Abstract
Heterozygous mutations in the gene encoding the pancreatic homeodomain transcription factor pancreatic duodenal homeobox 1 (PDX1) are associated with maturity onset diabetes of the young, type 4 (MODY4) and type 2 diabetes. Pdx1 governs the early embryonic development of the pancreas and the later differentiation of the insulin-producing islet β cells of the endocrine compartment. We derived a Pdx1 hypomorphic allele that reveals a role for Pdx1 in the specification of endocrine progenitors. Mice homozygous for this allele displayed a selective reduction in endocrine lineages associated with decreased numbers of endocrine progenitors and a marked reduction in levels of mRNA encoding the proendocrine transcription factor neurogenin 3 (Ngn3). During development, Pdx1 occupies an evolutionarily conserved enhancer region of Ngn3 and interacts with the transcription factor one cut homeobox 1 (Hnf6) to activate this enhancer. Furthermore, mRNA levels of all 4 members of the transcription factor network that regulates Ngn3 expression, SRY-box containing gene 9 (Sox9), Hnf6, Hnf1b, and forkhead box A2 (Foxa2), were decreased in homozygous mice. Pdx1 also occupied regulatory sequences in Foxa2 and Hnf1b. Thus, Pdx1 contributes to specification of endocrine progenitors both by regulating expression of Ngn3 directly and by participating in a cross-regulatory transcription factor network during early pancreas development. These results provide insights that may be applicable to β cell replacement strategies involving the guided differentiation of ES cells or other progenitor cell types into the β cell lineage, and they suggest a molecular mechanism whereby human PDX1 mutations cause diabetes.
Introduction
Insulin deficiency due to reduced pancreatic islet β cell number underlies the progression of both type 1 and type 2 diabetes, prompting efforts to develop β cell replacement therapies. The high hopes for human islet transplantation as such a therapy have been tempered by their limited availability and short-term function after transplantation, resulting in an intense focus on the development of alternate sources of β cells, through the guided differentiation of stem or progenitor cells or the transdifferentiation of more abundant mature cells. Transdifferentiation approaches have focused largely on the expression of a single or a combination of transcription factors that drive β cell development and differentiation and most have relied, at least in part, upon pancreatic duodenal homeobox 1 (Pdx1), a homeodomain transcription factor with critical regulatory roles in early pancreas development and in the mature β cell (1–5). Recent advances in the derivation of insulin-expressing β–like cells from ES cells (6–11) have also been guided by the principles of embryonic pancreas development (reviewed in ref. 12). Clinical translation of these approaches, however, will require improvements in efficiency, fidelity, and stability of generating the β cell phenotype, efforts which will likely be informed by the identification of novel factors and regulatory relationships in embryonic pancreas development.
The embryonic development of islet β cells is critically dependent on the function of the basic helix-loop-helix transcription factor neurogenin 3 (Ngn3). Genetic lineage-tracing studies and the phenotype of Ngn3-null mice demonstrate that all islet hormone-producing endocrine cell types (α, β, δ, pancreatic polypeptide, ε) derive from Ngn3-expressing progenitors (13–20). Ngn3 is first observed in a subpopulation of pancreatic epithelial cells at E9.5, and its expression is coordinated by the Notch signaling pathway (13, 18, 21) and a transcription factor coregulatory network comprised of SRY-box containing gene 9 (Sox9), one cut homeobox 1 (Hnf6), Hnf1b, and forkhead box A2 (Foxa2) (22). Pdx1 (also known as IPF1, IDX1, STF1, and XlhBox8) is a homeodomain transcription factor expressed throughout the pancreatic epithelium at E9.0 that is critically required for organogenesis in mice and humans (23–28).
The concept that Pdx1 is upstream of Ngn3 during embryonic development is based on genetic lineage-tracing studies, indicating that Pdx1-expressing progenitors give rise to both exocrine and endocrine lineages, while the progeny of Ngn3-expressing progenitors selectively populate the endocrine compartment (16, 29). Further support derives from the marked reduction or absence of the endocrine compartment in both Ngn3 and Pdx1 loss-of-function alleles as well as the broader phenotype of Pdx1 loss of function that includes arrested development of the exocrine compartment (13, 24, 25). However, close examination of these early embryonic phenotypes reveals that the endocrine compartment requires Ngn3 but is not fully dependent upon Pdx1. The arrested dorsal ductule in Pdx1–/– mice still contains the early hormone-expressing cells at E9.5, characteristic of the primary transition, whereas Ngn3–/– mice lack both first and second wave endocrine differentiation, suggesting that Pdx1 is specifically required only for the second wave (13, 25). Further, it is not clear from these studies whether Pdx1 is required directly for the emergence, maintenance, or differentiation of endocrine progenitors or only indirectly through its critical role in the formation of the pancreatic progenitor cells from which these cells emerge.
Heterozygous Pdx1 mutations are associated with early (MODY4) and late onset forms of type 2 diabetes (reviewed in ref. 30). Some of these mutations occur in the evolutionarily conserved but poorly characterized C terminus; these mutations are associated with modest decreases in transactivation (31–33). The C terminus is not essential for DNA binding but is required for normal activation of target gene promoters (34, 35), perhaps through its interactions with 2 recently identified protein partners, Pcif1 and Meis1a, whose roles in the pancreatic β cell have yet to be determined (36–38). Therefore, to determine the in vivo role of the Pdx1 C-terminal domain, we derived a Pdx1 hypomorphic allele harboring a premature truncation mutation of the C terminus (Pdx1ΔC) and thereby uncovered a direct role for Pdx1 in the specification of Ngn3+ endocrine progenitors. Pdx1 contributes to the specification of endocrine progenitors by regulating Ngn3 directly in concert with Hnf6, which we identified as a partner of the C terminus, and by participating in a cross-regulatory network during early pancreas development. These new insights into the role of Pdx1 to promote the transition from pancreatic progenitor to endocrine progenitor have the potential to impact on the generation of β cell replacements for regenerative medicine and islet transplantation.
Results
Derivation of hypomorphic Pdx1ΔC/ΔC mice.
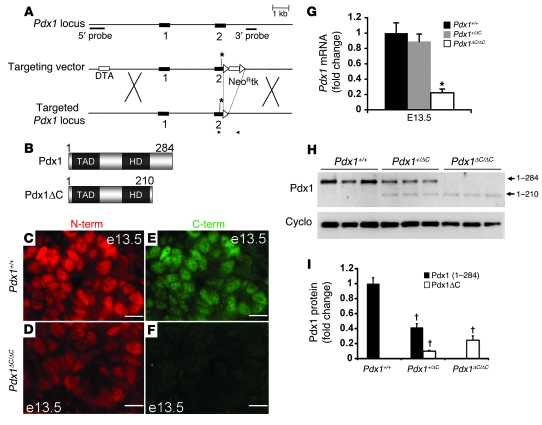
We engineered a Pdx1 targeting vector containing exon 1 and a mutated exon 2, in which the codon encoding Ser210 was replaced by a termination codon (Figure 1A). The resulting 210–amino acid Pdx1 protein, termed Pdx1ΔC (Figure 1B), was detected in pancreata from homozygous Pdx1ΔC/ΔC mice at E13.5 (Figure 1C) by immunostaining with N-terminal–specific antisera. The absence of Pdx1 staining using C-terminal–specific antisera confirmed that the Pdx1 C terminus is not translated in Pdx1ΔC/ΔC mice (Figure 1F). Pdx1 immunoreactivity was visibly reduced at E13.5 in homozygous mutant mice compared with that of littermate controls. Quantitative Western blot analysis indicated a 75% reduction in Pdx1 protein levels in E13.5 Pdx1ΔC/ΔC pancreata compared with that of littermate controls (Figure 1, H and I). Pdx1ΔC/ΔC mice displayed a similar reduction in pancreatic Pdx1 mRNA at E13.5 (Figure 1G), which may indicate a role for the C terminus in Pdx1 autoregulation (39, 40). Alternatively, the introduction of the nonsense mutation may have influenced Pdx1 mRNA stability, although this is less likely given the location of the mutation. Terminal exon nonsense mutations, such as the one we have generated, are generally resistant to nonsense-mediated RNA decay. By contrast, Pdx1+/– mice displayed 38% and 42% reduction in Pdx1 protein and transcript levels, respectively, compared with littermate controls (Supplemental Figure 1), similar to the 32% reduction in Pdx1 protein levels observed in adult islets isolated from these mice (41). Thus, hypomorphic Pdx1ΔC/ΔC mice express lower Pdx1 protein levels at E13.5 compared with Pdx1+/– mice.
Figure 1. Derivation of Pdx1ΔC/ΔC mice.
(A) Targeting strategy to replace the codon encoding Ser210 in the mouse Pdx1 locus with a termination codon (asterisk), thereby preventing translation of the C terminus. Schematic shows both exons (black rectangles), the 5′ diphtheria toxin A (DTA) gene, and the loxP-flanked 3′ neomycin resistance–thymidine kinase (NeoR-tk) cassettes (white rectangles). Black arrows indicate the location of genotyping primers. White arrows denote the location of loxP sites. X indicates sites of homologous recombination. (B) Predicted domain structure of the truncated Pdx1(1–210) protein, termed Pdx1ΔC, is represented below the wild-type full-length form of Pdx1(1–284). TAD, transactivation domain; HD, homeodomain. (C–F) Pdx1+/+ and Pdx1ΔC/ΔC E13.5 pancreas stained with antisera raised against either N-terminal (N-term; red) or C-terminal (C-term; green) Pdx1 epitopes. Scale bar: 10 μm. (G) Pdx1 mRNA levels in total pancreas from Pdx1+/+, Pdx1+/ΔC, and Pdx1ΔC/ΔC pancreata measured at E13.5 (n = 7–8 per genotype; *P = 0.0001). (H) Representative Western blot of E13.5 total pancreas protein from Pdx1ΔC/ΔC, Pdx1+/ΔC, and Pdx1+/+ littermate controls using N-terminal–specific Pdx1 (top panel) and cyclophilin B (cyclo; bottom panel) antisera. Both full-length Pdx1 and truncated Pdx1ΔC are indicated. Quantitation from 3 separate Western blots (n = 9 animals per group) is shown in I. †P < 0.00001.
Global defect in pancreatic endocrine development in Pdx1ΔC/ΔC mice.
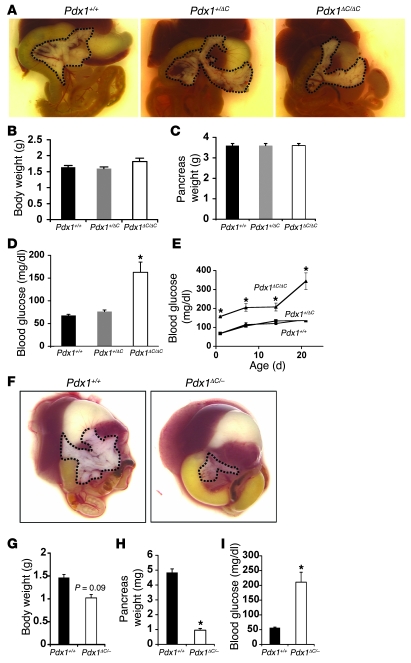
Pdx1ΔC/ΔC mice were born in the expected Mendelian ratios, with normal body and pancreatic weights and grossly normal pancreatic appearance (Figure 2, A–C), indicating that the C terminus is not required for pancreas organogenesis. To evaluate the influence of further reductions in gene dosage, we crossed Pdx1+/ΔC and Pdx1+/– mice. At birth, Pdx1ΔC/– progeny displayed a trend toward reduced body weight and a marked reduction in pancreatic size and weight (Figure 2, F–H), revealing an intermediate phenotype between Pdx1ΔC/ΔC and Pdx1–/– mice and indicating the threshold at which pancreatic organogenesis becomes sensitive to changes in gene dosage.
Figure 2. Dose-dependent regulation of pancreas organogenesis.
(A–E) Normal pancreas organogenesis in Pdx1ΔC/ΔC mice. (A) Foregut and accessory organs were dissected from newborn animals. The pancreas is highlighted by a black dotted line. (B) Body weights and (C) pancreatic weights were also measured at P1 (n = 6–15 per group). (D and E) Development of overt diabetes in Pdx1ΔC/ΔC mice. (D) Random blood glucose levels were measured at birth and (E) followed for 3 weeks in Pdx1ΔC/ΔC (triangles), Pdx1+/ΔC (squares), and Pdx1+/+ (circles) littermates (n = 6–21 per group; *P < 0.003 compared with both other groups). (F–I) Pancreatic hypoplasia in Pdx1ΔC/– mice. (F) Foregut and accessory organs were dissected from newborn animals. The pancreata from wild-type (left panel) and Pdx1ΔC/– (right panel) mice are highlighted by the black dotted line. (G) Body weights, (H) pancreatic weights, and (I) random blood glucose levels were measured in wild-type and Pdx1ΔC/– littermates at P1 (n = 5 per group; *P < 0.003).
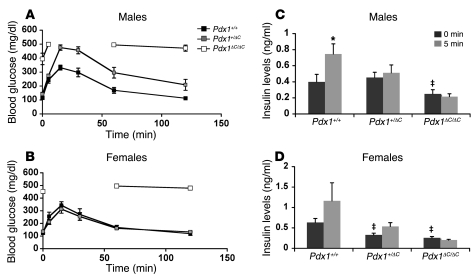
Similar to Pdx1ΔC/– animals, Pdx1ΔC/ΔC mice were mildly hyperglycemic at birth, with random blood glucose levels of 157 ± 3 mg/dl compared with 67 ± 3 mg/dl in heterozygous Pdx1+/ΔC mice and 68 ± 4 mg/dl in wild-type Pdx1+/+ mice (Figure 2, D and I). By 3 weeks of age, Pdx1ΔC/ΔC mice progressed to marked hyperglycemia, with random blood glucose levels of 344 ± 44 mg/dl compared with 141 ± 9 mg/dl in Pdx1+/ΔC and 136 ± 6 mg/dl in Pdx1+/+ mice (Figure 2E). At this age, total body and pancreatic weights in Pdx1ΔC/ΔC mice were unchanged compared with wild-type animals (data not shown). Intraperitoneal glucose tolerance tests performed in male and female mice at 3–4 weeks of age revealed overt diabetes in Pdx1ΔC/ΔC mice of both genders. Similar to Pdx1+/– mice (41, 42), heterozygous male Pdx1+/ΔC mice were glucose intolerant, whereas female Pdx1+/ΔC mice had normal glucose tolerance at this age (Figure 3, A and B). Consistent with their diabetic phenotype, male and female Pdx1ΔC/ΔC mice demonstrated a reduction in fasting plasma insulin and complete loss of the glucose-stimulated increase in plasma insulin. Male Pdx1+/ΔC mice showed a selective loss of the glucose-stimulated increase in plasma insulin, whereas female Pdx1+/ΔC mice had a reduction in fasting insulin levels but retained glucose responsiveness (Figure 3, C and D).
Figure 3. Impaired glucose tolerance and insulin secretion in Pdx1ΔC mice.
(A and B) Glucose tolerance in male (A) and female (B) mice. Intraperitoneal glucose tolerance tests using 2 g glucose/kg body weight performed on 3- to 4-week-old mice (n = 4–7 per group; ANOVA; male Pdx1+/+ versus Pdx1+/ΔC, P = 0.0037; male Pdx1+/+ versus Pdx1ΔC/ΔC, P < 0.0001; male Pdx1+/ΔC versus Pdx1ΔC/ΔC, P < 0.0001; female Pdx1+/+ versus Pdx1+/ΔC, P = NS; female Pdx1+/+ versus Pdx1ΔC/ΔC, P <0.0001; female Pdx1+/ΔC versus Pdx1ΔC/ΔC, P < 0.0001). Glucose levels in Pdx1ΔC/ΔC mice rose above 500 mg/dl, the limit of detection of the handheld glucometer. (C and D) Blunted acute insulin release in Pdx1ΔC/ΔC animals. Insulin levels were measured from whole blood collected at 0 and 5 minutes after intraperitoneal glucose injection (2 g glucose/kg body weight). Both male (C) and female (D) mice were 3–4 weeks of age (n = 4–7 animals per group). *P < 0.05 compared with level at 0 minutes after injection of same genotype; ‡P < 0.05 compared with level at 0 minutes after injection of wild-type mice.
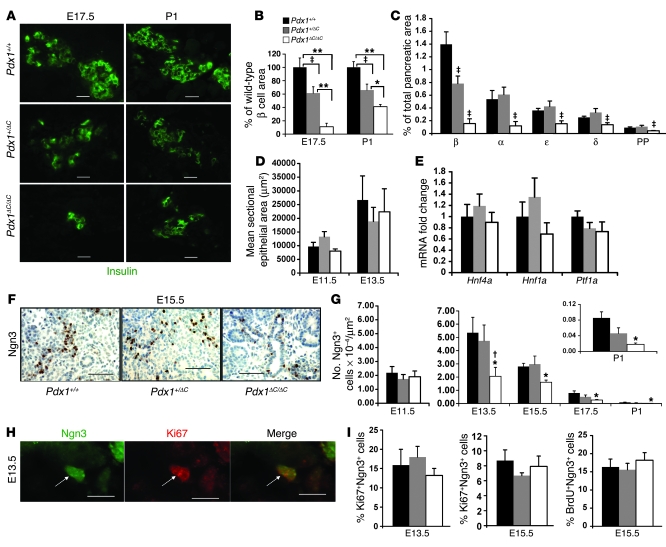
To determine whether a defect in β cell development could account for the hyperglycemia in Pdx1ΔC/ΔC mice, we first measured β cell area during embryonic development. At E17.5, a marked reduction in the pancreatic area occupied by β cells was observed in Pdx1ΔC/ΔC mice (Figure 4, A and B), which persisted in newborn animals. Heterozygous Pdx1+/ΔC mice also displayed a reduction in β cell area, revealing a gene dosage effect. We evaluated rates of β cell replication and apoptosis at E19.5 and found no differences among the genotypes (data not shown). To assess whether the decrease in β cell area at E17.5 reflected a β cell–specific or a general endocrine defect, we determined the area occupied by the other 4 endocrine cell types (α, δ, ε, pancreatic polypeptide). Interestingly, in Pdx1ΔC/ΔC mice, there was a global reduction in endocrine lineages, although β and α cells were more severely affected (Figure 4C). Only β cell area was reduced in Pdx1+/ΔC mice, indicating a greater sensitivity of the β cell lineage to Pdx1 expression level and function. Moreover, transcript levels for the endocrine lineage-specific hormones and transcription factors were all reduced at E13.5 during the secondary transition (Supplemental Figure 2). To exclude the possibility that an overall reduction in the pancreatic epithelium accounts for the reduction in the endocrine compartment in Pdx1ΔC/ΔC embryos, we measured epithelial area at E11.5 and E13.5 (Figure 4D). No differences were observed in Pdx1ΔC/ΔC embryos, which is consistent with the normal levels of Ptf1a and Hnf4a mRNA at E13.5 (Figure 4E) and with normal pancreatic weight at P1 (Figure 2C). Altogether, these data indicated a global defect in the development of the endocrine compartment, prompting us to examine the islet progenitor population in these mice.
Figure 4. Specific defect in endocrine progenitor specification in Pdx1ΔC/ΔC mice.
(A–C) Global reduction in endocrine lineages at E17.5 in Pdx1ΔC/ΔC mice. (A) Immunofluorescence for insulin (green). Scale bar: 20 μm. (B) Pancreatic area occupied by insulin-positive β cells. Values are expressed relative to wild-type mice (n = 5–9 per genotype; *P < 0.05, **P < 0.0005, ‡P < 0.05). (C) Percentage pancreatic area at E17.5 occupied by the 5 endocrine cell types (n = 6–8 animals per genotype, 3 sections per animal; ‡P < 0.05 compared with both other groups). PP, pancreatic polypeptide. (D and E) Normal epithelial area in Pdx1ΔC/ΔC mice. (D) Pancreatic epithelial area measured from 3 (E13.5) or 5 (E11.5) H&E-stained sections. (E) Relative mRNA levels of Hnf1a, Hnf4a, and Ptf1a in E13.5 pancreas (n = 7–9 per genotype). (F and G) Reduction in Ngn3+ pancreatic endocrine progenitor cells in Pdx1ΔC/ΔC mice. (F) Immunostaining for Ngn3 (brown) with hematoxylin counterstain (blue). Scale bar: 50 μm. (G) Number of Ngn3+ cells per unit of epithelial area (E11.5) or pancreatic area (E13.5–P1) quantified from 3 (E13.5–P1) or 5 (E11.5) tissue sections per animal (n = 4–8 per genotype; *P < 0.05 compared with wild-type mice; †P < 0.05 compared with Pdx1+/ΔC mice). (H and I) Ngn3+ endocrine progenitors replicate normally in Pdx1ΔC/ΔC mice. (H) Representative example of Ki67+Ngn3+ double-positive cell. Arrows point to the same cell in all panels. Scale bar: 10 μm. (I) Percentage of total Ngn3+ cells that are Ki67+ or BrdU+; more than 100 Ngn3+ cells per animal were counted (n = 4–5 per genotype).
Reduced numbers of Ngn3+ endocrine progenitors in Pdx1ΔC/ΔC mice.
All pancreatic endocrine cell types derive from progenitors that express Ngn3 (13, 16). Therefore, to determine whether the global reduction in endocrine cells was due to a defect in endocrine progenitors, we assessed Ngn3 immunostaining throughout embryonic development (E11.5–P1). In Pdx1ΔC/ΔC mice, a reduction in both number and staining intensity of Ngn3+ endocrine progenitors was present from E13.5 onward (Figure 4, F and G), indicating a role for Pdx1 in the formation, growth, or survival of the islet progenitor lineage during the secondary transition. The numbers of Ngn3+ cells present at E11.5, likely formed during the primary transition, were unchanged, suggesting that Pdx1 is required only during the secondary transition. In contrast, the numbers of Ngn3+ cells were unaffected in Pdx1+/– mice at all ages (Supplemental Figure 1). Pdx1+/ΔC mice also displayed normal numbers of Ngn3+ cells throughout development, indicating that Ngn3-independent mechanisms contributed to the reduction in β cell area observed in Pdx1+/ΔC mice (Figure 4, B and G).
To determine whether decreased proliferation of Ngn3-expressing progenitors accounts for the reduced numbers in Pdx1ΔC/ΔC animals, we performed double immunofluorescence for Ngn3 and Ki67 (Figure 4H). At E11.5, we did not observe any Ki67+Ngn3+ cells (data not shown), likely due to the very low number of Ngn3+ cells present at this age. At E13.5 and E15.5, proliferation rates of Ngn3+ cells were normal in Pdx1ΔC/ΔC and Pdx1+/ΔC mice (Figure 4I, left and center panels). Results were confirmed by BrdU incorporation at E15.5 (Figure 4I, right panel).
To determine whether decreased survival contributed to the reduction in Ngn3+ cell number, we performed double immunofluorescence for TUNEL and Ngn3. At E11.5 and E13.5, TUNEL+Ngn3+ cells were not detected in any group. At E15.5, no significant difference in apoptotic rates of Ngn3+ cells was observed in Pdx1ΔC/ΔC mice compared with that in littermate controls (Supplemental Table 1). Although the number of TUNEL+ cells at E19.5 was extremely low, we did observe an increase in the number of apoptotic Ngn3+ cells at this age in both Pdx1ΔC/ΔC and Pdx1+/ΔC animals (Supplemental Figure 3 and Supplemental Table 1), indicating that Pdx1 could be important in maintaining survival of the Ngn3 progenitor population during late gestation. This is in agreement with the more pronounced and progressive reduction in the number of Ngn3+ cells in Pdx1ΔC/ΔC mice relative to Pdx1+/+ mice, beginning at E17.5 (Figure 4, G and inset). Although Pdx1 was not coexpressed with Ngn3 at this age (Supplemental Figure 4), appearing to rule out a cell autonomous mechanism, it could regulate expression of a survival factor by neighboring cells. This is the first report to our knowledge indicating the Ngn3+ cells undergo apoptosis, albeit at a low rate.
Pdx1 directly regulates Ngn3.
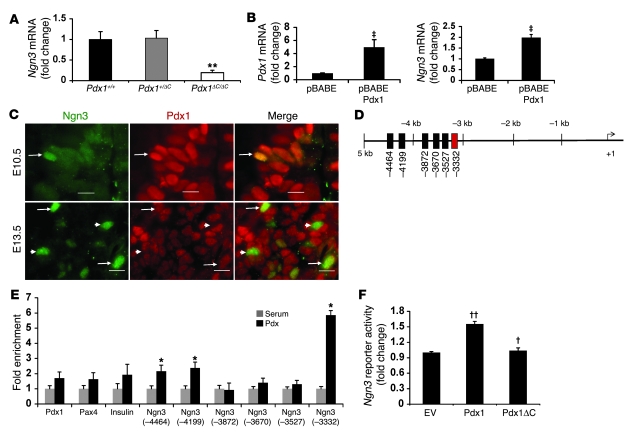
Since proliferation and survival rates of Ngn3+ cells were normal during early development and could not account for the general reduction in Ngn3+ cell number, we considered that Pdx1 might regulate the formation of islet progenitors. To determine whether Pdx1 could influence specification of endocrine progenitors by regulating Ngn3 at the transcriptional level, we measured Ngn3 mRNA levels at E13.5 and found Ngn3 expression reduced by 80% in Pdx1ΔC/ΔC mice (Figure 5A). In agreement with the observed reduction in Ngn3 staining intensity, Ngn3 transcript levels were reduced out of proportion to the reduction in cell number. Ngn3 mRNA levels were unaffected in Pdx1+/– mice (Supplemental Figure 1C), indicating a specific role for the C terminus or a critical threshold of Pdx1 protein that is required for Ngn3 expression. Conversely, a gain-of-function experiment demonstrated that Pdx1 is sufficient for activation of Ngn3. Overexpression of Pdx1 in mouse pancreatic ductal cells (PDCs) was sufficient to activate endogenous Ngn3 mRNA levels as compared with cells transduced with the empty vector (Figure 5B).
Figure 5. Pdx1 regulates Ngn3, at least in part, via its direct occupancy of a conserved upstream enhancer of Ngn3.
(A) Ngn3 mRNA levels in total pancreas of E13.5 animals (n = 5; **P = 0.005). (B) Expression of Ngn3 in PDCs transduced with pBABE or pBABE-Pdx1 retroviruses. Pdx1 and Ngn3 mRNA levels are expressed compared with empty vector (n = 6; ‡P < 0.007). (C) Colocalization of Pdx1 and Ngn3 during pancreas development. Double immunofluorescence for Ngn3 (green) and Pdx1 (red) in wild-type pancreata. Arrows indicate double-positive cells and arrowheads mark single-positive Ngn3 cells. Scale bar: 10 μm. (D) Diagram depicting conserved TT/AAT sites in the mouse Ngn3 promoter (black and red boxes). The region homologous to the previously described human Ngn3 Cluster 1 enhancer that contains binding sites for Hnf1, Foxa2, and Hnf6 is indicated in red. (E) ChIP with Pdx1 antisera performed on at least 65 pooled wild-type E13.5 pancreata per experiment (n = 3; *P < 0.05). (F) Promoter reporter assays were performed in HepG2 cells. Cells were transfected with expression vectors for Pdx1, Pdx1ΔC or empty vector, and the promoter reporter Ngn3(–3379 to –3227)-tkluc (n = 3; ††P < 0.05 versus empty vector, †P < 0.05 versus full-length Pdx1).
The reduction in Ngn3 protein and Ngn3 mRNA in Pdx1ΔC/ΔC mice as well as the upregulation of Ngn3 by Pdx1 overexpression in heterologous cells suggested that Ngn3 expression is regulated directly or indirectly by Pdx1. In considering the possibility that Pdx1 directly regulates Ngn3, we first determined whether and when Pdx1 and Ngn3 are coexpressed, because the evidence for colocalization has been conflicting (14, 15). At E10.5, few Ngn3+ cells were observed; however, all Ngn3+ cells at this stage also coexpressed Pdx1 (Figure 5C). By E13.5, a subpopulation of Ngn3+ cells no longer costained for Pdx1, suggesting that Pdx1 expression is not maintained in this population. Consistent with this notion, no Ngn3+Pdx1+ cells were observed at E15.5 and E19.5 (Supplemental Figure 4 and data not shown).
Having determined that Pdx1 expression marks the early Ngn3+ islet progenitor, we sought to assess whether Pdx1 could directly regulate Ngn3 transcription. We performed a genomic database search and identified 12 evolutionarily conserved putative Pdx1-binding sites (TT/AAT) clustered in 6 areas of 5 kb of the proximal mouse Ngn3 promoter (Figure 5D). To determine whether Pdx1 bound any of these sites in vivo, we performed Pdx1 ChIP using wild-type E13.5 pancreata. A 6-fold enrichment of Pdx1 occupancy was detected for Cluster 1 (–3,332; Figure 5E), a conserved enhancer region that was previously described in the human Ngn3 promoter to contain DNA-binding sites for Hnf1, Hnf6, and Foxa2 (43). No enrichment was detected at other previously described Pdx1-binding sites in the Pax4 and Pdx1 promoters (39, 40, 44, 45), suggesting that Pdx1 does not access the chromatin surrounding these promoters at E13.5. Moreover, few insulin-positive cells are present at this age, and the sensitivity of the assay may be insufficient to measure Pdx1 occupancy at the insulin promoter. To determine whether Pdx1 regulates the activity of this Ngn3 enhancer, we performed Ngn3 enhancer luciferase reporter assays in HepG2 cells. Pdx1 modestly transactivated the Ngn3 enhancer, while truncated Pdx1ΔC was unable to activate the enhancer (Figure 5F).
Pdx1 physically interacts and cooperates with Hnf6 to regulate Ngn3 enhancer activity.
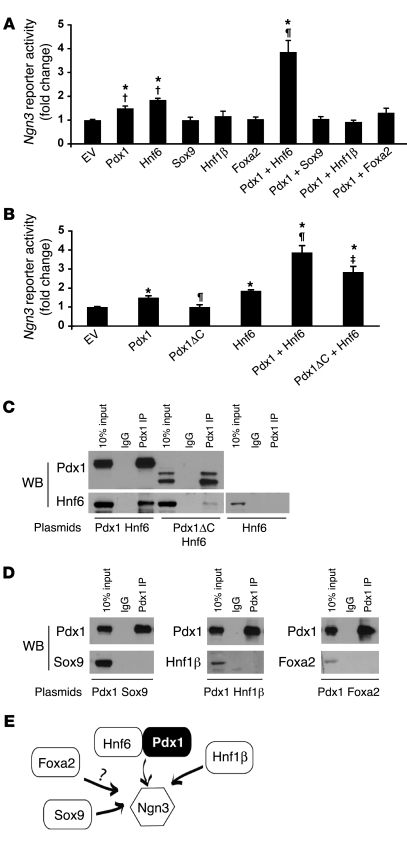
Previous reports demonstrate that Hnf6–/–, Hnf1b–/–, and Sox9+/– mice display decreased Ngn3 expression (46–48). Hnf6 and Sox9 individually and directly regulate the Ngn3 promoter; furthermore, the Ngn3 Cluster 1 enhancer contains potential Hnf6-, Hnf1β-, and Foxa2-binding sites (22, 43, 47, 48), altogether supporting roles for Hnf6, Hnf1β, Sox9, and Foxa2 in Ngn3 regulation. Thus, we sought to determine whether Pdx1 could function in concert with Hnf6, Hnf1β, Sox9, and/or Foxa2. Indeed, Pdx1 and Hnf6 together significantly increased Ngn3 enhancer transactivation, as compared with either factor alone (Figure 6A). In contrast, Sox9, Hnf1β, and Foxa2 failed to activate the Ngn3 enhancer individually or in concert with Pdx1. Moreover, Pdx1ΔC displayed a modest but significant reduction in the ability to function with Hnf6 at the Ngn3 enhancer (Figure 6B), suggesting a specific role for the Pdx1 C terminus in regulating Ngn3 expression.
Figure 6. Pdx1 cooperates with Hnf6 to regulate Ngn3.
HepG2 cells transfected with enhancer reporter Ngn3(–3379 to –3227)-tkluc and expression vectors for Pdx1, Hnf6, Sox9, Foxa2 (A) or Pdx1, Pdx1ΔC, and Hnf6 (B). *P < 0.05 versus empty vector; †P < 0.01 versus Pdx1 plus Hnf6; ¶P < 0.01 versus Pdx1; ‡P < 0.05 versus Pdx1 plus Hnf6. n = 4. (C and D) Pdx1 physically interacts with Hnf6. 293T cells were transfected with expression vectors for Pdx1 or Pdx1ΔC and Hnf6 (C) or Pdx1 and Sox9, Hnf1β, or Foxa2 (D). Cell lysates were immunoprecipitated with mouse monoclonal Pdx1 antisera or mouse IgG control. Western blots (WBs) using polyclonal antisera for Pdx1, Hnf6, Sox9, Hnf1β, and Foxa2 are shown. A representative example of 3 independent experiments is shown. (E) Model depicting the role of Pdx1 in the transcriptional regulation of Ngn3. The question mark denotes the absence of in vivo evidence to support the in vitro studies, suggesting Foxa2-mediated regulation of Ngn3.
To determine whether Pdx1 can interact with Hnf6, we cotransfected 293T cells with plasmids expressing Pdx1 and Hnf6. Immunoprecipitation with Pdx1 antisera, followed by Western blot analysis with Hnf6 antisera, revealed a physical association between Pdx1 and Hnf6 (Figure 6C). This interaction was attenuated by truncation of the C terminus, indicating a role for the C terminus in the Pdx1/Hnf6 association. The residual interaction between Pdx1ΔC and Hnf6 points to either the Pdx1 N terminus or the central homeobox as additional domains mediating this association. When Pdx1 was co-transfected with plasmids encoding Sox9, Hnf1β, or FoxA2, Pdx1 antisera did not immunoprecipitate any of these factors (Figure 6D), providing additional evidence for the specificity of the Pdx1/Hnf6 association. Thus, Pdx1 and Hnf6 could interact both physically and functionally to regulate Ngn3 promoter activity (Figure 6E), and this interaction was mediated at least in part by the Pdx1 C-terminal domain.
Pdx1 coordinates the transcriptional network of the developing pancreas.
We also measured the expression of other transcription factors and pathways previously described to regulate Ngn3 using quantitative real-time PCR. During early pancreas development, activation of the Notch signaling pathway in Pdx1-expressing progenitors stimulates hairy and enhancer of split 1 (Hes1) expression, which in turn inhibits expression of Ngn3, thereby promoting self-renewal of progenitor cells and inhibiting endocrine differentiation (18, 21, 49, 50). In parallel to the Notch pathway, Gdf11, a member of the TGF-β ligand family, also inhibits development of Ngn3+ islet progenitors (51, 52). Conversely, the transcription factors Hnf1β, Foxa2, Hnf6, and Sox9 have been identified as transcriptional activators of Ngn3 (22, 39, 43, 47, 53–58).
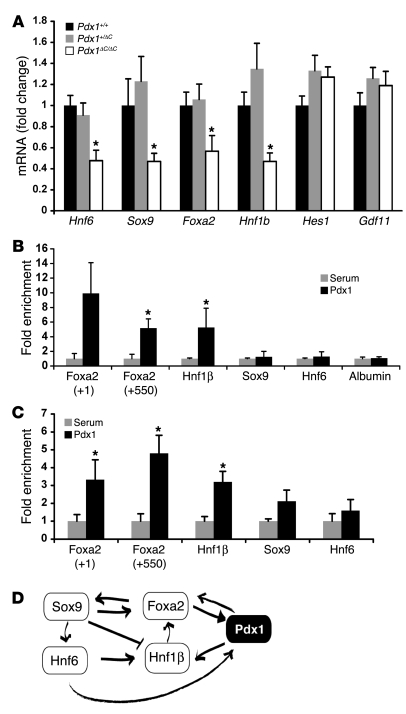
Transcript levels of Hes1 and Gdf11 were normal in Pdx1ΔC/ΔC mice at E13.5. In contrast, we observed an approximately 50% reduction in Hnf6, Sox9, Foxa2, and Hnf1b mRNA levels (Figure 7A), indicating that these genes are downstream of Pdx1. This is a surprising observation since both Foxa2 and Hnf6 are reported to be upstream regulators of Pdx1 (55, 57, 59). Thus, our results suggest that Pdx1 is a member of the recently described cross-regulatory network, involving Hnf6, Sox9, Hnf1β, and Foxa2 (22). To determine whether Pdx1 directly regulates Sox9, Hnf1b, Hnf6, or Foxa2, we examined the results of high-throughput DNA sequencing of MIN6 insulinoma cell chromatin (ChIPSeq) immunoprecipitated with anti-Pdx1 antiserum. Sites in the proximal promoters of the Hnf1b and Foxa2 genes were enriched, while no sites were enriched in the Sox9 and Hnf6 promoters (J. Yang and D.A. Stoffers, unpublished observations). These findings were confirmed in 3 independent MIN6 ChIP assays (Figure 7B). To determine whether Pdx1 also occupies these sequences during pancreas development, we performed Pdx1 ChIP assays using chromatin prepared from E13.5 pancreata. Pdx1 occupancy was enriched at the same conserved regulatory sequences of both the Hnf1b and Foxa2 genes (Figure 7C), suggesting that Pdx1 participated in the Hnf6, Sox9, Hnf1β, Foxa2 transcription factor cross-regulatory network by directly regulating Hnf1β and Foxa2 promoters in a progenitor pool at E13.5 (Figure 7D).
Figure 7. Pdx1 participates in a cross-regulatory network with Sox9, Hnf6, Foxa2, and Hnf1β.
(A) mRNA levels in total pancreas from Pdx1+/+, Pdx1+/ΔC, and Pdx1ΔC/ΔC pancreata at E13.5 (n = 7–9 per genotype; *P < 0.05). (B and C) ChIP with Pdx1 antisera of Min6 mouse insulinoma cells (B) or wild-type E13.5 pancreata (C). Two peaks of enrichment confirmed at +1 and +550 relative to the transcriptional start site of the Foxa2 gene and 1 peak at –31,087 relative to the transcriptional start site of the Hnf1b gene (n = 3; *P < 0.05). (D) Model depicting the role of Pdx1 in the cross-regulatory transcription factor network governing early pancreas development.
Taken together, these studies indicate that Pdx1 contributed to the specification of pancreatic endocrine progenitors by participating in the Hnf6, Sox9, Hnf1β, Foxa2 transcription factor cross-regulatory network and by regulating Ngn3 directly. Moreover, the Pdx1 C terminus may contribute to Ngn3 regulation independent of Pdx1 protein expression level through its role in the physical interaction of Pdx1 with Hnf6.
Discussion
In this study, we identified a direct role for Pdx1 in the specification of Ngn3+ endocrine progenitors. We discovered that Pdx1 contributes to the specification of endocrine progenitors by regulating Ngn3 directly and in concert with Hnf6 and by participating in a cross-regulatory transcription factor network during early pancreas development. Our data indicated that (a) Pdx1 plays a specific role in the emergence of pancreatic endocrine progenitors, in addition to being essential for overall pancreas organogenesis and (b) the role of Ngn3 in giving rise to hormone-expressing cells during the secondary transition is dependent upon Pdx1. Our results are supported by a recent study that describes the absence of Ngn3+ cells specifically during the secondary transition in Pdx1–/– mice (60).
We identified Pdx1 as a member of the cross-regulatory network involving Hnf6, Foxa2, Hnf1β, and Sox9. The combined defects in expression of Hnf6, Foxa2, Hnf1β, Sox9, and Pdx1 observed in Pdx1ΔC/ΔC mice likely influence Ngn3 expression in this model. We determined that Pdx1 directly occupies sequences in both Hnf1b and Foxa2 genes at E13.5. Thus, the position of Pdx1 both upstream of Foxa2, Hnf6, Sox9, and Hnf1β and downstream of Foxa2 and Hnf6 indicates that Pdx1 participates in the recently described cross-regulatory network involving these transcription factors (22). In contrast, at this stage of pancreas development, Pdx1 does not participate in the network involving Hnf1α and Hnf4α (61), since mRNA levels of Hnf1a and Hnf4a were not altered in Pdx1ΔC/ΔC mice. The occupancy of the Ngn3 promoter by Pdx1 adjacent to potential binding sites for Hnf1, Foxa2, and Hnf6 is similar to enhancer regions that regulate expression of Pdx1 (39, 45, 53, 54), suggesting the existence of conserved transcriptional regulatory modules that direct gene expression in the endocrine pancreas.
In addition to identifying a direct regulation of Foxa2 and Hnf1b by Pdx1, we identified Hnf6 as a protein partner for Pdx1, highlighting the complex role of Pdx1 in the transcriptional network of endocrine progenitors. Our data suggested that Pdx1 and Hnf6 cooperate to regulate Ngn3 expression. The importance of the Pdx1 C terminus in this interaction is demonstrated by the attenuated functional and physical interaction observed when the C terminus is truncated. However, the Pdx1 C-terminal deletion did not abolish the interaction between Pdx1 and Hnf6, indicating that other Pdx1 domains are also required. These findings suggested that the Pdx1 C terminus enhances the Pdx1/Hnf6 association by directly interacting with Hnf6 and/or by changing the conformation of other interacting regions. Alternatively, additional C-terminal–interacting protein partners may be required for Ngn3 regulation. Truncation of the C terminus is also predicted to disrupt the interaction of Pdx1 with Pcif1 and Meis2, nuclear factors whose roles in pancreas development are yet to be determined (36, 37). Several human C-terminal mutations are associated with reduced transactivation in promoter/reporter assays. One of these mutations, E224K, is located in the PCIF1 interaction motif and abrogates PCIF1 interaction in reporter assays (38), but the remaining C-terminal mutations lie outside of the PCIF1 interaction motif. Future experiments involving the derivation and characterization of mice harboring C-terminal mutations that selectively disrupt interaction with specific protein partners as well the identification and characterization of novel protein partners will advance our understanding of the mechanism whereby Pdx1 regulates endocrine progenitors.
Our finding that Pdx1 directly regulated Ngn3 is consistent with several reports in which induction of Pdx1 expression results in increased Ngn3 expression. Pdx1 overexpression in ES cells or human bone marrow–derived mesenchymal cells induces Ngn3 expression and promotes differentiation into insulin-producing cells (8, 62). Furthermore, recent protocols for in vitro differentiation of human ES cells into insulin-producing cells reveal increased Ngn3 expression following Pdx1 induction (8, 10). Our results provide a molecular explanation for this regulatory relationship, thereby providing insight into the requirements for manipulating multipotent cells to generate insulin-producing cells.
Recognition of the regulatory network of endocrine progenitors may be critical for guiding the differentiation of ES cells into robust β cells as well as for the transdifferentiation of mature cell types for eventual translational application. Similar strategies could be envisioned for fostering the transition of induced pluripotent stem cells into β cells. Our studies directly linking Pdx1 with Ngn3 expression and the transcription factor network of endocrine progenitors also suggest a mechanism whereby human Pdx1 mutations lead to diabetes.
Methods
Animals.
To generate Pdx1ΔC/ΔC mice, the targeting vector was constructed by subcloning 2 XbaI fragments (8.0 kb and 2.8 kb) containing 4 kb of the 5′ flanking region, exon 1, intron 1, exon 2, and 3′ sequences (25) of the Pdx1 locus from a mouse SV129 λFIXII bacteriophage clone. The Ser210 to termination codon mutation, along with a diagnostic BclI site, was introduced by overlap PCR. Neomycin resistance–thymidine kinase (NeoR-tk) and diphtheria toxin A selection cassettes were inserted (Figure 1A). Targeted ES cell clones were injected into blastocysts by the University of Pennsylvania Transgenic and Chimeric Mouse Facility. Chimeric males were bred to SV129 females to achieve germ-line transmission. The resulting offspring were bred to C57BL/6, and all studies were performed on F3 offspring. To detect BrdU incorporation, pregnant mothers were pulsed with BrdU for 1 hour (0.5 mg/kg body weight). Blood glucose levels were measured using a handheld glucometer (FreeStyle). Pdx1+/– mice on a Black Swiss genetic background were obtained from C. Wright (Vanderbilt University, Nashville, Tennessee, USA) and backcrossed for 9 generations onto a C57BL/6 background. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Intraperitoneal glucose tolerance tests were performed after a 6-hour fast using 2 g glucose/kg body weight. Insulin levels were measured from whole blood collected at 0 and 5 minutes after intraperitoneal glucose injection (2 g glucose/ kg body weight). Plasma insulin was measured by ELISA (Crystal Chem Inc.).
Cell culture.
All cell lines were cultured in a humidified chamber at 37°C and 5% CO2. Baby hamster kidney cells were grown in DMEM medium (Invitrogen) with 10% FBS (Gemini) and 1X penicillin-streptomycin (Invitrogen). HepG2 cells were cultured in MEM media (Invitrogen) with 10% FBS (Gemini), 1X penicillin-streptomycin (Invitrogen), and nonessential amino acids (Invitrogen). PDCs were grown as described previously (36). Pdx1 was overexpressed by retroviral transduction in primary mouse PDCs, as described previously (63).
RNA isolation and analysis.
Total RNA was isolated from more than 10 cultured islets or individual E13.5/P1 pancreata using TRIzol (Invitrogen) and the RNeasy kit (Qiagen). Integrity was confirmed on an Agilent Bioanalyzer. cDNA was generated using the SuperScript II Reverse Transcriptase kit (Invitrogen), and SYBR Green–based Real-Time Quantitative PCR (iQ5 iCycler and reagents; Bio-Rad) was used to measure mRNA levels. All values were normalized to HPRT mRNA levels and expressed as fold change compared with wild-type mice. Primer sequences are included in Supplemental Table 2.
Western blotting.
Individual pancreata from E13.5 animals were sonicated in lysis buffer (50 mM Tris-Cl, pH 7.8, 2% SDS, 10% glycerol, 10 mM Na4P2O7, 100 mM NaF, 6 M urea, 10 mM EDTA). Protein (1 μg) was resolved on 4%–12% NuPage Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes. After blocking in 5% milk, membranes were incubated with rabbit anti-Pdx1 (1:10,000; a gift from C. Wright), followed by HRP-conjugated anti-rabbit antibody (1:10,000; Santa Cruz Biotechnology Inc.) and ECL Plus (GE Healthcare). Protein quantitation was determined using Bio-Rad ChemiDoc XRS and Quantity One software. Linearity was verified using a standard curve generated with varying amounts of wild-type E13.5 pancreas protein.
Immunostaining.
Samples were fixed in 4% paraformaldehyde overnight at 4°C and were paraffin-embedded, and 6 μm-thick sections were generated. After dewaxing, rehydrating, and blocking, slides were incubated with primary antibodies overnight at 4°C at the following dilutions: guinea pig anti-insulin (1:1,000; Linco), rabbit anti-glucagon (1:1,000; Biodesign), goat anti-glucagon (1:500; Santa Cruz Biotechnologies Inc.), goat anti-somatostatin (1:1,000; Santa Cruz Biotechnology Inc.), rabbit anti-pancreatic polypeptide (1:15,000; Linco), rabbit anti-ghrelin (1:2,000; Phoenix Pharmaceuticals Inc.), sheep anti-BrdU (1:750; US Biological), rabbit anti-Ki67 (1:3,000; Vector Laboratories), rabbit anti-Pdx1 N terminus (1:10,000; a gift from C. Wright), goat anti-Pdx1 (1:10,000; a gift from C. Wright), rabbit anti-Pdx1 C terminus (1:1,000; a gift from J. Habener, Massachusetts General Hospital, Boston, Massachusetts, USA), goat anti-Pdx1 A17 (1:500; Santa Cruz Biotechnology Inc.), rabbit anti-Ngn3 (1:250; ref. 64), and mouse monoclonal anti-Ngn3 (1:500–1:3,000; Developmental Studies Hybridoma Bank). For light microscopy, HRP-conjugated secondary antisera (Vector Laboratories) were used and visualized with diaminobenzidine and a hematoxylin counterstain. For immunofluorescence, the signal was visualized with Cy2- and Cy3-conjugated secondary antisera (Jackson ImmunoResearch Laboratories Inc.). For TUNEL staining, the ApopTag detection kit (catalog no. S7100; Chemicon) was used with Cy3-conjugated mouse anti-digoxigenin (Jackson ImmunoResearch Laboratories Inc.). Images were captured using a Nikon Eclipse E600 microscope with a CoolSNAP camera (Photometrics). All area measurements and cell counting were performed with IVision Software (BioVision Inc.).
ChIP.
ChIP assay and its analysis was performed as described previously (36), with a few modifications. For E13.5 pancreas ChIP, 60–100 pancreata were isolated from CD1 E13.5 embryos and crosslinked immediately in 1% formaldehyde. Following quenching in 125 mM glycine, samples were resuspended in cell lysis buffer and were homogenized and spun. The pellet was resuspended in nuclear lysis buffer and sonicated with a Bioruptor to generate 200–300 bp DNA fragments. Relative enrichment was determined using quantitative real-time PCR. Values were compared with input serial dilutions and are expressed as fold enrichment compared with isotype matched nonimmune serum samples.
Promoter reporter assays.
The Ngn3 enhancer (–3,379 to –3,227) was amplified using PCR from mouse genomic DNA and cloned into the BamHI and SacI sites of a thymidine kinase minimal promoter luciferase reporter plasmid (a gift from E. Suh, University of Pennsylvania). Reporter assays were performed as described previously (37). Ngn3-minimal thymidine kinase promoter-luciferase (Ngn3-tkluc) or (TAAT1)5 -65 somatosatin promoter-chloramphenicol acetyltransferase (SMS-CAT) and expression plasmids pCMX-Pdx1, pCMX-Pdx1(1–210), pHD-Foxa2 (a gift from K. Kaestner), pCMV4-Hnf6 (gift from M. Gannon, Vanderbilt University), pCB6-Hnf1β (a gift from M. Sander, UCSD, San Diego, California, USA), and pCDNA3-Sox9 (a gift from M. Sander), and a β-galactosidase internal control plasmid were cotransfected into HepG2 or baby hamster kidney cells with Lipofectamine 2000 (Invitrogen), and the cells were harvested 48 hours later. Luciferase (Promega) or CAT activity was measured and normalized to β-galactosidase activity. Protein expression of Pdx1 and Pdx1(1–210) was measured by Western blotting to quantitate expression levels. Normalized reporter activity from cells expressing equivalent levels of Pdx1 and Pdx1(1–210) protein were compared.
Coimmunoprecipitations.
293T cells were seeded in 100-mm culture dishes and transfected with 500 ng of pCMX-Pdx1, pCMX-Pdx1(1–210), pCMV-Hnf6, Hnf1β, or Sox9 plasmids using Lipofectamine 2000 (Invitrogen). Cell were harvested 2 days later in lysis buffer (50 mM Tris-Cl, pH 7.5, 1% NP40, 100 mM NaCl, and protease inhibitor cocktail [Chemicon]). Lysate was precleared by rotating at 4°C for 2 hours with Protein G agarose beads (GE Healthcare). Following a brief centrifugation to remove beads, lysates were incubated at 4°C overnight with 2 μg of Pdx1 monoclonal antibody (R&D Systems) or mouse IgG control (Santa Cruz Biotechnology Inc.), followed by a 2-hour, 4°C incubation with 50 μl of Protein G agarose beads. Beads were briefly centrifuged and washed with fresh lysis buffer 5 times. Bound proteins were eluted with 2X SDS-PAGE buffer and analyzed by Western blotting using the following antibodies: rabbit anti-Pdx1 N terminus (1:10,000; a gift from C. Wright), rabbit anti-Hnf6 (1:500; Santa Cruz Biotechnology Inc.), rabbit anti-Sox9 (1:2,000; Chemicon), goat anti-Hnf1β (1:500; Santa Cruz Biotechnology Inc.), and goat anti-Foxa2 (1:500; Santa Cruz Biotechnology Inc.).
Statistics.
Values are expressed as mean ± SEM and were compared using 2-tailed Student’s t tests. P values of less than 0.05 are considered statistically significant.
Supplementary Material
Acknowledgments
We thank U. Jhala, C.L. May, K. Tremblay, and members of the Stoffers laboratory for critically reading this manuscript. We are grateful to C. Wright for providing Pdx1+/– mice, Pdx1 antisera, and helpful discussions. We thank G. Swain (Morphology Core, University of Pennsylvania School of Medicine) for his morphology expertise, D. Groff for breeding and genotyping, L. Sussel and J. LeLay for ChIP advice, and M. Wescott and A.J. von Burstin in the Rustgi laboratory for providing PDCs. Ngn3 antibody was obtained from the Developmental Studies Hybridoma Bank (National Institute of Child Health and Human Development and The University of Iowa). This work was supported by NIH grants R01 DK068157 (to D.A. Stoffers), P01 DK49210 (to D.A. Stoffers and K.H. Kaestner), and R01 DK060694 (to A.K. Rustgi and D.A. Stoffers) and NIH predoctoral National Research Service Award 5F31HL071273 (to J.M. Oliver-Krasinski) and was facilitated by the Morphology Core of the Center for Molecular Studies in Digestive and Liver Disease, University of Pennsylvania (NIH P30 DK50306).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Ngn3, neurogenin 3; PDC, pancreatic ductal cell; Pdx1, pancreatic duodenal homeobox 1.
Citation for this article: J. Clin. Invest. 119:1888–1898 (2009). doi:10.1172/JCI37028
Michael F. Crutchlow’s present address is: Merck Research Laboratories, Merck & Co. Inc., North Wales, Pennsylvania, USA.
References
- 1.Ferber S., et al. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat. Med. 2000;6:568–572. doi: 10.1038/75050. [DOI] [PubMed] [Google Scholar]
- 2.Horb M.E., Shen C.N., Tosh D., Slack J.M. Experimental conversion of liver to pancreas. Curr. Biol. 2003;13:105–115. doi: 10.1016/S0960-9822(02)01434-3. [DOI] [PubMed] [Google Scholar]
- 3.Kojima H., et al. NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat. Med. 2003;9:596–603. doi: 10.1038/nm867. [DOI] [PubMed] [Google Scholar]
- 4.Sapir T., et al. Cell-replacement therapy for diabetes: Generating functional insulin-producing tissue from adult human liver cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7964–7969. doi: 10.1073/pnas.0405277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips B.W., et al. Directed differentiation of human embryonic stem cells into the pancreatic endocrine lineage. Stem Cells Dev. 2007;16:561–578. doi: 10.1089/scd.2007.0029. [DOI] [PubMed] [Google Scholar]
- 7.Shim J.H., et al. Directed differentiation of human embryonic stem cells towards a pancreatic cell fate. Diabetologia. 2007;50:1228–1238. doi: 10.1007/s00125-007-0634-z. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J., et al. Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells. 2007;25:1940–1953. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- 9.D’Amour K.A., et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 10.D’Amour K.A., et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 11.Kroon E., et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 12.Oliver-Krasinski J.M., Stoffers D.A. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwitzgebel V.M., et al. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 15.Jensen J., et al. Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation. Diabetes. 2000;49:163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- 16.Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 17.Johansson K.A., et al. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev. Cell. 2007;12:457–465. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Apelqvist A., et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 19.Prado C.L., Pugh-Bernard A.E., Elghazi L., Sosa-Pineda B., Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller R.S., et al. Genetic determinants of pancreatic epsilon-cell development. Dev. Biol. 2005;286:217–224. doi: 10.1016/j.ydbio.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 21.Jensen J., et al. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 22.Lynn F.C., et al. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 25.Offield M.F., et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 26.Schwitzgebel V.M., et al. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J. Clin. Endocrinol. Metab. 2003;88:4398–4406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- 27.Fujitani Y., et al. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland A.M., Hale M.A., Kagami H., Hammer R.E., MacDonald R.J. Experimental control of pancreatic development and maintenance. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera P.L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 30. Ham, J.N., and Stoffers, D.A. 2009. Role of Pdx1 in beta cell growth. InIslet cell growth factors . R.N. Kulkarni, editor. Landes Bioscience. Austin, Texas, USA. In press. [Google Scholar]
- 31.Weng J., et al. Functional consequences of mutations in the MODY4 gene (IPF1) and coexistence with MODY3 mutations. Diabetologia. 2001;44:249–258. doi: 10.1007/s001250051608. [DOI] [PubMed] [Google Scholar]
- 32.Hani E.H., et al. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J. Clin. Invest. 1999;104:R41–R48. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cockburn B.N., et al. Insulin promoter factor-1 mutations and diabetes in Trinidad: identification of a novel diabetes-associated mutation (E224K) in an Indo-Trinidadian family. J. Clin. Endocrinol. Metab. 2004;89:971–978. doi: 10.1210/jc.2003-031282. [DOI] [PubMed] [Google Scholar]
- 34.Lu M., Miller C., Habener J.F. Functional regions of the homeodomain protein IDX-1 required for transactivation of the rat somatostatin gene. Endocrinology. 1996;137:2959–2967. doi: 10.1210/en.137.7.2959. [DOI] [PubMed] [Google Scholar]
- 35.Peers B., Leonard J., Sharma S., Teitelman G., Montminy M.R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol. Endocrinol. 1994;8:1798–1806. doi: 10.1210/me.8.12.1798. [DOI] [PubMed] [Google Scholar]
- 36.Deramaudt T.B., et al. The PDX1 homeodomain transcription factor negatively regulates the pancreatic ductal cell-specific keratin 19 promoter. J. Biol. Chem. 2006;281:38385–38395. doi: 10.1074/jbc.M605891200. [DOI] [PubMed] [Google Scholar]
- 37.Liu A., Desai B.M., Stoffers D.A. Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol. Cell. Biol. 2004;24:4372–4383. doi: 10.1128/MCB.24.10.4372-4383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu A., Oliver-Krasinski J., Stoffers D.A. Two conserved domains in PCIF1 mediate interaction with pancreatic transcription factor PDX-1. FEBS Lett. 2006;580:6701–6706. doi: 10.1016/j.febslet.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 39.Gerrish K., Cissell M.A., Stein R. The role of hepatic nuclear factor 1 alpha and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 2001;276:47775–47784. doi: 10.1074/jbc.M109244200. [DOI] [PubMed] [Google Scholar]
- 40.Chakrabarti S.K., James J.C., Mirmira R.G. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J. Biol. Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 41.Brissova M., et al. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J. Biol. Chem. 2002;277:11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 42.Johnson J.D., et al. Increased islet apoptosis in Pdx1+/– mice. . J. Clin. Invest. 2003;111:1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.C., et al. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- 44.Smith S.B., Watada H., Scheel D.W., Mrejen C., German M.S. Autoregulation and maturity onset diabetes of the young transcription factors control the human PAX4 promoter. J. Biol. Chem. 2000;275:36910–36919. doi: 10.1074/jbc.M005202200. [DOI] [PubMed] [Google Scholar]
- 45.Gerrish K., Van Velkinburgh J.C., Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol. Endocrinol. 2004;18:533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 46.Haumaitre C., et al. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1490–1495. doi: 10.1073/pnas.0405776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacquemin P., et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell. Biol. 2000;20:4445–4454. doi: 10.1128/MCB.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seymour P.A., et al. A dosage-dependent requirement for Sox9 in pancreatic endocrine cell formation. Dev. Biol. 2008;323:19–30. doi: 10.1016/j.ydbio.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hald J., et al. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev. Biol. 2003;260:426–437. doi: 10.1016/S0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 50.Murtaugh L.C., Stanger B.Z., Kwan K.M., Melton D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dichmann D.S., Yassin H., Serup P. Analysis of pancreatic endocrine development in GDF11-deficient mice. Dev. Dyn. 2006;235:3016–3025. doi: 10.1002/dvdy.20953. [DOI] [PubMed] [Google Scholar]
- 52.Harmon E.B., et al. GDF11 modulates NGN3+ islet progenitor cell number and promotes beta-cell differentiation in pancreas development. Development. 2004;131:6163–6174. doi: 10.1242/dev.01535. [DOI] [PubMed] [Google Scholar]
- 53.Gerrish K., et al. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 2000;275:3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 54.Samaras S.E., et al. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in Pancreatic beta cells: role for hepatocyte nuclear factor 3 beta and Pax6. Mol. Cell. Biol. 2002;22:4702–4713. doi: 10.1128/MCB.22.13.4702-4713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu K.L., et al. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poll A.V., et al. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes. 2006;55:61–69. doi: 10.2337/diabetes.55.01.06.db05-0681. [DOI] [PubMed] [Google Scholar]
- 57.Wilding L., Gannon M. The role of pdx1 and HNF6 in proliferation and differentiation of endocrine precursors. Diabetes Metab. Res. Rev. 2004;20:114–123. doi: 10.1002/dmrr.429. [DOI] [PubMed] [Google Scholar]
- 58.Jacquemin P., Lemaigre F.P., Rousseau G.G. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev. Biol. 2003;258:105–116. doi: 10.1016/S0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 59.Lee C.S., et al. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- 60.Burlison J.S., Long Q., Fujitani Y., Wright C.V., Magnuson M.A. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev. Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boj S.F., Parrizas M., Maestro M.A., Ferrer J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14481–14486. doi: 10.1073/pnas.241349398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyazaki S., Yamato E., Miyazaki J. Regulated expression of pdx-1 promotes in vitro differentiation of insulin-producing cells from embryonic stem cells. Diabetes. 2004;53:1030–1037. doi: 10.2337/diabetes.53.4.1030. [DOI] [PubMed] [Google Scholar]
- 63.Deramaudt T.B., et al. N-cadherin and keratinocyte growth factor receptor mediate the functional interplay between Ki-RASG12V and p53V143A in promoting pancreatic cell migration, invasion, and tissue architecture disruption. Mol. Cell. Biol. 2006;26:4185–4200. doi: 10.1128/MCB.01055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee C.S., De León D.D., Kaestner K.H., Stoffers D.A. Regeneration of pancreatic islets after partial pancreatectomy in mice does not involve the reactivation of neurogenin-3. Diabetes. 2006;55:269–272. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.