Abstract
Notch signaling is vital for proper cardiovascular development and function in both humans and animal models. Indeed, mutations in either JAGGED or NOTCH cause congenital heart disease in humans and NOTCH mutations are associated with adult valvular disease. Notch typically functions to mediate developmental interactions between adjacent tissues. Here we show that either absence of the Notch ligand Jagged1 or inhibition of Notch signaling in second heart field tissues results in murine aortic arch artery and cardiac anomalies. In mid-gestation, these mutants displayed decreased Fgf8 and Bmp4 expression. Notch inhibition within the second heart field affected the development of neighboring tissues. For example, faulty migration of cardiac neural crest cells and defective endothelial-mesenchymal transition within the outflow tract endocardial cushions were observed. Furthermore, exogenous Fgf8 was sufficient to rescue the defect in endothelial-mesenchymal transition in explant assays of endocardial cushions following Notch inhibition within second heart field derivatives. These data support a model that relates second heart field, neural crest, and endocardial cushion development and suggests that perturbed Notch-Jagged signaling within second heart field progenitors accounts for some forms of congenital and adult cardiac disease.
Introduction
Development of the outflow tract (OFT) of the heart is complex, involving input from several embryologically distinct cell populations including cardiac progenitors from the second heart field, cardiac neural crest cells, and endothelial cells. The second (or anterior) heart field is a name given to a group of mesoderm-derived cardiac precursor cells that reside in the anterior pharynx and migrate into the heart after initial heart tube formation. These cells give rise to myocardium of the OFT, the right ventricle, interventricular septum, portions of the atria, and vascular smooth muscle at the base of the outflow vessels (1–6). Disruption of this population of cells results in cardiac defects primarily affecting the right ventricle and OFT. The LIM homeodomain transcription factor Islet1 is a marker of this cell population and is expressed in the pharyngeal mesenchyme and the distal OFT in mid-gestation embryos (7). The coordinated proliferation and differentiation of these cells into the appropriate adult structures is likely to be regulated by intrinsic factors and signals from surrounding tissues in the pharynx, such as endoderm and neural crest cells. However, the interaction between these different tissue types during OFT development remains poorly understood.
In addition to playing a critical role in development, Islet1-expressing precursor cells can be isolated from fetal hearts and expanded in vitro and are capable of differentiating into cardiomyocytes, smooth muscle cells, and endothelial cells (8, 9). Notch is an excellent candidate to regulate the development of cells in the second heart field. This signaling pathway is known to regulate multiple different stem and progenitor cell populations (10). In the context of cardiac development, Notch has been shown to inhibit the differentiation of embryonic stem cells into cardiomyocytes in vitro (11–13). In addition, studies in Drosophila and Xenopus have demonstrated that Notch can inhibit cardiomyocyte differentiation in vivo (14, 15). We have previously observed the expression of multiple Notch signaling components in the cardiac OFT myocardium, including the receptor Notch2, the ligand Jagged1 (Jag1), and downstream target genes Hrt2 and Hrt3 (16). These observations suggest that Notch may play a critical role in the development of second heart field–derived structures. The importance of these studies are highlighted by Alagille syndrome, a human disorder caused by JAG1 or NOTCH2 mutations that is characterized by right-sided OFT defects as well as disorders in the skeletal, ocular, renal, and hepatic organ systems (17). JAG1 mutations have also been implicated in cases of tetralogy of Fallot and pulmonary artery stenosis (18, 19). Moreover, disruption of appropriate Notch signaling is suggested to be important in the development of aortic valve disease, including bicuspid aortic valve disease (20). This disease has an incidence of up to 2% of the total population (21). A more detailed understanding of the molecular pathways underlying cardiogenesis and congenital heart disease is required to develop safe and efficient diagnostics and therapeutics for such disease processes. These insights will also affect the ability to manipulate induced pluripotent stem cells for therapeutic use.
In this study we demonstrate, through a variety of in vitro, ex vivo, and genetic studies, an unanticipated mechanism for Notch signaling in the second heart field during OFT development with direct implications for human Notch-associated cardiovascular disorders. Deletion of the Notch ligand Jag1, or inhibition of Notch signaling in the second heart field results in downregulation of 2 critical factors involved in OFT development: Fgf8 and bone morphogenetic protein 4 (Bmp4). We show that this pathway within the second heart field promotes the development of neighboring tissues, including the migration of cardiac neural crest cells and endothelial-mesenchymal transition (EMT) in the endocardial cushions. Furthermore, we provide evidence that exogenous Fgf8, normally expressed by second heart field-derived myocardium, is sufficient to rescue the defects in EMT of endocardial cushions observed after Notch inhibition. Together, these results outline a pathway by which Jag1-Notch signaling in the second heart field promotes OFT development via regulation of Fgf8 expression and cross-talk with adjacent tissues.
Results
Inhibition of Notch in the second heart field results in cardiovascular defects.
To elucidate the role of Notch in the second heart field, we took advantage of the fact that all 4 mammalian Notch receptors (Notch1–4) interact with mastermind-like proteins after activation and translocation to the nucleus. A dominant-negative truncated form of mastermind-like (DNMAML) is able to interact with intracellular Notch but cannot recruit transcriptional coactivators. This construct has been shown to be an effective and specific inhibitor of Notch signaling (16, 22). We used mice in which a fusion of DNMAML and GFP has been inserted into the Rosa26 locus downstream of a floxed PGK Neo cassette such that Notch can be inhibited in a tissue-specific manner after activation by Cre recombinase (22). DNMAML mice were crossed with 2 different Cre recombinase lines that each express in the second heart field. The first of these lines contains Cre knocked into the Islet1 locus (Islet1Cre) (23). The second is a transgenic line in which Cre is under control of a promoter fragment from the Mef2c gene, which has been shown to drive expression specifically in the anterior/second heart field (Mef2c-AHF-Cre) (1). Activation of DNMAML with either of these Cre lines resulted in fully penetrant neonatal lethality associated with severe cardiac defects (Table 1 and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI38922DS1). At late gestation, mutant embryos were found at approximately the expected Mendelian ratios. However, no mutant offspring survived after 1 day of age from either cross (Supplemental Table 1).
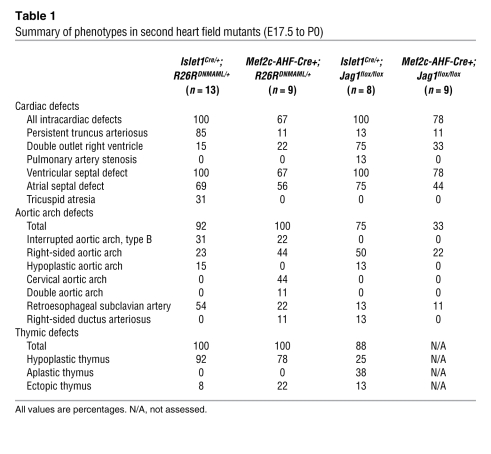
Table 1 .
Summary of phenotypes in second heart field mutants (E17.5 to P0)
Analysis of the hearts of late-gestation and neonatal Islet1Cre/+;DNMAML mutants demonstrated a number of severe cardiac abnormalities (Table 1). All of the mutants had OFT defects, including persistent truncus arteriosus or double outlet right ventricle (Figure 1, B and C). In addition, nearly all mutants had aortic arch defects including right-sided aortic arch, hypoplastic aortic arch, or retroesophageal subclavian arteries (Table 1 and Figure 1, B and I). We also observed several vascular rings, a potentially lethal congenital malformation in which vascular structures surround and constrict the trachea and esophagus (Figure 1C). Sectioning through the Islet1Cre/+;DNMAML hearts also revealed multiple intracardiac defects. Three of 10 hearts demonstrated severe right ventricular hypoplasia and tricuspid valve atresia (Figure 1M). All 3 of these mutants had atrial septal defects allowing passage of blood from the right to left atrium (Figure 1N). All Islet1Cre/+;DNMAML mutants displayed ventricular septal defects (Figure 1O). In addition to cardiovascular defects, the mutants also showed disruption of other pharyngeal structures, including the thymus (Table 1). These results demonstrate a critical role for Notch in the development of the cardiac OFT and right ventricle, the cardiac inflow tract, and pharyngeal structures including the aortic arch and the thymus. Islet1Cre/+ mice had no discernible cardiac defects (refs. 23, 24; data not shown), indicating neither Islet1 haploinsufficiency nor Cre expression could account for the phenotype in the Islet1Cre/+;DNMAML mutants.
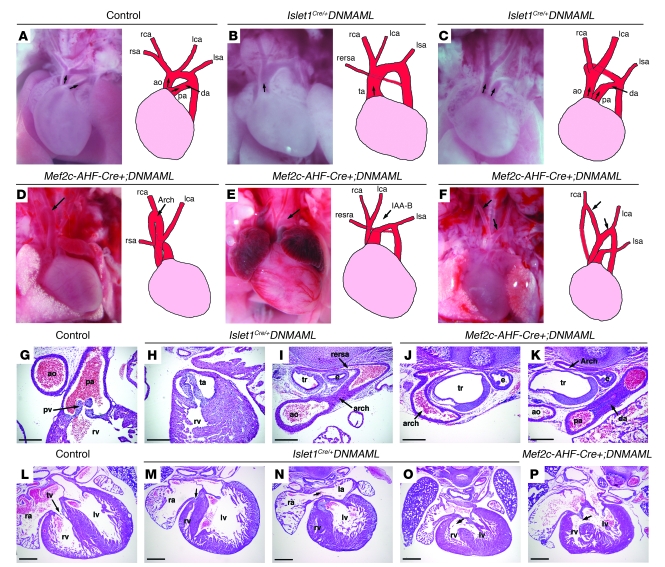
Figure 1. Cardiovascular defects in Notch second heart field mutants.
E17.5–P0 control (A), Islet1Cre/+;DNMAML (B and C), and Mef2c-AHF-Cre;DNMAML (D–F) mutants. (A) Arrows indicate pulmonary artery and aorta. (B) A single OFT vessel (arrow) and retroesophageal right subclavian artery. (C) Double outlet right ventricle (arrows) with a right aortic arch and left ductus arteriosus producing a vascular ring. (D) Cervical right aortic arch (arrow). (E) Interruption of the aortic arch, type B (IAA-B, arrow), and retroesophageal right subclavian artery. (F) Double aortic arch (arrows). (G–P) H&E sections of E17.5–E18.5 hearts. (G) Control with separate aorta and pulmonary artery. (H) Islet1Cre/+;DNMAML mutant with single OFT arising from right ventricle. (I) Islet1Cre/+;DNMAML mutant with retroesophageal right subclavian artery dorsal to the trachea. (J and K) Sections from a Mef2c-AHF-Cre+;DNMAML mutant showing a vascular ring. A right-sided circumflex arch (J) runs posterior to the trachea and esophagus en route to a left-sided descending aorta and left ductus arteriosus (K). (L) Control with normal tricuspid valve (arrow). (M and N) Islet1Cre/+;DNMAML with tricuspid atresia (arrow, M), right ventricular hypoplasia, and atrial septal defect (arrow in N). (O and P) Islet1Cre/+;DNMAML (O) and Mef2c-AHF-Cre+;DNMAML (P) hearts showed ventricular septal defects (arrows). ao, aorta; da, ductus arteriosus; e, esophagus; IAA-B, interruption of the aortic arch, type B; la, left atrium; lca, left carotid artery; lsa, left subclavian artery; lv, left ventricle; pa, pulmonary artery; pv, pulmonic valve; ra, right atrium; rca, right carotid artery; rersa, retroesophageal right subclavian artery; rsa, right subclavian artery; rv, right ventricle; ta, truncus arteriosus; tr, trachea; tv, tricuspid valve. Original magnification: ×30 (A–F). Scale bars: 250 μm (G–K), 500 μm (L–P).
The hearts of Mef2c-AHF-Cre+;DNMAML mutants demonstrated similar defects to those seen in the Islet1Cre/+;DNMAML mice (Table 1 and Figure 1, D–F), though the OFT defects were slightly less severe. All of the mutants analyzed had aortic arch defects. The most common defects seen in the Mef2c-AHF-Cre+;DNMAML mutants were right-sided aortic arch and cervical aortic arches, extending high in the neck (Figure 1D). Complete interruption of the aortic arch (Figure 1E), double aortic arch (Figure 1F), aberrant subclavian arteries (Figure 1, D, E, J, and K) and vascular rings (Figure 1, J and K) were also observed. Sectioning through the hearts of Mef2c-AHF-Cre+;DNMAML mutants revealed ventricular septal defects in 100% of mutants (Figure 1P and Table 1).
In order to determine which tissues were expressing DNMAML in Islet1Cre/+;DNMAML and Mef2c-AHF-Cre;DNMAML mutants, we performed immunostaining for GFP to detect the DNMAML-GFP fusion protein (Supplemental Figure 1). Mid-gestation Islet1Cre/+;DNMAML embryos showed DNMAML-GFP expression in the pharyngeal mesenchyme and the pharyngeal endoderm, while Mef2c-AHF-Cre;DNMAML embryos showed DNMAML-GFP expression in the mesenchyme only (Supplemental Figure 1, A and B). At late gestation, Islet1Cre/+;DNMAML embryos expressed DNMAML-GFP in derivatives of the second heart field, including the base of the truncus arteriosus, myocardium of the OFT and right ventricle, a small subset of cells in the left ventricle, the inflow tract, and both atria, as well as endoderm derivatives (Supplemental Figure 1, C and D). A similar pattern was seen when Islet1Cre and Mef2c-AHF-Cre mice were crossed with R26RLacZ to generate a fate map (Supplemental Figure 1, E and F). In summary, Islet1Cre resulted in Cre-mediated recombination broadly throughout the heart including both the OFT and inflow tract, as well as in the pharyngeal endoderm derivatives. Mef2c-AHF-Cre mice had more restricted Cre-mediated recombination in the right ventricle and proximal OFT. Therefore, it is possible that the atrial septal defects in Islet1Cre/+;DNMAML mice are, in part, due to defects of inflow tract formation. Importantly, neither Cre line results in significant recombination in the aortic arch or its major branches. These results are consistent with other reports using these mouse lines (1, 25). The broader expression pattern of the Islet1Cre/+ line, as well as the fact that Mef2c is downstream of Islet1 (26) and therefore likely activated later, may explain the more severe cardiac phenotypes observed in the Islet1Cre/+;DNMAML mutants when compared with the Mef2c-AHF-Cre;DNMAML embryos (Table 1).
Cardiac neural crest cell migration is altered by Notch inhibition in the second heart field.
The spectrum of aortic arch defects in both the Islet1Cre/+;DNMAML and Mef2c-AHF-Cre+;DNMAML mice suggests a defect in the formation or remodeling of the aortic arch arteries earlier in development. To investigate this, we analyzed the aortic arch arteries of Islet1Cre/+;DNMAML mutants at E10.5. To visualize the endothelium of the aortic arch arteries, we performed immunostaining for the endothelial marker PECAM on sections through the pharyngeal arches of control and mutant embryos (Figure 2, A–D). In control embryos the lumens of the third, fourth, and sixth aortic arch arteries were large and clearly patent (Figure 2, A and C). However, Islet1Cre/+;DNMAML mutants showed generalized hypoplasia of the fourth pharyngeal arch and severe narrowing or complete absence of the fourth and sometimes the sixth aortic arch arteries (Figure 2, B and D). Similar findings were also present in the Mef2c-AHF-Cre;DNMAML mutants (data not shown). The fourth embryonic aortic arch arteries are the precursors of the main segment of aortic arch and the proximal region of the right subclavian artery. Therefore, defective formation of the fourth arch artery is consistent with the types of aortic arch defects that were observed in the late-gestation embryos, such as interrupted or right-sided aortic arches and aberrant subclavian arteries.
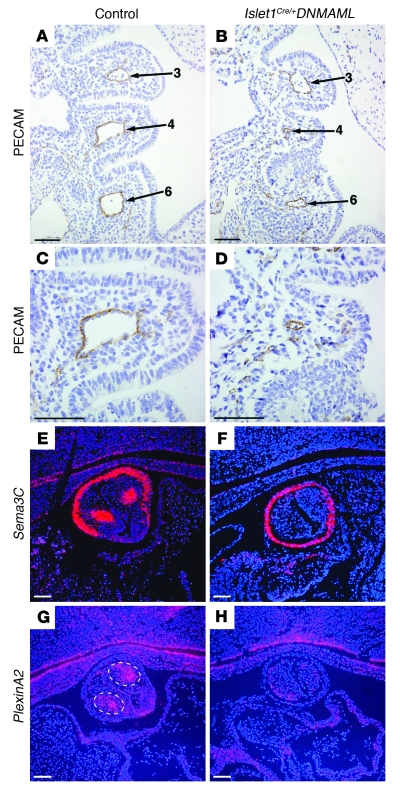
Figure 2. Notch inhibition in the second heart field results in abnormal patterning of the pharyngeal arches and impaired cardiac neural crest cell migration.
(A–D) PECAM immunostaining of coronal sections through E10.5 embryos to visualize the endothelium of the aortic arch arteries (numbered). Note the hypoplasia of the fourth pharyngeal arch and narrowing of the fourth and sixth aortic arch arteries in the mutant (B). (C and D) Higher-magnification views of the fourth aortic arch artery in control (C) and Islet1Cre/+;DNMAML (D) embryos show severe hypoplasia in the mutant. (E–H) In situ hybridization for the neural crest markers Sema3C (E and F) and PlexinA2 (G and H) of coronal sections through the OFT of E11.5 control (E and G) and mutant (F and H) embryos. Dotted lines in G highlight neural crest cells within the outflow endocardial cushions expressing PlexinA2. Control genotype was Islet1+/+;DNMAML. Scale bars: 50 μm (A–D), 100 μm (E–H).
Cardiac neural crest cells populate the aortic arches and cardiac OFT and play critical roles in patterning of the aortic arch arteries and in septation of the truncus arteriosus. We examined the expression of markers of cardiac neural crest by in situ hybridization in the developing OFTs of wild-type and mutant embryos. PlexinA2 and Sema3C were both expressed by post-migratory cardiac neural crest. In wild-type embryos, expression is evident in 2 clusters of cells within the outflow endocardial cushions that represent the leading edge of the 2 prongs of cardiac neural crest invading the OFT (27). However, in Islet1Cre/+;DNMAML mutants, expression in this region was lost, though myocardial expression of Sema3C was preserved (Figure 2, F and H). We examined indexes of apoptosis and proliferation at E9.5–E10. No significant difference in the percentage of phospho–histone H3–positive neural crest cells was apparent in the ventral pharynx between E9.5 Islet1Cre/+;DNMAML (3.91% ± 1.19%, n = 6) and Islet1+/+;DNMAML (2.90% ± 2.46%, n = 6) embryos (P = 0.39). In addition, we did not detect a significant difference in apoptosis (as assayed by TUNEL staining) of neural crest cells populating the ventral pharynx (Islet1Cre/+;DNMAML: 1.05% ± 0.90%, n = 6, versus Islet1+/+;DNMAML: 1.57% ± 1.28%, n = 4; P = 0.47) (Supplemental Figure 2). These results suggest that inhibition of Notch in the secondary heart field affects migration of cardiac neural crest cells into the OFT, suggesting that a Notch-dependent signal from the second heart field can modulate cardiac neural crest behavior.
The Notch ligand Jag1 is required in the second heart field.
The expression pattern of Jag1 suggests that it is a candidate to act as an important Notch ligand in the second heart field. At E10.5 and E11.5, Jag1 is expressed in the pharyngeal endoderm, pharyngeal mesenchyme, and the OFT of the heart (Figure 3, A and B). In addition, it is expressed in the endothelium and smooth muscle of the aortic arch arteries (Figure 3B). To understand the role of Jag1 in the second heart field, we crossed Islet1Cre mice with mice harboring a conditional allele of Jag1 (Jag1flox). Immunostaining for Jag1 at E11.5 revealed efficient deletion in pharyngeal mesenchyme and the OFT of Islet1Cre/+;Jag1flox/flox, while expression was maintained in the region of the aortic arch arteries (Figure 3, C and D).
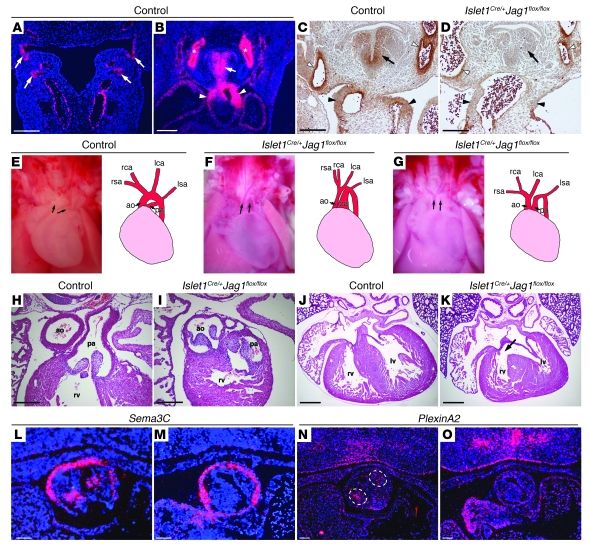
Figure 3. Jag1 is an essential Notch ligand in the second heart field.
(A and B) In situ hybridizations for Jag1 in control embryos. (A) Jag1 expression in pharyngeal endoderm (arrows, E10.5 embryo). (B) Jag1 expression in pharyngeal mesenchyme (arrow), OFT (arrowheads), and aortic arch arteries (asterisks, E11.5 embryo). (C and D) Jag1 immunostaining through E11.5 control (C) and mutant (D) embryos demonstrated loss of Jag1 expression in pharyngeal mesenchyme (arrow) and OFT (black arrowheads) of the mutant. Expression was maintained in the aortic arch arteries (white arrowheads). (E–G) Photographs and diagrams of hearts from E18.5 control (E) and mutant (F and G) embryos. (E) Arrows indicate pulmonary artery and aorta. (F) Double outlet right ventricle (arrows). (G) Double outlet right ventricle (arrows) and an interrupted aortic arch. (H–K) H&E-stained sections of E18.5 control (H and J) and mutant (I and K) hearts, demonstrating a double outlet right ventricle and a ventricular septal defect (arrow). (L–O) In situ hybridization of E10.5 control (L and N) and mutant (M and O) embryos. Control sections (L and N) demonstrated normal neural crest expression of Sema3C (L) and PlexinA2 (N) in the 2 clusters of cells in the center of the developing OFT cushions (dotted circles in N), while mutants (M and O) demonstrated reduced expression. Original magnification, ×30 (E–G). Scale bars: 250 μm (A, C, D, H, and I), 500 μm (B, J, and K), 100 μm (L–O).
Islet1Cre/+;Jag1flox/flox mutants died in the neonatal period (Supplemental Table 1). Analysis of late-gestation embryos revealed severe cardiac and aortic arch defects in Islet1Cre/+;Jag1flox/flox mutants similar to those seen in the second heart field DNMAML mutants, and included persistent truncus arteriosus, double outlet right ventricle, ventricular septal defects, atrial septal defects, and aortic arch defects such as right-sided aortic arch and interrupted aortic arch (Figure 3, F–K, and Table 1). Similar cardiovascular defects were seen when Mef2c-AHF-Cre was used to delete Jag1 (Table 1). Expression of the neural crest markers Sema3C and PlexinA2 is deficient in the OFT cushions of Islet1Cre/+;Jag1flox/flox embryos (Figure 3, L–O). Thus, second heart field–specific Jag1 deletion phenocopies the second heart field DNMAML mutants, suggesting that Jag1 functions as an essential Notch ligand in the second heart field.
Notch inhibition diminishes Fgf8 expression in the developing OFT.
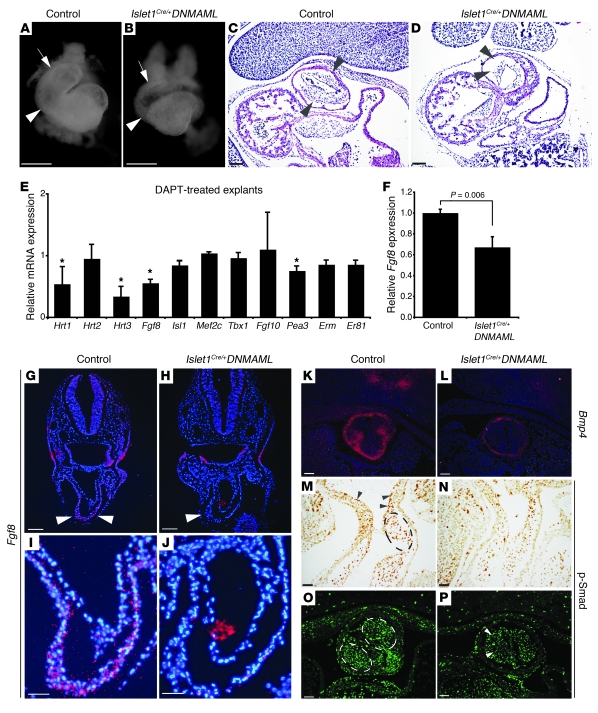
We examined the hearts of Isl1Cre/+;DNMAML embryos at E9.5, a relatively early stage, when the second heart field contributes a substantial number of progenitors to the developing right ventricle and OFT. At this time, the mutants showed significant hypoplasia of the right ventricle and OFT when compared with littermate controls (Figure 4, A and B). These results show that Notch plays a critical role in the early patterning of second heart field–derived structures. H&E staining of sections through the OFT at E10.5 demonstrated a statistically significant reduction in the cellularity of the endocardial cushions in the mutant embryos (Figure 4, C and D, and Supplemental Figure 3). The clusters of condensed mesenchyme of neural crest cells within the endocardial cushions, which can be discerned with H&E staining, were not evident in the mutant when compared with wild-type control (Figure 4, C and D).
Figure 4. Notch regulates Fgf8 expression in the second heart field.
(A and B) Light microscopy of E9.5 hearts of control (A) and Islet1Cre/+;DNMAML mutants (B). The mutant OFT (arrow) and right ventricle (arrowhead) were hypoplastic. (C and D) H&E-stained sections of E10.5 OFT cushions in control (C) and Islet1Cre/+;DNMAML mutant (D). The mutant OFT cushions (arrowheads) were hypocellular (D). (E and F) Quantitative RT-PCR of DAPT-treated cultured pharyngeal explants (E) or tissue directly isolated from E10.5 embryos (F), expressed relative to controls. (E) The data represent an average of 4 independent experiments, with error bars indicating 1 standard deviation. Asterisks indicate gene products that showed statistically significant changes in all 4 experiments (P < 0.02). (F) Fgf8 expression levels in the anterior pharynx and OFT of E10.5 control and mutant embryos. The data represent the average of 3 pools of 3–4 embryos per genotype. (G–J) In situ hybridizations showing Fgf8 expression in the OFT of E9.5 control (G and I) and mutant embryos (H and J). Arrowheads point to developing OFT myocardium. (K and L) In situ hybridization for Bmp4 at E11.5 in control (K) and mutant (L) embryos. (M and N) Immunohistochemistry for phospho-Smad in the OFT at E10.5 of control (M and O) and mutant (N and P) embryos showed robust phospho-Smad expression in the OFT cushions (circled) and endothelium (arrowheads) of controls with decreased expression in mutants. Control genotype was Islet1+/+;DNMAML. Scale bars: 100 μm (C–H, K–P), 250 μm (A, B, I, and J).
In order to identify downstream molecules involved in Notch signaling in the second heart field, we utilized a pharyngeal explant assay. In this assay, a region of the pharynx and the distal OFT was dissected from E9.5 mouse embryos and cultured in vitro. This region of the embryo is enriched for Islet1-positive progenitor cells that reside in the anterior pharynx and cardiac OFT and also includes surrounding tissues including the pharyngeal endoderm, pharyngeal ectoderm, and neural crest cells (28). Wild-type pharyngeal explants were treated with the γ-secretase inhibitor DAPT to inhibit Notch signaling. The explants were then analyzed by quantitative RT-PCR for the expression of a number of candidate genes involved in OFT development. Of several candidate genes examined, Fgf8 was notable for being altered in 4 of 4 independent experiments (Figure 4E). We also examined the Ets family transcription factors Pea3, Erm, and Er81, which have been implicated downstream of Fgf8. Pea3 was significantly downregulated in the DAPT-treated explants. Erm and Er81 also trended downward, but the decrease was not statistically significant. We also observed decreased expression of the Notch target genes Hrt1 and Hrt3, indicating that DAPT treatment effectively inhibited Notch signaling. Interestingly, Hrt2 expression was unchanged, consistent with several reports suggesting that regulation of this gene in the heart may not be strictly Notch dependent (29–31). Additional candidates that were tested using the explant assay but that did not reveal any statistically significant differences included Islet1, Mef2c, Tbx1, and Fgf10 (Figure 4E). The relative preservation of expression of these and other markers of cardiac OFT development suggests that the downregulation of Fgf8 is a specific effect of γ-secretase inhibition, rather than a nonspecific result of gross perturbations in patterning of the cardiac mesoderm.
Based upon the results of the explant assays, we examined Fgf8 mRNA expression by quantitative RT-PCR on tissue directly dissected from the pharyngeal region and cardiac OFT of wild-type and Islet1Cre/+;DNMAML mutant embryos at E10.5. These experiments revealed a statistically significant reduction in Fgf8 expression in mutants when compared with littermate controls (Figure 4F). Furthermore, in situ hybridization on sections through the OFT of E9.5 embryos demonstrated a clear reduction in Fgf8 mRNA in the developing OFT myocardium in the Islet1Cre/+;DNMAML mutants (Figure 4, G–J). In order to determine whether this reduction was due to a change in the number of Islet1-expressing cells in the OFT of mutant embryos, we performed immunohistochemistry and counted positively staining cells on serial sections of E10.5 Islet1Cre/+;DNMAML embryos compared with Islet1Cre/+ embryos, but we did not detect a significant difference (Islet1Cre/+;DNMAML: 325.4 ± 110.3 cells, n = 5, versus Islet1Cre/+: 347.3 ± 33.5 cells, n = 4; P = 0.72). Similar results were found when comparing E10.0 Islet1Cre/+;Jag1flox/+ (226.4 ± 26.8 cells, n = 6) with Islet1Cre/+;Jag1flox/flox littermates (257.0 ± 72.2 cells, n = 5) littermates (P = 0.503) (Supplemental Figure 4).
We also used in situ hybridization to examine the expression of Bmp4, a downstream effector of Fgf8 signaling in the cardiac OFT (24, 32, 33). Bmp4 expression was dramatically reduced in the OFT cushions and surrounding myocardium of Islet1Cre/+;DNMAML mutants compared with wild-type embryos (Figure 4, K and L). Bmp signaling results in Smad phosphorylation, and immunohistochemistry revealed decreased phospho-Smad1/5/8 levels in the cardiac mesenchyme populating the OFT cushions and the endothelium lining the OFT of Islet1Cre/+;DNMAML mutants compared with wild-type (Figure 4, M–P). Similar abnormalities of endocardial cushion development, and reductions in Fgf8, Bmp4, and phospho-Smad1/5/8 expression, were observed in the Islet1Cre/+;Jag1flox/flox embryos (Supplemental Figure 5). Thus, Notch inhibition or Jag1 deletion in the second heart field results in abnormal endocardial cushions, reduced myocardial Fgf8 in the OFT, and reduced Bmp signaling.
Notch signaling in the second heart field mediates Fgf8-dependent EMT in the OFT cushions.
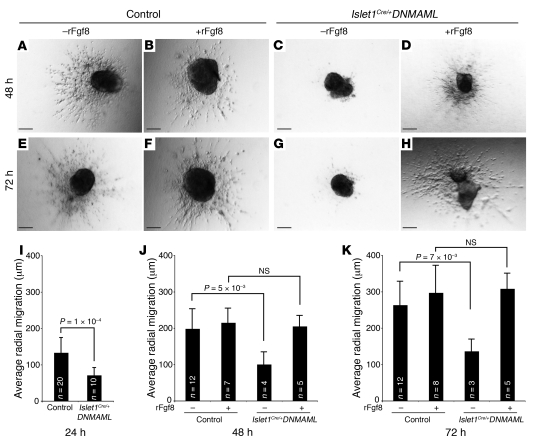
Endocardial cushions form through a process of EMT in which endothelial cells lining the anlagen of the cardiac valves respond to signals from overlying myocardium to invade the extracellular matrix of the cardiac jelly (34). The hypocellularity of the endocardial cushions in Islet1Cre/+;DNMAML mutants suggested that EMT might have been altered. Therefore, we used an established explant assay system to investigate EMT in vitro (35). Microdissected E10.5 OFT explants, including endothelium and myocardium derived from the second heart field in the region of the forming endocardial cushions, were placed on collagen gels and cultured for 24–72 hours. At all time points examined (24, 48, and 72 hours), wild-type explants exhibited elongated cells migrating away from the beating myocardium and invasion of the collagen gel (Figure 5, A and E data not shown). Many fewer cells were evident in explants derived from Islet1Cre/+;DNMAML mutants at all time points examined (Figure 5, C and G; data not shown). Moreover, the cells that did migrate away from the explant were circular and of abnormal morphology (Figure 5, C and G). Quantification of explants pooled from 3 independent experiments resulted in a nearly 50% reduction in average radial migration at 24, 48, and 72 hours (Figure 5, I–K).
Figure 5. Defective EMT in Islet1Cre/+;DNMAML mutants is rescued by rFgf8.
(A–D) Representative images of outflow explants taken 48 hours (24 hours after treatment with rFgf8 or vehicle) after plating. Control explants treated without (A) or with (B) rFgf8 underwent EMT and invasion into the collagen gel. (C) Islet1Cre/+;DNMAML explants displayed deficient EMT. (D) Addition of rFgf8 to mutant explants rescued EMT. (E–H) Similar results were seen at 72 hours. (I–K) Quantitative analysis was performed on samples pooled from 3 independent experiments at 24, 48, and 72 hours. Error bars represent 1 standard deviation. Control genotype was Islet1+/+;DNMAML. Scale bars: 100 μm.
These results demonstrate a tissue-specific requirement for Notch in second heart field–derived myocardium during EMT, suggesting that Notch may regulate a soluble signal from the myocardium that modulates EMT in the adjacent endothelial cells. Our finding of reduced Fgf8 expression in Islet1Cre/+;DNMAML mutants led us to test whether soluble Fgf8 could rescue EMT in mutant explants. Twenty-four hours after initial plating, we added recombinant Fgf8 (rFgf8) (50 ng/ml) or vehicle to wild-type and mutant explant cultures, and we analyzed the ability of explants to undergo EMT. While rFgf8 had little effect on wild-type explants (Figure 5, B and F), mutant explants showed a restoration of cellular morphology (Figure 5, D and H) and ability to invade the collagen matrix at both 48 and 72 hours (Figure 5, J and K). These results suggest that decreased Fgf8 observed in second heart field–derived myocardium of Islet1Cre/+;DNMAML mutants is at least partially responsible for defective EMT and hypocellular outflow endocardial cushions in these embryos, though changes in proliferation cannot be excluded.
Discussion
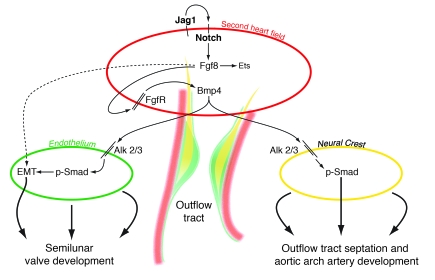
This study reveals an essential role for Notch and one of its ligands, Jag1, in the second heart field during cardiac OFT and aortic arch development. Inhibition of Notch or deletion of Jag1 in the second heart field using either Islet1Cre or Mef2c-AHF-Cre resulted in cardiac outflow and aortic arch artery defects that closely resembled those seen in humans with congenital heart disease. Development of the OFT involved complex interactions among multiple cell types including myocardium, endothelium, endocardial cushions, and neural crest. Information regarding the signaling pathways that mediate these interactions remains poorly described. We show that deletion of Jag1 or inhibition of Notch results in downregulation of Fgf8 in second heart field–derived myocardium, hypocellular endocardial cushions, and neural crest defects. These results demonstrate what we believe is a novel link between 2 signaling pathways critical in many developmental and disease processes and delineates a pathway by which different tissue types interact during cardiac OFT development (Figure 6).
Figure 6. Model depicting tissue-tissue interactions during OFT development.
The model depicts second heart field myocardium (red), endothelium undergoing EMT (green), and cardiac neural crest (yellow). Jag1/Notch signaling in the second heart field is proposed to stimulate Fgf8, which functions within the second heart field to regulate downstream cascades including Bmp4, which in turn signals to endothelium and neural crest.
Prior studies support our conclusions by providing evidence for gene expression in the second heart field that affects neighboring tissues. In particular, manipulation of Fgf8 and Fgf receptors within the second heart field has been shown to affect endocardial cushion development and neural crest invasion in agreement with our findings (24, 32). In addition to promoting endocardial cushion development, Fgf8 also plays a role in the development of cardiac neural crest–dependent structures and is essential for OFT septation and aortic arch artery development (25, 33, 36–40). Fgf8 mutants show hypoplasia of the fourth pharyngeal arch and defective development of the fourth aortic arch artery, a phenotype that is very similar to what we observed in Islet1Cre/+;DNMAML and Mef2c-AHF-Cre;DNMAML mutants (38, 39). Fgf8 is expressed by pharyngeal endoderm, ectoderm, and second heart field mesenchyme. The neural crest patterning defects observed in Fgf8 mutants are probably not due to a direct role of Fgf8 signaling on neural crest, as deletion of known Fgf receptors in neural crest does not recapitulate cardiovascular defects (24, 32). Rather, accumulated evidence suggests that Fgf signaling functions as part of an autocrine loop within myocardium, an interpretation strengthened by the fact that overexpression of the secreted Fgf inhibitor Sprouty1 in the second heart field produces structural cardiovascular defects resembling those seen in Fgf8 mutants (24). Taken together with the present study, these data suggest that Notch/Jag functions in the second heart field to regulate Fgf8, and that Fgf8 signals within the second heart field to modulate downstream pathways, which in turn affect endocardial cushions and neural crest. The exact mechanism by which Jag1/Notch regulates Fgf8 expression in the second heart field remains to be elucidated. The Fgf8 locus contains several conserved binding sites for CSL, the DNA binding component of the Notch transcriptional complex. However, preliminary experiments from our laboratory failed to show evidence for direct Notch-dependent activation of these elements in vitro (our unpublished observations). It is also possible that Notch may indirectly affect Fgf8 expression via an intermediate factor. The Hrt family of genes, which have been implicated downstream of Notch and are expressed in the developing heart, are unlikely to function as direct mediators of Fgf8 expression, as they generally function as transcriptional repressors rather than activators.
A likely candidate for mediating downstream effects of Fgf8 in OFT development is Bmp4. Bmp4 has been shown previously to function downstream of Fgf8 (33), and our studies demonstrate a reduction of Bmp4 expression in the OFT cushions and myocardium upon Notch inhibition in the second heart field. A recent study demonstrated that conditional loss of Bmp4 in the second heart field results in persistent truncus arteriosus and abnormal OFT cushion development (41). Furthermore, deletion of BMP receptors Alk2 or Alk3 in the cardiac neural crest results in OFT defects (42, 43), suggesting that Bmp4 may signal directly to neural crest. Bmps also are likely to directly affect endocardial cushion development; Bmp receptors are required for EMT, and the Bmp antagonist Noggin modulates endocardial cushion formation (25, 34, 41, 44–46). Our data are consistent with this model, since Notch inhibition in the second heart field results in a reduction in phospo-Smad expression in endocardial cushion mesenchyme. It is important to note that various cells contribute to the endocardial cushions, including endothelial cells, neural crest cells, and a population derived from Islet1-positive cells. However, it is unclear whether the Islet1-positive cells migrate into the cushions or are added to the cushions as a result of EMT from Islet1-positive endocardial cells (47).
It is likely that Notch plays different roles during development at different time periods, and our study highlights this fact. Notch has been implicated in the differentiation of embryonic stem cells into cardiomyocytes in vitro (11, 12, 48). However, as discussed above, we are unable to detect a difference in the number of Islet1 precursor cells in our mutants. This suggests that the critical time period for Notch-dependent activation of cardiac precursor cell differentiation is prior to the expression of Islet1. In contrast, later inhibition of Notch with DNMAML using SM22α-Cre, which is active in second heart field myocardium only after migration into the heart proper, does not recapitulate structural cardiac disorders (16, 49). Therefore, the critical time period for Notch activity in the second heart field for appropriate OFT patterning must be before SM22α-Cre can result in the effective expression of DNMAML. Thus, a relatively early function of Notch in second heart field precursors is relayed through time and space to exert significant morphogenetic defects later in development to modulate endocardial cushion and aortic arch maturation.
Elucidation of the role of Notch/Jag in cardiovascular development is directly relevant to human disease. Alagille syndrome, caused by mutations in JAG1 or NOTCH2 (50, 51), is associated with right-sided cardiac defects resembling those seen in our second heart field mutant mouse models. Our results suggest that many of these cardiac malformations are likely to be caused by defective patterning of the second heart field as a result of perturbed Jag1/Notch signaling. Human mutations in NOTCH1 have also been implicated in aortic valve disease (20). Previous studies have established a cell-autonomous role for Notch in endothelial cells during EMT and valve development (52, 53). In contrast, our results demonstrate a non–cell-autonomous pathway by which Notch affects EMT and valve formation by regulation of Fgf8 in the second heart field. This offers an additional mechanism by which human Notch mutations may result in valve malformations. Given the results of our studies, and the accumulating literature summarized here, it is likely that many of the cardiovascular manifestations of these human disorders are caused by abnormalities in tissue-tissue interactions and non–cell-autonomous defects resulting from perturbation of Notch. It also seems likely that additional human disorders related to abnormal cardiac valve development and aortic arch remodeling may be related to subtle or severe abnormalities of Notch/Jag1 function during development and to alterations of tissue-tissue communication during the complex process of OFT morphogenesis.
Methods
Mice.
All mice were maintained on mixed genetic backgrounds. Islet1Cre (23) and Mef2c-AHF-Cre (1) mice were genotyped using Cre-specific primers. DNMAML (22) mice were genotyped using primers specific for the ROSA26 locus. Jag1flox mice were genotyped as previously described (16). All animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Histology, immunohistochemistry, and in situ hybridization.
Samples were fixed overnight (E9.5 to E11.5 embryos) or for 48 hours (E17.5–E18.5 embryos and neonates) with 4% paraformaldehyde and dehydrated through an ethanol series. Samples were then paraffin-embedded and sectioned. Antibodies used for immunostaining were anti-GFP rabbit polyclonal (catalog A11122; Invitrogen), anti-Jag1 rabbit polyclonal H-114 (catalog SC-8303; Santa Cruz Biotechnology Inc.), anti-PECAM (CD31) rat monoclonal MEC 13.3 (catalog 553370; BD Biosciences — Pharmingen), anti-AP2α mouse monoclonal (catalog 5E4; Developmental Studies Hybridoma Bank), anti–phospho-Smad1/5/8 rabbit polyclonal (catalog 9511L; Cell Signaling Technology), and anti–phospho–histone H3 mouse monoclonal (catalog 9706L; Cell Signaling Technology). Radioactive in situ hybridizations were performed using previously described probes for PlexinA2 (54), Sema3C (55), Jag1 (16), Fgf8 (40), and Bmp4 (43). Immunohistochemistry and in situ hybridization images were analyzed using Adobe Photoshop. Control and mutant images were treated identically in all cases where brightness and contrast were altered.
H&E staining was performed using standard protocols. Quantification of OFT cushion nuclei was performed by analyzing H&E-stained sections from 3 sets of littermate embryos. The cells in the cushions and ventral pharynx of the embryos were manually counted. Quantification of Islet1-positive cells was performed by NIH ImageJ software.
Pharyngeal explant assays.
The anterior pharynx from the level of the second pharyngeal arch to the cardiac inflow tract, along with the distal region of the cardiac OFT, was microdissected from E9.5 embryos isolated from timed pregnant wild-type CD1 females (Charles River Laboratories). Glass tissue culture slides were pre-coated with a solution containing 3 mg/ml rat tail collagen and 100 μg/ml fibronectin, and the explants were grown for 48 hours in DMEM supplemented with 5% horse serum. DAPT (5 μM; γ-secretase inhibitor X; Calbiochem) or vehicle (DMSO) was added to the medium upon culture.
Quantitative RT-PCR.
For the pharyngeal explant assays, pools of 12–24 explants per condition were used for each RT-PCR experiment, and all results represent an average of 3 or 4 independent experiments. For direct analysis of Islet1Cre/+;DNMAML embryos, the anterior pharynx from the second pharyngeal arch to the level of the cardiac inflow tract, plus the distal aspect of the cardiac OFT, was microdissected from each embryo. Tissue from 3–4 embryos per genotype (Islet1Cre/+;DNMAML or Cre-negative littermate controls) was pooled for each experiment, and the results represent an average of 3 independent experiments.
RNA was isolated using TRIzol reagent (Invitrogen), and cDNA was generated using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR for Fgf8 and Islet1 was performed using the Taqman Gene Expression Assay (Applied Biosystems) and normalized against GAPDH levels. Expression levels of the remaining genes was determined using SYBR Green PCR Master Mix (Applied Biosystems), and all values were normalized against GAPDH. Primer sequences used for SYBR Green PCR assay are as follows: GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′; GAPDH reverse, 5′-GAAGTCACAGGAGACAACCTGGTC-3′; Hrt1 forward, 5′-GAAGCGCCGACGAGACCGAATCAA-3′; Hrt1 reverse, 5′-CAGGGCGTGCGCGTCAAAATAACC-3′; Hrt2 forward, 5′-CGACGTGGGGAGCGAGAACAAT-3′; Hrt2 reverse, 5′-GGCAAGAGCATGGGCATCAAAGTA-3′; Hrt3 forward, 5′-GGTCCCCACTGCCTTTGAGA-3′; Hrt3 reverse, 5′-TAGCTGACTGCTCAGGGAAGGCAA-3′; Mef2c forward, 5′-CCAGTGTCCAGCCATAACAGTTTG-3′; Mef2c reverse, 5′-AGATTCATAGGGGGAGGAGATTTG-3′; Tbx1 forward, 5′-ACCGGTATGCTTTCCATAGCTCCT-3′; Tbx1 reverse, 5′-AATCTGTTTCATCCACTGTGCGCC-3′; Fgf10 forward, 5′-CAGTAAGACACGCAAGCATTTACTG-3′; Fgf10 reverse, 5′-AATCTGATCCAATTCTTCCATGGT-3′; Pea3 forward, 5′-ACGTGTACAAGTTTGTGTGCGAGC-3′; Pea3 reverse, 5′-ATCCAAGTGGGACAAAGGGACTGT-3′; Erm forward, 5′-TCCAGAACCTGGATCACAGCAACA-3′; Erm reverse, 5′-GGCTTTCAGGCATCATCTTTGGCA-3′; Er81 forward, 5′-AAATTTGTGTGTGACCCGGAAGCC-3′; Er81 reverse, 5′-TGTGACGTTCCATGTCCGTCTTCA-3′.
OFT cushion and EMT explant assays.
The cardiac OFT was microdissected from E10.5 embryos isolated from pregnant homozygous DNMAML females impregnated by Islet1Cre/+ males. The explants were dissected and grown on collagen gels as previously described (35). At 24 hours, the medium was replaced with either medium containing 50 ng/ml recombinant mouse Fgf8b (R&D Systems) or medium containing vehicle solution (0.1 mg/ml BSA in PBS). The explant cushions were scored at 24, 48, and 72 hours. Cell migration from the edge of the explant was measured at 8 different locations (every 45°) and then averaged. Samples were pooled from 3 independent experiments and used for quantitative analysis.
Statistics.
The Student’s 2-tailed t test analysis was used to evaluate differences between groups in all histological, RT-PCR, and EMT experiments. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We would like to thank Brian Black and Sylvia Evans for providing reagents, and members of the Epstein laboratory for thoughtful discussions and critical reading of the manuscript. This work was supported by funding from the University of Pennsylvania Medical Scientist Training Program (to F.A. High), an American Heart Association Physician-Scientist/Postdoctoral Fellowship (to R. Jain), and grants from the NIH and funds from the W.W. Smith Endowed Chair for Cardiovascular Research (to J.A. Epstein).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Bmp4, bone morphogenetic protein 4; DNMAML, dominant-negative truncated form of mastermind-like; EMT, endothelial-mesenchymal transition; OFT, outflow tract; rFgf8, recombinant Fgf8.
Citation for this article: J. Clin. Invest. 119:1986–1996 (2009). doi:10.1172/JCI38922
References
- 1.Verzi M.P., McCulley D.J., De Val S., Dodou E., Black B.L. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. . 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 2.Kelly R.G., Brown N.A., Buckingham M.E. The arterial pole of the mouse heart forms from Fgf10-Expressing cells in pharyngeal mesoderm. Dev. Cell. 2001;1:435–440. doi: 10.1016/S1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 3.Waldo K.L., et al. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 4.Mjaatvedt C.H., et al. The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 5.Zaffran S., Kelly R.G., Meilhac S.M., Buckingham M.E., Brown N.A. Right ventricular myocardium derives from the anterior heart field. Circ. Res. 2004;95:261–268. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
- 6.Waldo K.L., et al. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Cai C.L., et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laugwitz K.L., et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moretti A., et al. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 10.Chiba S. Concise review: Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder T., et al. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech. Dev. 2006;123:570–579. doi: 10.1016/j.mod.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Nemir M., Croquelois A., Pedrazzini T., Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ. Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder T., et al. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4018–4023. doi: 10.1073/pnas.0438008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rones M.S., McLaughlin K.A., Raffin M., Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- 15.Han Z., Bodmer R. Myogenic cell fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development. 2003;130:3039–3051. doi: 10.1242/dev.00484. [DOI] [PubMed] [Google Scholar]
- 16.High F.A., et al. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J. Clin. Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warthen D.M., et al. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum. Mutat. 2006;27:436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- 18.Krantz I.D., et al. Jagged1 mutations in patients ascertained with isolated congenital heart defects. Am. J. Med. Genet. 1999;84:56–60. doi: 10.1002/(SICI)1096-8628(19990507)84:1<56::AID-AJMG11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 19.Eldadah Z.A., et al. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum. Mol. Genet. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- 20.Garg V., et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman J.I.E., Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. doi: 10.1016/S0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 22.Tu L., et al. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L., et al. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park E.J., et al. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park E.J., et al. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dodou E., Verzi M.P., Anderson J.P., Xu S.M., Black B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 27.Engleka K.A., et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Cohen E.D., et al. Wnt/{beta}-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe Y., et al. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 2006;133:1625–1634. doi: 10.1242/dev.02344. [DOI] [PubMed] [Google Scholar]
- 30.Kokubo H., Tomita-Miyagawa S., Hamada Y., Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- 31.Rutenberg J.B., et al. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., et al. Frs2{alpha}-deficiency in cardiac progenitors disrupts a subset of FGF signals required for outflow tract morphogenesis. Development. 2008;135:3611–3622. doi: 10.1242/dev.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ilagan R., et al. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima Y., Yamagishi T., Hokari S., Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP). Anat. Rec. 2000;258:119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Lakkis M.M., Epstein J.A. Neurofibromin modulation of ras activity is required for normal endocardial-mesenchymal transformation in the developing heart. Development. 1998;125:4359–4367. doi: 10.1242/dev.125.22.4359. [DOI] [PubMed] [Google Scholar]
- 36.Hutson M.R., et al. Cardiac arterial pole alignment is sensitive to Fgf8 signaling in the pharnyx. Dev. Biol. 2006;295:486–497. doi: 10.1016/j.ydbio.2006.02.052. [DOI] [PubMed] [Google Scholar]
- 37.Macatee T.L., et al. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu-Issa R., Smyth G., Smoak I., Yamamura K., Meyers E.N. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 39.Frank D.U., et al. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown C.B., et al. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev. Biol. 2004;267:190–202. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 41.McCulley D.J., Kang J.O., Martin J.F., Black B.L. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev. Dyn. 2008;237:3200–3209. doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stottmann R.W., Choi M., Mishina Y., Meyers E.N., Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaartinen V., et al. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- 44.Choi M., Stottmann R.W., Yang Y.P., Meyers E.N., Klingensmith J. The bone morphogenetic protein antagonist noggin regulates mammalian cardiac morphogenesis. Circ. Res. 2007;100:220–228. doi: 10.1161/01.RES.0000257780.60484.6a. [DOI] [PubMed] [Google Scholar]
- 45.Gaussin V., et al. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J., et al. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev. Biol. 2005;286:299–310. doi: 10.1016/j.ydbio.2005.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snarr B.S., Kern C.B., Wessels A. Origin and fate of cardiac mesenchyme. Dev. Dyn. 2008;237:2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- 48.Schroeder T., et al. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4018–4023. doi: 10.1073/pnas.0438008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proweller A., et al. Notch signaling in vascular smooth muscle cells is required to pattern the cerebral vasculature. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16275–16280. doi: 10.1073/pnas.0707950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 51.Oda T., et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 52.Timmerman L.A., et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niessen K., et al. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008;182:315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown C.B., et al. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- 55.Gitler A.D., Lu M.M., Epstein J.A. PlexinD1 and semaphorin signaling are required in endothelial cells for cardiovascular development. Dev. Cell. 2004;7:107–116. doi: 10.1016/j.devcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.