences). After sorting, cells were placed into culture in 96-well plates with 10:1 autologous, peptide-pulsed PBMCs and 50 U/ml IL-2. After a recovery period of 8 days in culture, T cells were used in assays as described.
ELISPOT.
Nitrocellulose plates (Millipore), coated overnight with 5 μg/ml anti–IFN-γ or IL-13 antibody (MabTech), were blocked for 2 hours with 10% FBS at 37°C. CD8+ T cells were plated with 25,000 peptide-pulsed target cells and incubated for 20 hours at 37°C. Cytokine-producing cells were detected using biotinylated secondary antibodies (MabTech), and SFCs were developed by incubation with streptavidin-horseradish peroxidase (Vector Laboratories) and AEC substrate (Sigma-Aldrich). All conditions were performed in duplicate or triplicate, and SFCs were enumerated by an outside consultant (Zellnet Consulting).
Cytotoxicity assay.
Sorted CD8+ T cells were incubated with 51chromium-loaded peptide-pulsed T2 cells for 4 hours. Supernatants were harvested and counted in a TopCount Counter (Packard). The percentage of specific lysis was calculated using the following formula: (specific cpm – medium only cpm)/(maximum lysis – medium only cpm) ×100. Spontaneous release was determined by counting 51chromium in the medium of each target, and maximum lysis was determined by adding 1% SDS to each target. All conditions were done in at least triplicate, and error bars represent SD at each point.
Multiplex analysis of cytokines.
Supernatants were taken from sorted or unsorted T cells, cultured with T2 cells pulsed with HIV (negative control), M1, or Hu157 peptide for 24 hours. Samples were analyzed neat and at a 1:10 dilution using the Biosource Human Cytokine 25-Plex kit (Invitrogen) according to the manufacturer’s instructions and a Perkin-Elmer Luminex 200 reader.
Intracellular FACS staining.
Tetramer-sorted T cells were incubated at a 1:1 ratio, with HIV (negative control), M1, or Hu157 peptide-pulsed T2 stimulator cells in 96-well plates, for either 6 hours (CD107a and cytokines) or 18 hours (IL-13 only staining) with CD107a antibody, 10 μg/ml brefeldin A, and 6 μg/ml monensin in R10 medium. Cells were washed 3 times with PBS, stained in FACS buffer with CD8 antibody for 20 minutes at 4°C, and washed, followed by 30 minutes at 4°C with Cytofix/Cytoperm (BD Biosciences — Pharmingen). After washing, cells were stained in Perm/Wash buffer (BD Biosciences) with IFN-γ, IL-13, and IL-4 antibodies for 30 minutes at 4°C, followed by washing and acquisition on a BD FACSCaliber.
Microarray analysis.
Tetramer-sorted T cells specific for each antigen were incubated with mDCs pulsed with relevant peptide or irrelevant peptide, or in anti-CD28– and tetramer-coated wells for 18 hours. Samples of culture supernatants were taken, and the cells were lysed with buffer RLT (RNAeasy kit; Qiagen) containing RNAsin (Promega). Probes for genechips were synthesized by double amplification (Ambion) and hybridized onto Affymetrix HG133 Plus 2.0 chips. Data were normalized using GeneSpring software (Agilent) gene chip-robust multiarray averaging (GC-RMA) normalization, followed by application of a filtering signal of more than 100. Fold change of relevant peptide response compared with irrelevant peptide response was determined for each T cell group.
In vitro skewing of T cells.
Naive T cells were purified from normal donor ER+ cells using the Miltenyi Biotec Naive T Cell Isolation kit, followed by positive selection for CD4+ and CD8+ T cells, according to the manufacturer’s instructions. One million T cells were plated in anti-CD3/CD28 coated 24-well plates with 12.5 μg/ml IL-4 (Th1) or 2.5 ng/ml IL-12 (R&D Systems) and 5 μg/ml anti–IL-4 (BioLegend) (Th2) for 4 days and then rested in the same cytokine conditions for 7 days. T cells were then harvested and stimulated with 50 ng/ml PMA and 1 μg/ml ionomycin for 4 hours before intracellular staining.
Statistics.
To analyze the significance of changes in cytokine levels, a 2-tailed paired Student’s t test was used. P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank The Rockefeller University Flow Cytometry Core Facility for expert help in sorting tetramer-positive cells, the Translational Immunomonitoring Resource Center of RUH Center for Clinical and Translational Science (CTSA) for help with Luminex analysis, James Okano for generating the AdenoHuD construct, and members of the Darnell laboratory for critical discussion and review of the manuscript. This work was supported by the NIH grant (R01 CA85784 to R.B. Darnell) and RUH CTSA grant (UL1 RR024143) from the National Center for Research Resources at the NIH. R.B. Darnell is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: CSF, cerebrospinal fluid; M1, influenza matrix protein; mDC, mature DC; PND, paraneoplastic neurologic disorder; RUH, The Rockefeller University Hospital; SFC, spot-forming cell; SCLC, small cell lung cancer.
Citation for this article: J. Clin. Invest. 119:2042–2051 (2009). doi:10.1172/JCI36131
References
- 1.Darnell R.B. Onconeural antigens and the paraneoplastic neurologic disorders: at the intersection of cancer, immunity and the brain. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4529–4536. doi: 10.1073/pnas.93.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darnell R.B., Furneaux H.M., Posner J.B. Antiserum from a patient with cerebellar degeneration identifies a novel protein in Purkinje cells, cortical neurons, and neuroectodermal tumors. J. Neurosci. 1991;11:1224–1230. doi: 10.1523/JNEUROSCI.11-05-01224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell R.B., Posner J.B. Paraneoplastic syndromes involving the nervous system. N. Engl. J. Med. 2003;349:1543–1554. doi: 10.1056/NEJMra023009. [DOI] [PubMed] [Google Scholar]
- 4.Darnell R.B., Posner J.B. Paraneoplastic syndromes affecting the nervous system. Semin. Oncol. 2006;33:270–298. doi: 10.1053/j.seminoncol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Graus F., Elkon K.B., Cordon-Cardo C., Posner J.B. Sensory neuronopathy and small cell lung cancer; antineuronal antibody that also reacts with the tumor. Am. J. Med. 1986;80:45–52. doi: 10.1016/0002-9343(86)90047-1. [DOI] [PubMed] [Google Scholar]
- 6.Roberts W.K., Darnell R.B. Neuroimmunology of the paraneoplastic neurological degenerations. Curr. Opin. Immunol. 2004;16:616–622. doi: 10.1016/j.coi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Szabo A., et al. HuD, a paraneoplastic encephalomyelitis antigen contains RNA-binding domains and is homologous to Elav and sex lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-Z. [DOI] [PubMed] [Google Scholar]
- 8.Ma W.J., Cheng S., Campbell C., Wright A., Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 9.Myer V.E., Fan X.H.C., Steitz J.A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okano H.J., Darnell R.B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert M.L., et al. Tumor-specific killer cells in paraneoplastic cerebellar degeneration. Nat. Med. 1998;4:1321–1324. doi: 10.1038/3315. [DOI] [PubMed] [Google Scholar]
- 12.Albert M.L., Austin L.M., Darnell R.B. Detection and treatment of activated T cells in the cerebrospinal fluid of patients with paraneoplastic cerebellar degeneration. Ann. Neurol. 2000;47:9–17. doi: 10.1002/1531-8249(200001)47:1<9::AID-ANA5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 13.Darnell R.B., Albert M.L. cdr2-specific CTLs are detected in the blood of all patients with paraneoplastic cerebellar degeneration analyzed. Ann. Neurol. 2000;48:270–271. doi: 10.1002/1531-8249(200008)48:2<270::AID-ANA25>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 14.Santomasso B.D., et al. A T-cell receptor associated with naturally occurring human tumor immunity. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19073–19078. doi: 10.1073/pnas.0704336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Beukelaar J.W., et al. No evidence for circulating HuD-specific CD8+ T cells in patients with paraneoplastic neurological syndromes and Hu antibodies. Cancer Immunol. Immunother. 2007;56:1501–1506. doi: 10.1007/s00262-007-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plonquet A., et al. Peptides derived from the onconeural HuD protein can elicit cytotoxic responses in HHD mouse and human. J. Neuroimmunol. 2003;142:93–100. doi: 10.1016/S0165-5728(03)00269-8. [DOI] [PubMed] [Google Scholar]
- 17.Rousseau A., et al. T cell response to Hu-D peptides in patients with anti-Hu syndrome. J. Neurooncol. 2005;71:231–236. doi: 10.1007/s11060-004-1723-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka M., et al. Cytotoxic T cell activity against peptides of Hu protein in anti-Hu syndrome. J. Neurol. Sci. 2002;201:9–12. doi: 10.1016/S0022-510X(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 19.Dalmau J., Furneaux H.M., Gralla R.J., Kris M.G., Posner J.B. Detection of the anti-Hu antibody in the serum of patients with small cell lung cancer--a quantitative western blot analysis. Ann. Neurol. 1990;27:544–552. doi: 10.1002/ana.410270515. [DOI] [PubMed] [Google Scholar]
- 20.Darnell R.B., Posner J.B. Observing the invisible: successful tumor immunity in humans. Nat. Immunol. 2003;4:201. doi: 10.1038/ni0303-201. [DOI] [PubMed] [Google Scholar]
- 21.Bachinsky M.M., et al. Mapping and binding analysis of peptides derived from the tumor-associated antigen survivin for eight HLA alleles. Cancer Immun. 2005;5:6. [PubMed] [Google Scholar]
- 22.Weichold F.F., et al. Impact of MHC class I alleles on the M. tuberculosis antigen-specific CD8+ T-cell response in patients with pulmonary tuberculosis. Genes Immun. 2007;8:334–343. doi: 10.1038/sj.gene.6364392. [DOI] [PubMed] [Google Scholar]
- 23.Newberg M.H., et al. Importance of MHC class 1 alpha2 and alpha3 domains in the recognition of self and non-self MHC molecules. J. Immunol. 1996;156:2473–2480. [PubMed] [Google Scholar]
- 24.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat. Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 25.Sad S., Marcotte R., Mosmann T.R. Cytokine-induced differentiation of precursor mouse CD8+ T cells into cytotoxic CD8+ T cells secreting Th1 or Th2 cytokines. Immunity. 1995;2:271–279. doi: 10.1016/1074-7613(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee A., et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauer S., Andreou I. Tumor progression and serum anti-HuD antibody concentration in patients with paraneoplastic neurological syndromes. Eur. Neurol. 2002;47:189–195. doi: 10.1159/000057897. [DOI] [PubMed] [Google Scholar]
- 28.Seder R.A., et al. CD8+ T cells can be primed in vitro to produce IL-4. J. Immunol. 1992;148:1652–1656. [PubMed] [Google Scholar]
- 29.Salgame P., et al. Differing lymphokine profiles of functional subsets of human CD4 and CD8 T cell clones. Science. 1991;254:279–282. doi: 10.1126/science.1681588. [DOI] [PubMed] [Google Scholar]
- 30.Maggi E., et al. Th2-like CD8+ T cells showing B cell helper function and reduced cytolytic activity in human immunodeficiency virus type 1 infection. J. Exp. Med. 1994;180:489–495. doi: 10.1084/jem.180.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi N., et al. Increased IL-13-producing T cells in ALS: positive correlations with disease severity and progression rate. J. Neuroimmunol. 2007;182:232–235. doi: 10.1016/j.jneuroim.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Koebel C.M., et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 33.Dunn G.P., Old L.J., Schreiber R.D. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 34.Coyle A.J., et al. Virus-specific CD8+ cells can switch to interleukin 5 production and induce airway eosinophilia. J. Exp. Med. 1995;181:1229–1233. doi: 10.1084/jem.181.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Punnonen J., Yssel H., de Vries J.E. The relative contribution of IL-4 and IL-13 to human IgE synthesis induced by activated CD4+ or CD8+ T cells. J. Allergy Clin. Immunol. 1997;100:792–801. doi: 10.1016/S0091-6749(97)70276-8. [DOI] [PubMed] [Google Scholar]
- 36.Sad S., et al. Cytotoxicity and weak CD40 ligand expression of CD8+ type 2 cytotoxic T cells restricts their potential B cell helper activity. Eur. J. Immunol. 1997;27:914–922. doi: 10.1002/eji.1830270417. [DOI] [PubMed] [Google Scholar]
- 37.Mosmann T.R., Sad S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol. Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 38.Firat H., et al. H-2 class I knockout, HLA-A2.1-transgenic mice: a versatile animal model for preclinical evaluation of antitumor immunotherapeutic strategies. Eur. J. Immunol. 1999;29:3112–3121. doi: 10.1002/(SICI)1521-4141(199910)29:10<3112::AID-IMMU3112>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.He T.C., et al. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.