SYNOPSIS
The understanding of the mechanism, oxidant(s) involved, and how/what protein radicals are produced during the reaction of wild type Cu, Zn-superoxide dismutase (SOD1) with H2O2 and their fate is incomplete, but a better understanding of the role of this reaction is needed. We used immuno-spin trapping and mass spectrometry analysis to study the protein oxidations driven by human (h) and bovine (b) SOD1 when reacting with H2O2 using human serum albumin (HSA) and mouse brain homogenate (mBH) as target models. In order to gain mechanistic information about this reaction, we considered both copper- and carbonate radical anion-initiated protein oxidation. We chose experimental conditions that clearly separated SOD1-driven oxidation via CO3 •− from that initiated by copper released from the SOD1 active site. In the absence of (bi)carbonate, site-specific radical-mediated fragmentation is produced by SOD1 active-site copper. In the presence of (bi)carbonate and DTPA (to suppress copper chemistry), CO3 •− radical produced distinct radical sites in both SOD1 and HSA, which caused protein aggregation without causing protein fragmentation. The CO3 •− produced by reaction of hSOD1 with H2O2 also produced distinctive DMPO nitrone adduct positive protein bands in the mBH. Finally, we propose a biochemical mechanism to explain CO3 •− production from carbon dioxide, enhanced protein radical formation, and protection by (bi)carbonate against H2O2-induced fragmentation of the SOD1 active site. Our study is important for establishing experimental conditions for studying the molecular mechanism and targets of oxidation during the reverse reaction of SOD1 with H2O2; these results are the first step in analyzing the critical targets of SOD1-driven oxidation during such pathological processes as neuroinflammation.
Keywords: superoxide dismutase, peroxymonocarbonate, carbonate radical anion, Fenton-like chemistry, protein radical, nitrone adduct, immuno-spin trapping
INTRODUCTION
Much of the mechanistic biochemical evidence regarding Cu,Zn-superoxide dismutase (SOD1)1-driven pathogenic oxidative mechanisms has been obtained by studying the reaction between bovine or human SOD1 and H2O2 [1–4]. The reaction of SOD1 and H2O2 may be particularly important in sites of inflammation, particularly in peroxisomes, which are known to have high local concentrations of H2O2 and SOD1, and where the degradation of damaged proteins occurs [5]. The study of radical pathways induced by H2O2/SOD1-driven peroxidation may be important to the understanding of the molecular mechanisms of tissue damage during neuroinflammation as occurs, for example, in familial amyotrophic lateral sclerosis (fALS).
Mechanistically, the reaction of SOD1 with H2O2 proceeds in several sequential steps (Reactions 1 and 2) that end with the formation of an enzyme-bound oxidant [i.e., (CuO)1+ ↔ (Cu-•OH)2+ ↔ Cu3+]at the enzyme active site [6, 7]. This species promotes oxidation of one or more histidine residues at the SOD1-active site [8, 9], partial copper release [10, 11], enzyme inactivation [6], and SOD1 fragmentation at its active site [10, 12].
| Reaction 1 |
| Reaction 2 |
The copper-bound oxidant at the enzyme-active site (reactions 1 and 2) is generally proposed to oxidize (bi)carbonate to the diffusible carbonate radical anion (CO3 •−) [7, 13–16]. Recently, CO2 has been postulated to be the chemical entity in (bi)carbonate buffer that reacts with the strong oxidant at the SOD1 active site to give CO3 •− [14]; however, the chemical mechanism of this reaction remains controversial [7, 17–19]. Much of our current knowledge of SOD1-driven oxidation has been gained by studying the oxidation of chemical compounds (e.g., NADPH [20], dichlorofluoresceine [11], peroxidase substrates [12, 21], ethanol, spin trap compounds [21, 22], formate and azide [13], etc.), the loss of superoxide dismutase activity, or the activity or the structure of protein or nucleic acid targets [23]. Because of its diffusibility and powerful oxidative nature, CO3 •− may form distinct radicals in SOD1 itself or tissue proteins, which may be trapped with 5,5-dimethyl-1-pyrroline N-oxide (DMPO) to form stable nitrone adducts for further characterization [12, 24]. However, in cells and tissues it is difficult to study the formation of protein radicals using electron spin resonance (ESR) with or without spin trapping because of their rapid decay and the severe overlap of their spectra.
We have developed a new biochemical tool with which to study protein [25–27] and DNA radicals [24, 28] called immuno-spin trapping [26, 27] that helps to identify protein radicals induced by SOD1-driven oxidation in cells and tissues. Immuno-spin trapping [29] involves the trapping of protein radicals with the spin trap DMPO in situ and in real time and the further detection of the protein radical-DMPO nitrone adducts with an anti-DMPO serum [25] by using heterogeneous immunoassays and also by mass spectrometry [30]. In principle, this technology allows the simultaneous detection of more than one protein radical at the same time and in the same reaction system, as they are formed during tissue oxidative damage [24, 29].
We have previously shown that (bi)carbonate, but not DTPA, protects SOD1 against H2O2-induced fragmentation at its active site and that, in the presence of DTPA, (bi)carbonate is required in order to observe SOD1 nitrone adducts with immuno-spin trapping [12]. Here we have used immuno-spin trapping and mass spectrometry to understand the mechanism of protein radical formation induced by the bovine and human SOD1/HSA or mouse brain homogenate (mBH)/H2O2 system. To accomplish this goal, we chose experimental conditions that would separate the two major radical pathways of protein modification by H2O2-induced, SOD1-driven oxidation: copper (both active site and released)-and CO3 •−-triggered radical reactions. Further, we have analyzed the way in which these two different initiators of radical reactions contribute to oxidizing target proteins and SOD1 itself using HSA and mBH as models. In this study, we also propose a novel mechanism to explain how (bi)carbonate blocks the fragmentation of the SOD1 active site by the enzyme-bound oxidant (reaction 2).
EXPERIMENTAL
Materials
Bovine Cu,Zn-superoxide dismutase (bSOD1, from bovine erythrocytes) and beef liver catalase were purchased from Roche Applied Science (Indianapolis, IN). Sodium bicarbonate (99.7–100.3%) was purchased from Alfa Aesar (Ward Hill, MA). The spin trap DMPO was purchased from Alexis Biochemicals (San Diego, CA), purified twice by vacuum sublimation at room temperature, and stored under an argon atmosphere at −80 °C until use. The DMPO concentration was measured at 228 nm, assuming a molar absorption coefficient of 7,800 M−1 cm−1. Reagent grade 30% H2O2 was obtained from Fisher Scientific Co. (Fair Lawn, NJ). The H2O2 concentration was verified using UV-vis absorption at 240 nm (ε240nm = 43.6 M−1 cm−1). Recombinant human SOD1 (expressed in E. coli) was from BioVision (Cat#4802, Mountain View, CA). Erythrocyte human SOD1 (hSOD1, Cat#S-9636) and all other reagents were purchased from Sigma Chemical Co. (St. Louis, MO). Buffers were treated with Chelex® 100 ion-exchange resin (Bio-Rad Laboratories, Hercules, CA) to remove transition metals usually found as contaminants. The pH of the buffer solutions containing bicarbonate was adjusted to 7.4 by bubbling with a gas mixture of 5% CO2 and 95% N2.
Cu-Zn superoxide dismutase, human serum albumin, and mouse brain homogenate preparation
Our bSOD1 and hSOD1 (from erythrocytes and recombinant) solutions were prepared and determined to be free of detectable unbound copper as previously described [31]. SOD1 concentration is expressed as micromolar (µM), determined by measuring the absorbance at 258 nm (ε258 nm = 10.3 mM−1 cm−1) at pH 7.4 [32]. HSA solutions were prepared as recently described [31, 33]. Our HSA preparations were free of detectable copper contamination, and the free sulfhydryl /albumin ratio was similar to that reported [31, 33]. The HSA concentration was determined by using the BCA assay (Pierce) or assuming its UV-visible extinction coefficient of ε280 nm= 35,700 M−1 cm−1. Male C576J/BL mice (25–30 g body weight) were euthanized according to institutionally approved protocols, and brains were collected in ice-cold 10 mM sodium phosphate buffer, pH 7.4, washed in the same buffer and homogenized (1g/ml). The homogenate was centrifuged at 11,700×g and the supernatant was dialyzed (3 kDa cut-off) against 10 mM sodium phosphate buffer, pH 7.4. The protein concentration in the dialyzed mouse brain homogenate (mBH) was determined using the BCA assay.
Chemical reactions
Typically, the reactions of 15 µM SOD1, 7.5 µM (0.5 mg/ml) HSA or 0.5 mg/ml mBH and 0.1 mM H2O2 were carried out in the presence of 100 mM DMPO in 100 mM chelexed sodium (bi)carbonate buffer, pH 7.4 (BB), or 100 mM sodium phosphate buffer, pH 7.4 (PB), or in PB containing a physiological concentration of (bi)carbonate (i.e., ~ 25 mM) and with or without 0.1 mM DTPA. Solutions were incubated at 37 °C for 1 h and the reaction stopped by adding 10 IU catalase to eliminate excess H2O2. The pH of the reaction mixtures after the reactions was completed was between 7.3–7.6.
Measurement of H2O2-induced, bathocuproine-assisted reduction of released Cu2+
The H2O2-induced Cu2+ release from bSOD1 or hSOD1 and its reduction to Cu1+ was monitored using the specific Cu1+ chelator BCDS [34]. Cu1+ binds to BCDS and produces a complex (Cu1+(BCDS)2)−3 that exhibits a characteristic absorbance maximum at 480 nm, with ε480 = 12,540 M−1 cm−1
Measurement of SOD1 activity
Bovine and human SOD1 activity was measured using the ferricytochrome c reduction assay [11, 32]. For controls, the ratio of ferricytochrome c reduction was measured in samples containing 15 µM active or heat-inactivated SOD1 (incubation for 40 min at 75 °C) [32].
Anti-DMPO serum
A rabbit antiserum against the nitrone form of DMPO was obtained in our laboratory [25] and used to develop immuno-spin trapping [26, 27, 29]; this antiserum has been successfully used to detect protein [27, 29] and DNA radicals [24, 28]. The anti-DMPO serum is commercially available from Alexis Biochemicals, Cayman Chemicals, AbCam, Chemicon International, and Oxford Biomedical Research.
Immuno-spin trapping assays
DMPO-protein radical-derived nitrone adducts were determined using a standard enzyme-linked immunosorbent assay (ELISA) and Western blot as previously described [12, 29]. Briefly, the reaction mixture was separated by reducing SDS-PAGE (1.2 µg proteins/ lane). After the separation of protein, gels were stained using Coomassie blue, or the proteins were blotted to a nitrocellulose membrane, and the nitrone adducts were detected by Western blot. Briefly, immuno-complexes were detected by exposing the membrane to NBT/BCIP One Step reagent from Pierce (Rockford, IL.) for 15 min or, when indicated, by enhanced chemiluminescence (ECL) using a CDP-Star II (Roche Molecular Biochemicals) / Nitro Block II (Tropix, Bradford, MA) system. Where indicated, a MagicMark™ XP Western Protein Standard (Invitrogen) was used as a molecular weight marker that glows after Western blot development [24].
Measurement of H2O2-induced, SOD1-driven oxidation of guaiacol
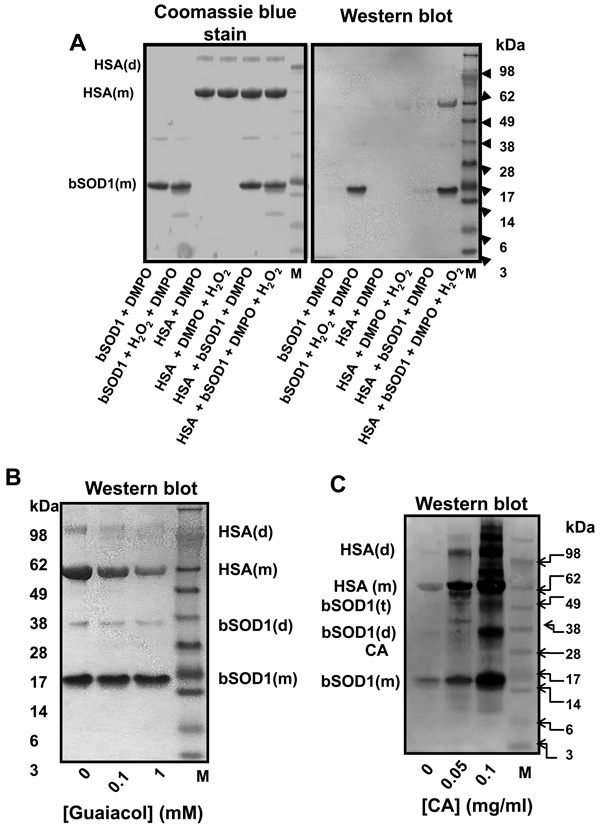
The peroxidative activity of bSOD1 or hSOD1/H2O2 with or without (bi)carbonate or DTPA was measured using guaiacol as a substrate. The reaction was initiated by adding 5 µM SOD1 to 100 mM chelexed buffers, pH 7.4, containing 0.5 mM guaiacol and 1 mM H2O2 (final volume, 1 ml). Oxidation of guaiacol to tetraguaiacol was measured spectrophotometrically in a Beckman DU® 640 spectrometer at 470 nm and at room temperature for 15 min. An extinction coefficient of 26.6 mM−1 cm−1 was used to quantify tetraguaiacol formation.
Mass spectrometric identifications from gel bands
In-gel tryptic digestion
The protein bands were manually excised from the gel, cut into small pieces, and transferred into a 96-well microtiter plate. Gel pieces were subjected to automatic tryptic digestion using an Investigator™ Progest protein digestion station (Genomic Solutions, Ann Arbor, MI). Briefly, gel bands were sequentially washed twice with 25 mM ammonium bicarbonate buffer (pH 7) and acetonitrile, dehydrated, rehydrated with 25 µL of the enzyme solution, and digested at 37 °C for 8 hrs. The enzyme solution used was sequencing grade modified trypsin (Promega Corporation, Madison, WI) at a concentration of 0.01 mg/ml in 25 mM ammonium bicarbonate buffer (pH 7). Resulting tryptic peptides were extracted from the gel, lyophilized, and stored at −80 °C. Prior to mass spectrometric analysis, the peptides were reconstituted in 40 µl of a 97:3 solution of water:acetonitrile (0.1% formic acid).
ESI Mass Spectrometry
For the nanoLC/ESI/MS/MS analyses, we used an Agilent XCT Ultra ion trap (Agilent Technologies, Inc., Santa Clara, CA) equipped with an HPLC-Chip Cube MS interface and an Agilent 1100 nanoLC system. Injections of 30 µl of the peptide digests were made onto a 40 nl enrichment column followed by a 43 mm × 75 µm analytical column packed with ZORBAX 300SB C18 particles. Peptides were separated and eluted using a linear gradient of 3–50% acetonitrile (0.1% formic acid) over 40 min, followed by a linear gradient of 50–95% acetonitrile over 7 min at a flow rate of 500 nl/min. The ion trap mass spectrometer was operated in the positive ion mode, standard enhanced mode using the following settings: capillary voltage, −2150 V; mass range, 300–1500; ICC smart target (number of ions in the trap prior to scan out), 100,000 or 200 milliseconds of accumulation; and MS/MS fragmentation amplitude, 1.0 V. During the LC/MS/MS analyses, automated data-dependent acquisition software was employed with the six most abundant ions (threshold requirement of 10,000 counts) from each spectrum selected for MS/MS analysis.
Following the analyses, the MS/MS data were extracted and analyzed using Spectrum Mill MS Proteomics software (Agilent Technologies, Inc). To generate peak lists, the raw data files were processed using the Data Extractor function with the following parameters: deconvoluted ions of 300–6,000 Da and a retention time of 10 to 60 min. MS scans with the same precursor m/z were merged based on a +/− 1.4 m/z window and a +/− 15 sec retention time window. Using the extracted data, searches were performed against the NCBI nonredundant protein database using the MS/MS search function. Parameters used for the searches included: precursor mass tolerance, +/− 1.5 Da; product mass tolerance, +/− 1 Da; enzyme specificity, trypsin, with maximum two missed cleavage sites; variable modifications, oxidized methionine and N-terminal pyroglutamic acid; and at least 2 unique peptides matched peak intensity, 80%; species, mouse. Proteins with a summed MS/MS search score of 30 or greater were considered for validation. At this scoring threshold, the false positive rate was essentially 0% as determined by searching against a reversed sequence database. All MS/MS sequence assignments used for protein identifications were manually validated.
RESULTS
Parallel SOD1-centered radical and enzyme inactivation induced by H2O2
We observed H2O2-induced bovine and human erythrocyte SOD1 inactivation in (bi)carbonate buffer (Fig. 1A). In agreement with our previous report [12] and other authors [11, 18], we did not find any effect on H2O2-induced SOD1 inactivation when the (bi)carbonate concentration was 25 mM or less in phosphate buffer with DTPA.
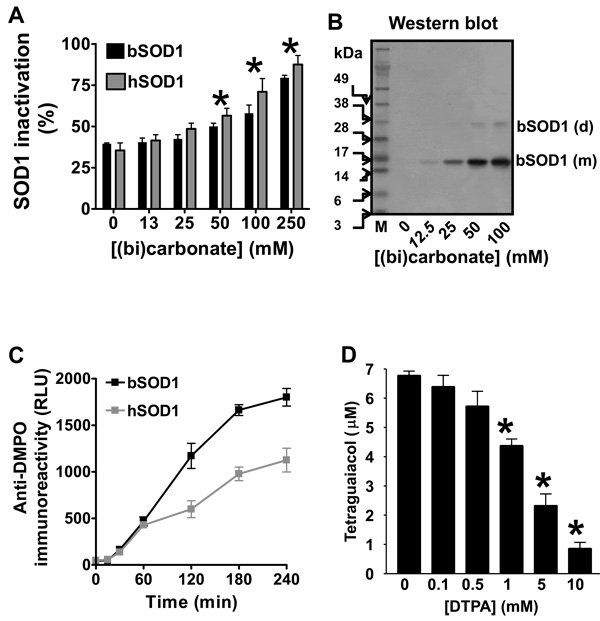
Figure 1. Parallel SOD1 inactivation and SOD1-centered radical formed by carbonate radical anion and the effect of DTPA.
A,SOD1 activity determined after reacting 15 µM erythrocyte human (h) or erythrocyte bovine (b) SOD1 with 0.1 mM H2O2 for 1 h at 37 °C in 100 mM phosphate buffer containing different concentrations of (bi)carbonate, pH 7.4, and 0.1 mM DTPA. Asterisk indicates P<0.05 with respect to reactions with no (bi)carbonate added. B, Western blot of nitrone adducts produced when 15 µM bSOD1, 100 mM DMPO, and 0.1 mM H2O2 were reacted for 1 h at 37 °C in fresh, argon-bubbled (to purge carbon dioxide) 100 mM sodium phosphate buffer (PB) containing different concentrations of (bi)carbonate, pH 7.4, with 0.1 mM DTPA. Reactions were stopped by removing the excess H2O2 with 10 IU catalase. M indicates molecular weight marker, (m) monomer, and (d) dimer. C, ELISA analysis of nitrone adducts produced from reactions of 15 µM bSOD1 or hSOD1 with 0.1 mM H2O2 and carried out in 100 mM phosphate buffer containing 25 mM (bi)carbonate, pH 7.4. Reactions were stopped at different times with 10 IU catalase. RLU is relative light units. D, Tetraguaiacol formed from reactions that contained the same reagent concentrations, except that the H2O2 concentration was 1 mM, and buffer as B, but the reaction buffer contained different concentrations of DTPA. The oxidation of guaiacol (0.5 mM) to tetraguaiacol was determined at 15 min as indicated in Experimental. Values significantly different (P<0.05) from the sample without DTPA are indicated with an asterisk. Data shown are from a representative experiment or the mean ± S. E. from three experiments, each in triplicate.
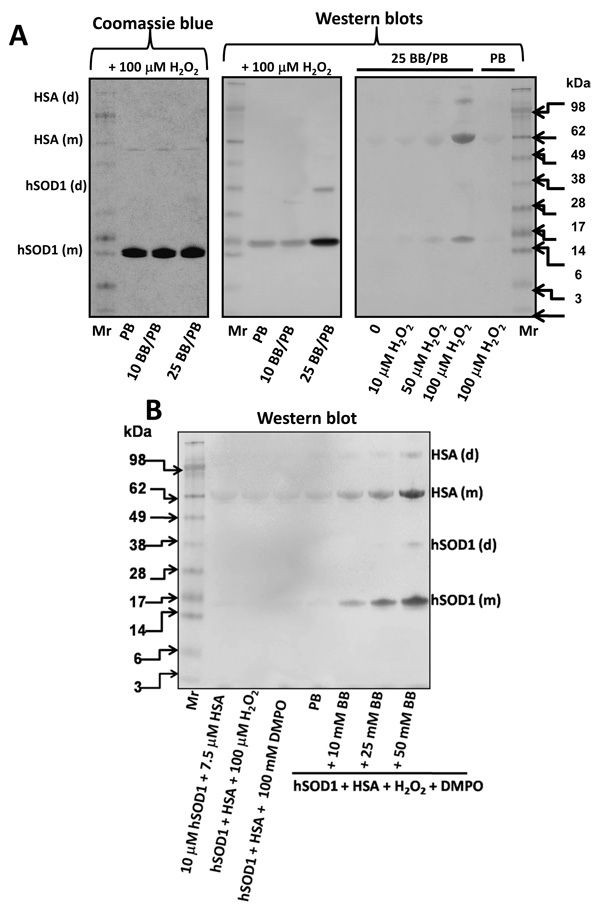
When (bi)carbonate was added to a bSOD1/H2O2 or hSOD1/H2O2 system in fresh argon-bubbled (to purge carbon dioxide) phosphate buffer (PB) containing the copper chelator DTPA, the amount of H2O2-induced bSOD1-centered radicals observed as monomers and dimers increased, as seen by Western blot (Fig. 1B). The hSOD1 enzyme produced similar results (data not shown). Although bSOD1-centered radicals were detected at the physiological (bi)carbonate concentration of 25 mM, higher (bi)carbonate concentrations led to more nitrone adduct formation, demonstrating that the (bi)carbonate-dependent mechanism was not saturated under physiological conditions. We could not find any H2O2-induced SOD1 nitrone adducts in argon-purged, freshly prepared chelexed phosphate buffer, pH 7.4, containing 0.1 mM DTPA in a closed system, but we could detect them by Western blot when phosphate buffer was equilibrated with air at room temperature for 15 min, presumably due to the absorption of CO2 (data not shown). In phosphate buffer with 0.1 mM DTPA, the SOD1 nitrone adduct (SOD1-self oxidized) formation was totally dependent on (bi)carbonate. It increased with incubation time (Fig. 1C) and was completely prevented by cyanide (data not shown), suggesting that (bi)carbonate and copper redox cycling at the enzyme active site is essential for producing SOD1 nitrone adducts.
In Figs. 1A–C, the copper chelator DTPA was added to eliminate the contribution of H2O2-induced, free copper-mediated SOD1 oxidation; however, it is known that CO3 •− reacts relatively quickly with DTPA (rate constant ~ 1.7 × 107 M−1 s−1 [35]), and, thus, the concentration of DTPA was carefully adjusted to avoid underestimating CO3 •−-triggered oxidations. Indeed, we observed that concentrations of DTPA higher than 0.5 mM inhibited CO3 •−-mediated guaiacol oxidation (Fig. 1D), suggesting that high concentrations of DTPA scavenge CO3 •−. Accordingly, in subsequent experiments shown in this report, we used 0.1 mM DTPA, which is over three-fold higher than the copper content of the SOD1 solutions used in our experiments.
Albumin protects bovine and human SOD1 activity by acting as an alternative target for radical modification
In order to investigate the potential for the induction of SOD1-driven oxidation in surrounding proteins, we used human serum albumin (HSA) as a model target. We observed that HSA blocked H2O2-induced SOD1 self-inactivation in (bi)carbonate buffer, but in phosphate buffer it had no effect (Fig. 2A). This suggested that HSA protects SOD1 activity by scavenging the diffusible CO3 •− that enhances H2O2-induced SOD1 inactivation. When we separated the reaction mixture in SDS-PAGE and stained the gels with Coomassie blue, we observed that both proteins were extensively fragmented when the reaction was carried out in phosphate buffer (Fig. 2B, left panel). DTPA protected HSA against fragmentation, but did not affect SOD1 fragmentation at its active site as assessed by the ~5 and ~10 kDa SOD1 fragments [10, 12] (Fig. 2B, left panel). However, when we performed the reaction in (bi)carbonate buffer with DTPA, the active site of SOD1 was protected against site-specific fragmentation (Fig. 2B, right panel, [12]), but extensive fragmentation at other sites of both SOD1 and HSA still occur until DTPA is added. Although active-site fragmentation is not occurring, copper must still be released presumably as a result of histidine oxidation (Table 1). This released copper must be causing protein fragmentation through Fenton-type chemistry. Similar results were obtained when erythrocyte bSOD1 was replaced by hSOD1 (data not shown).
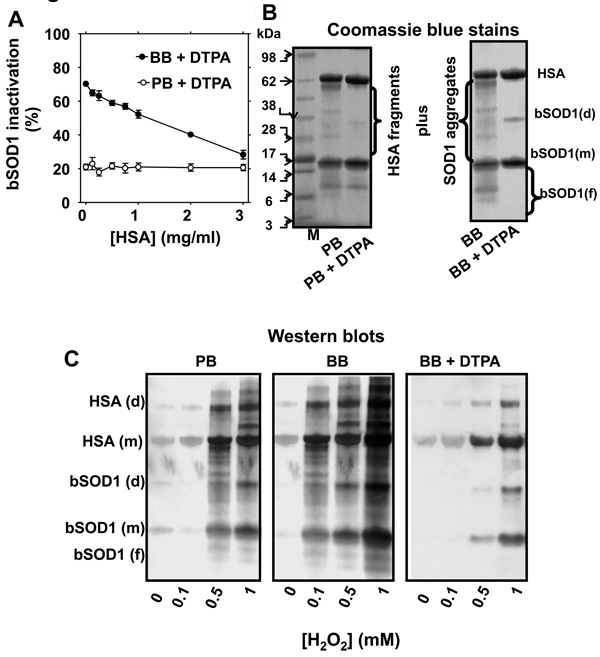
Figure 2. Structural and functional modification of SOD1 by H2O2 and the effect of HSA.
A, SOD1 activity determined in a reaction mixture that contained 15 µM bSOD1, 0.1 mM H2O2 and different concentrations of human serum albumin (HSA). Reactions were carried out at pH 7.4 in 100 mM (bi)carbonate buffer (BB) or 100 mM phosphate buffer (PB), with 0.1 mM DTPA for 1 h. B, Coomassie blue-stained gels of reaction mixtures containing 15 µM bSOD1 and 7.5 µM HSA incubated as in A, but carried out in PB (left panel) or BB (right panel) with or without 0.1 mM DTPA. The reactions were started by adding 0.1 mM H2O2 and carried out at 37 °C then stopped with 10 IU catalase. C, Western blots produced when 15 µM SOD1, 7.5 µM HSA, and 100 mM DMPO were mixed either in 100 mM phosphate buffer (PB), 100 mM (bi)carbonate buffer (BB), or BB containing 0.1 mM DTPA (BB+DTPA), pH 7.4, and reacted with different concentrations of H2O2. Reaction mixtures were incubated for 1 h at 37 °C and stopped by adding 10 IU catalase. M, molecular weight marker; (f), (m) and (d) indicate fragment, monomer and dimer, respectively. SOD1 activity assay and SDS-PAGE/Coomassie blue stains were performed as described under Experimental. Data show mean values ± S.E. or representative results.
Table 1.
Approximate rate constants (M−1 s−1) for the reactions of hydroxyl radical and carbonate radical anion with some amino acids at pH~7.4*
| •OH | CO3•− | |
|---|---|---|
| Tyrosine | 1 × 1010 | 5 × 107 |
| Cysteine | 2 × 1010 | 5 × 107 |
| Tryptophana | 1 × 1010 | 7 × 108 |
| Histidine | 5.7 × 109 | 5.6 × 106 |
| Methionine | 9 × 109 | 1 × 108 |
Chemical kinetic rate constants are from http://www.rcdc.nd.edu/solnkin2/
human SOD1 contains Trp instead of Tyr as bovine SOD1
As modulated by the copper chelator DTPA and (bi)carbonate, two structural consequences of free radical chemistry can be delineated in the formation of H2O2-induced, SOD1-driven protein radicals: fragmentation and aggregation. Fragmentation other than at the active site appears to be mediated by copper released from the active site of SOD1. In the absence of DTPA and (bi)carbonate, we saw evidence of both pathways, as protein nitrone adducts (both fragments and intact proteins) increased with the concentration of H2O2 (Fig. 2C, left panel). At the same H2O2 concentration, the number of radical sites was higher when the reaction was carried out at pH 7.4 in 100 mM (bi)carbonate buffer (BB) rather than in 100 mM phosphate buffer (PB) (Fig. 2C, central panel). But in (bi)carbonate buffer with 0.1 mM DTPA, we observed distinct protein nitrone adducts of monomers and aggregates from both proteins, but not of fragments (Fig. 2C, B). These results suggest that DTPA inhibits the Fenton-like chemistry triggered by copper released from the SOD1, which results in protein fragmentation. Once the released copper rebinds to specific residues on SOD1 and HSA, in the presence of excess H2O2 these residues can also act as additional sites for CO3 •− generation (see below). It is noteworthy that this amplifying copper-dependent radical chemistry occurs at protein Cu-binding sites other than the active site of SOD1.
SOD1-driven, copper-dependent radical damage
We further examined SOD1-driven, copper-triggered radical chemistry in proteins by reacting SOD1 and HSA with H2O2 in phosphate buffer without DTPA (Fig. 3). Under these conditions, HSA and SOD1 itself were targets of copper-mediated, site-specific fragmentation (Fig. 3A, left panel). H2O2-induced fragmentation of SOD1 at its active site was evidenced by its ~5 and ~10 kDa fragments [10, 12] and was not prevented by DTPA, although DTPA did strongly inhibit nitrone adduct formation of the protein (Fig. 3A, right panel). This result suggests that the formation of H2O2-induced, copper-bound oxidant at the SOD1 active site precedes release of copper. Released copper would then be rebound to HSA and other sites in SOD1 itself to react with excess H2O2 and induce free radical-mediated, site-specific fragmentation. To test this hypothesis, we measured H2O2-induced copper release from the bSOD1 active site by BCDS-assisted reduction of Cu2+ to Cu1+ as its red complex (Fig. 3B). Depending on the concentration of H2O2, fifteen minutes of reaction was enough to release a significant amount of copper, an amount that would be sufficient to produce protein radicals, as we previously found by immuno-spin trapping in the HSA/Cu2+/H2O2 system [31]. After two hours, 5 mM H2O2 released approximately one-half of the available copper at the active site.
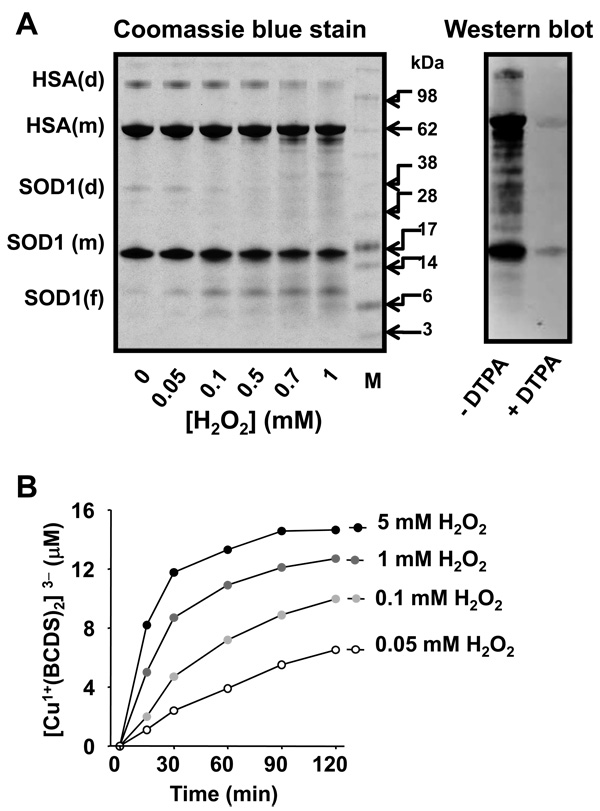
Figure 3. Copper-mediated oxidative modification to SOD1 itself and HSA.
A, left panel, Gel stained with Coomassie blue; 15 µM bSOD1, 7.5 µM HSA and different concentrations of H2O2 were reacted in 100 mM sodium phosphate buffer, pH 7.4, for 1 h at 37 °C. Right panel,Anti-DMPO Western blot analysis of a reaction mixture containing bSOD1 and HSA and incubated with 0.1 mM H2O2 and 100 mM DMPO in the same buffer as in left panel with or without DTPA. B, Copper released and measured as (Cu1+(BCDS)2)3− from bSOD1 when 15 µM bSOD1 was treated with different concentrations of H2O2 in 100 mM sodium phosphate buffer, pH 7.4.
To study the effect of phosphate, (bi)carbonate, and DTPA on oxidation of substrates too bulky to access the SOD1 active site, we examined the oxidation of guaiacol to tetraguaiacol (Fig. 4A). We did not observe guaiacol oxidation when SOD1 was reacted with H2O2 in argon-purged (to eliminate CO2) phosphate buffer with or without DTPA. The strongest oxidation of guaiacol was observed when the reaction was carried out in 100 mM (bi)carbonate buffer; this oxidation was inhibited by almost 30% with 0.1 mM DTPA and by over 50% with 100 mM phosphate. Phosphate may compete with (bi)carbonate for copper binding and also for access to the positively charged channel to the SOD1 active site. We obtained similar results when bSOD1 was replaced with hSOD1 (data not shown). Other peroxidase substrates such as ABTS and NADPH were used with similar results (data not shown). We also observed that H2O2-induced loss of copper from the SOD1 active site was enhanced by (bi)carbonate and inhibited by phosphate (Fig. 4B). Presumably, the oxidation of histidine residues at the SOD1 active site by CO3 •− is responsible for this copper release, which occurs without SOD1-active site fragmentation.
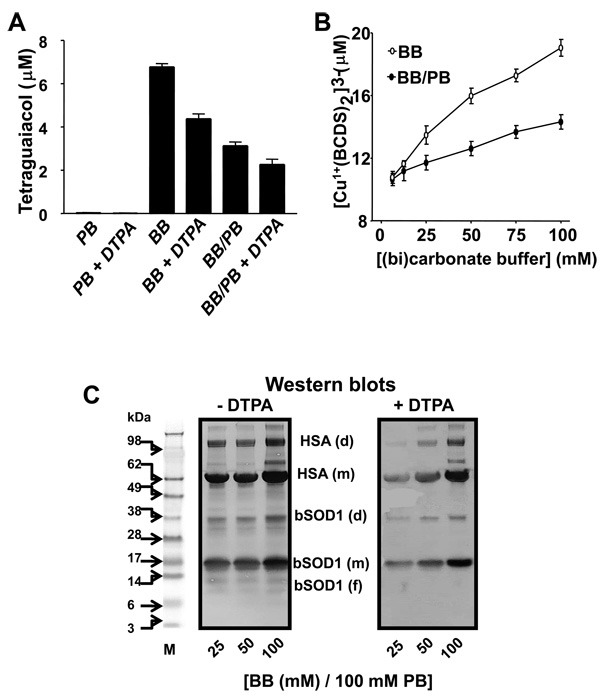
Figure 4. Effect of (bi)carbonate on SOD1-driven, copper- and carbonate radical anionmediated oxidations.
A, Guaiacol (0.5 mM) oxidation when 15 µM SOD1 was reacted with 1 mM H2O2 either in 100 mM PB, 100 mM BB, or 100 mM PB containing 100 mM BB (BB/PB), pH 7.4, with or without 0.1 mM DTPA. Tetraguaiacol production was determined after a 15-min incubation at 37 °C. B, Copper released from the bSOD1 active site when 15 µM bSOD1 was added to (bi)carbonate diluted either in distilled water (BB) or 100 mM phosphate buffer (BB/PB), pH 7.4, with 1 mM BCDS. The reaction was started with 0.1 mM H2O2, and the formation of (Cu1+(BCDS)2)3− was determined after 1 h as described in the Experimental section. C, Anti-DMPO Western blots produced when 15 µM bSOD1, 7.5 µM HSA, 100 mM DMPO and 0.1 mM H2O2 were incubated for 1 h at 37 °C in 100 mM phosphate buffer (PB) containing different amounts of (bi)carbonate buffer (BB). Solutions were without (left panel) or with (right panel) the copper chelator DTPA. M, molecular weight marker; (f), (m) and (d) indicate fragments, monomer and dimer, respectively. Graph shows results from a representative experiment.
With or without DTPA, (bi)carbonate was observed to enhance the production of H2O2-induced SOD1 and HSA nitrone adducts (Fig. 4C). However, protein radicals were higher in the absence of DTPA than in its presence. Products of copper-mediated, site-specific modification of SOD1 and HSA were observed when the reaction was carried out without DTPA, but they disappeared when DTPA was included (compare smears in Fig. 4C, left vs right panel).
Radical chemistry induced by SOD1-driven carbonate radical anion oxidations
Thus, (bi)carbonate enhances H2O2-induced loss of copper from the enzyme active site presumably by oxidizing active site histidine residues, and this copper then re-binds to other SOD1 sites or to target proteins such as HSA. The re-bound copper acts as additional sites for peroxymonocarbonate reduction to CO3 •− [31] (see also scheme 1), thus preventing site-specific fragmentation. Accordingly, in the following experiments we used a physiological concentration of (bi)carbonate (i.e., ~25 mM) in 100 mM sodium phosphate buffer with 0.1 mM DTPA in order to specifically study CO3 •− -triggered free radicals in SOD1 itself and HSA (Fig. 5A). Under these conditions, SOD1 and HSA radicals were observed as their monomer and dimer and required the addition of all components (Fig. 5A, right panel). This result confirms the proposal of Bonini at al. that the CO3 •− formed at the active site of SOD1 can diffuse out the channel to react with bovine serum albumin forming diverse radicals [4]. CO3 •− -triggered protein radicals increased with incubation time (data not shown). The oxidation profile of HSA (Fig. 5A) was independent of whether bSOD1 or hSOD1 was used, establishing that the tryptophan radical formed from hSOD1 has little role in HSA oxidation (see Fig. 6). In addition, SOD1-driven, CO3 •− -triggered oxidation of HSA, but not of SOD1 itself, was partially prevented by adding guaiacol to the reaction mixture (Fig. 5B) with concomitant oxidation to tetraguaiacol (data not shown). These results suggest that by scavenging CO3 •−, guaiacol protected HSA against SOD1-driven oxidation.
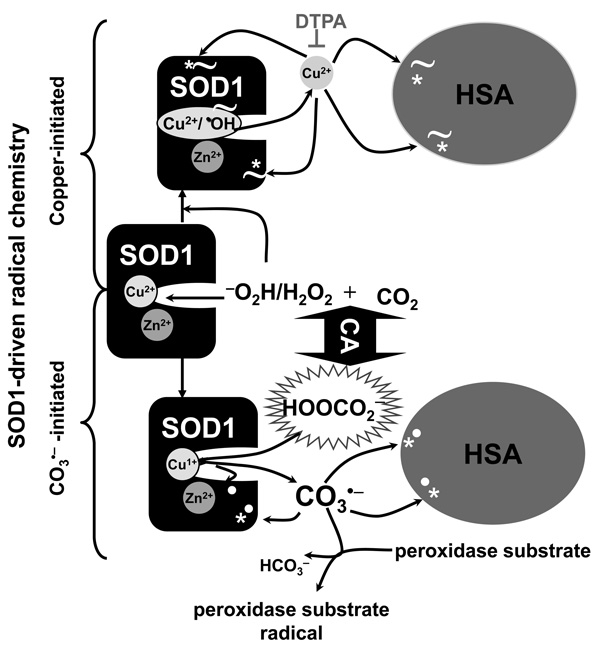
Scheme I. Mechanisms of H2O2-induced, SOD1-driven, copper- and carbonate radical anioninitiated protein radicals.
The reaction between H2O2 and SOD1 involves the formation of a strongly bound oxidant (indicated as Cu/•OH) at the enzyme active site (see reactions 1 and 2). H2O2-induced, SOD1-driven oxidations are: i) Copper-initiated: This oxidizing species oxidizes key histidine residues at the enzyme active site and releases Cu2+. Released Cu2+ is reduced to Cu1+ by excess H2O2. Cu1+ rebinds to proteins and redox-cycles with H2O2, producing site-specific fragmentation (indicated with the symbol ~). DTPA chelates Cu2+ and prevents its redox-cycling and binding to proteins, thus preventing copper-catalyzed site-specific fragmentation of proteins. ii) Carbonate radical anion (CO3•−)-initiated: H2O2 reacts with CO2 to produce another oxidizing species, the peroxymonocarbonate anion (HOOCO2 −). Peroxymonocarbonate anion is an adduct between the deprotonated form of H2O2, −O2H, and CO2 [37, 38]. Peroxymonocarbonate can be reduced to CO3 •− by metal centers [12, 31, 37, 43, 46]. Peroxymonocarbonate is a small anionic species that we propose can diffuse through the anionic channel to the enzyme active site and be reduced to CO3 •− by the product of reaction 1 (Cu1+-SOD1). As a result, reaction 2 does not occur and the bound oxidant formation is prevented. CO3 •− oxidizes amino acid side-chains (marked with bold white dots), does not appear to fragment proteins [31]. Although CO3 •− is highly reactive it is of course more diffusible than the copper-bound oxidant. CO3 •− oxidizes peroxidase substrates like guaiacol or proteins such as HSA. Thus, SOD1 drives CO3 •−-mediated oxidation that promotes its own and other proteins’ oxidation. Carbonic anhydrase (CA) enhances CO3 •−-mediated, protein-centered radicals, including that of CA, that can be trapped by the nitrone spin trap DMPO. DMPO traps radical sites produced by copper- and CO3 •−-mediated oxidations (marked with *). Moreover, CO3 •− oxidizes key residues for the activity of the enzyme and promotes an enhanced release of copper presumable by oxidizing the Cu2+-coordinating histidines.
Figure 5. SOD1-driven, carbonate radical anion-mediated oxidation of proteins.
A, Coomassie blue-stained gel (left) and Western blots (right) produced when reaction mixtures containing the components indicated in each lane and their final concentrations were the same as Fig. 4C, but reactions were performed in 25 mM (bi)carbonate added to 100 mM phosphate buffer containing 0.1 mM DTPA. Reactions were stopped after 1 h with 10 IU catalase. B, Western blot produced from reactions containing the same components as in A (last lane), but different concentrations of guaiacol. The reaction mixture was incubated for 1 h and stopped by adding 10 IU catalase. C, Western blot produced from reaction mixtures containing the same components as in A (last lane) at different concentrations of carbonic anhydrase (CA). Incubations were stopped after 1 h with 10 IU catalase. The analyses of the reaction mixtures by Coomassie blue stain and Western blots for the detection of protein nitrone adducts were carried out as described under Experimental. Data show results from representative experiments. M indicates molecular weight marker; (m), (d) and (t) indicate monomer, dimer and trimer, respectively.
Figure 6. Human Cu, Zn-superoxide dismutase (hSOD1, isolated from erythrocytes)-driven and bicarbonate-dependent protein radical formation.
A-left panel shows a protein staining of the reaction mixture containing 10 µM hSOD1, 100 mM DMPO and 100 µM H2O2 in 100 mM chelexed-phosphate buffer, pH 7.4 containing 100 µM DTPA (PB), and without or with 10 or 25 mM sodium bicarbonate (BB). After incubation for 1 h at 37 °C the reaction was stopped by adding catalase. The final pH of the reaction mixture was between 7.3–7.6. Central panel, same as the left panel, but the nitrone adducts where detected by Western blot with the anti-DMPO antibody. Right panel, Western blot of the reaction mixtures containing the same reagents and concentrations as in the left panel, but 7.5 µM HSA was included in the reaction mixture and the reactions performed in PB or in PB containing 25 mM BB (25 BB/PB), and different concentrations of H2O2 were added. Mr, indicates molecular weight marker (SeaBlue, Invitrogen). B, Western blot with the anti-DMPO antiserum of the reaction mixtures containing similar concentrations of reagents and performed as in A, except that the effect of different concentrations of BB was evaluated. Data are representative of three independent experiments.
Gaining further insights into the species oxidized by the (bi)carbonate-dependent species formed at the active site of SOD1, we observed that carbonic anhydrase/dehydratase (CA) enhanced H2O2-induced SOD1 and HSA nitrone adducts and also was a target for its own catalyzed reactions, producing CA nitrone adducts (Fig. 5C). Figure 6A shows a protein staining and a Western blot of hSOD1, isolated from human erythrocytes, reacted with H2O2 and DMPO in the absence of copper chemistry. Fig. 6A central panel, also shows the (bi)carbonate-dependent formation of a protein radical in the enzyme. The right panel in Fig. 6A shows that the formation of hSOD1 and HSA-centered radicals are dependent on (bi)carbonate and H2O2. Control experiments and dose-dependent effect of (bi)carbonate on hSOD1-driven protein radical production are shown in Fig. 6B. Human SOD1 isolated from transformed E. coli produced results similar to those of the human and bovine erythrocyte enzymes (data not shown). Once again, our results show the importance of a (bi)carbonate-dependent and diffusible species capable of generating DMPO-trappable radical sites in SOD1 itself and in any other protein in the reaction microenvironment when SOD1 reacts with H2O2 in the presence of DTPA.
Protein radical formed by the hSOD1-driven, carbonate radical anion-mediated oxidations in a mouse brain homogenate
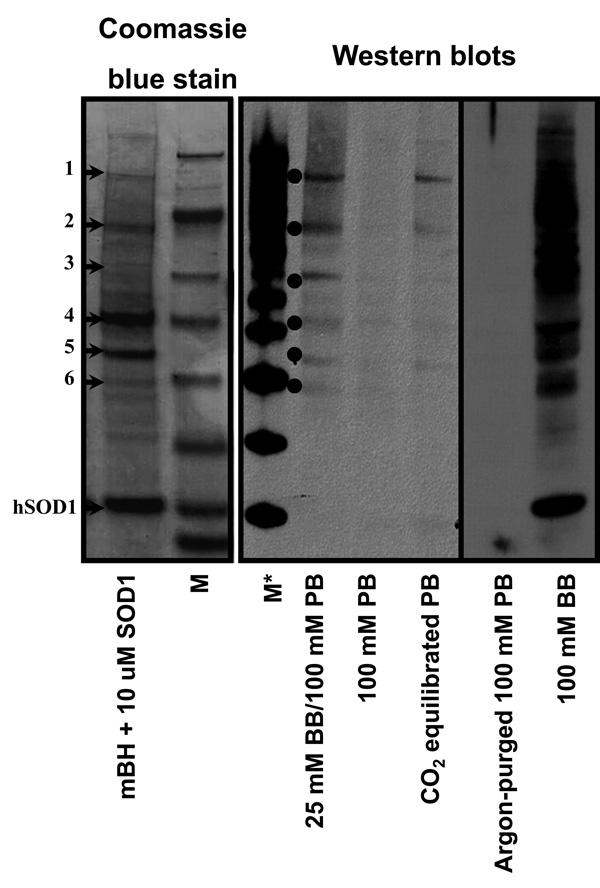
We have devised experimental conditions to study protein oxidation driven by the bovine and human SOD1/H2O2 systems that are important to understanding the mechanism of possible protein modifications in vivo. Here we used the SOD1/H2O2/DTPA/(bi)carbonate system to generate CO3 •− without copper-derived hydroxyl radical-like-induced oxidations. As a target protein mixture for CO3 •−, we used a mouse brain homogenate (mBH) (Fig. 7). We partially characterized the corresponding Coomassie blue stained gel bands using LC/ESI/MS/MS analyses. The Coomassie stained protein bands (corresponding to the anti-DMPO bands observed by Western blot) were excised from the gel, digested, and then subjected to LC/MS/MS analyses for protein identification.
Figure 7. hSOD1-driven, carbonate radical-triggered protein oxidations in mouse brain homogenate (mBH).
Coomassie blue stain and Western blot showing the pattern of protein oxidations induced by the hSOD1/H2O2/DTPA (20 µM)/(bi)carbonate system in mBH. Left panel shows a Coomassie blue stain of a mixture of 20 µM hSOD1 with 500 µg/ml of mBH in 100 mM phosphate buffer (PB). The numbers on the left indicate LC/MS/MS assignments (see Table 1) based on positive bands detected by the anti-DMPO Western blot. The central panel shows an anti-DMPO Western blot of a reaction mixture containing 2 µM hSOD1, 500 µg/ml mBH, 50 µM DTPA, in either 25 mM sodium bicarbonate (BB), 100 mM of sodium phosphate buffer (PB) or PB equilibrated with CO2 (gas). Right panel shows a Western blot of a reaction mixture between 10 µM bSOD1, 500 µg/ml mBH, and 50 µM DTPA in argon-purged (to eliminate dissolved CO2) PB or in 100 mM BB. For Western blot analysis 50 mM DMPO was added. In all cases the reaction was performed at pH 7.4, started by adding 100 µM H2O2, and stopped by extensive dialysis against 50 mM ammonium bicarbonate, pH 8.0, to eliminate excess reagents for MS analysis [30]. M indicates Sea Blue® Plus2 Prestained molecular marker; M* indicates MagicMark™ Western Standard. The stains shown are representative from three separate experiments.
Following LC/MS/MS analyses, the raw data were extracted and searched against the NCBI nonredundant database (species: mouse only) (Table 2). Only those proteins with a summed MS/MS score greater than 30 and at least two distinct tryptic peptides are listed. The immuno-spin trapping analysis detected 6 distinct protein bands. As shown in Table 2, many proteins were identified in each band of the Western blot. One or more of the identified proteins in each band may be a target of CO3 •−-triggered modification, leading to formation of protein radicals that are trapped by DMPO. These protein radicals can be assigned to mitochondrial proteins, heat shock proteins, and cell cytoskeleton proteins (Table 2). The above results exhibit the utility of the combined approaches of immuno-spin trapping and mass spectrometry to detect potential targets for oxidation by CO3 •− in a complex system such as mouse brain homogenate.
Table 2.
LC/MS/MS Protein Identifications1 from Coomasie Blue Stained Gel Bands (Figure 7) which Correspond to anti-DMPO Positive Bands in Mouse Brain Homogenate Exposed to hSOD1-driven, Carbonate Radical Anion-initiated Oxidations
| Band2 | Protein | Number of Unique Peptides3 | Protein Score4 | Percent Sequence Coverage | Accession Number5 | Entry Name6 |
|---|---|---|---|---|---|---|
| Band 1 | 1 | 17 | 274.34 | 40 | 13242237 | heat shock 70kD protein 8 |
| 2 | 9 | 104.82 | 18 | 6754256 | heat shock protein, A | |
| 3 | 6 | 86.31 | 11 | 20837174 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | |
| 4 | 5 | 46.37 | 16 | 8567410 | synapsin II | |
| 5 | 5 | 43.03 | 8 | 16877778 | neurochondrin | |
| 6 | 3 | 39.01 | 8 | 29124477 | immunoglobulin superfamily, member 8 | |
| 7 | 4 | 38.67 | 6 | 1339938 | glycerol-3-phosphate dehydrogenase | |
| 8 | 3 | 37.18 | 7 | 6678359 | transketolase | |
| Band 2 | 1 | 16 | 276.99 | 42 | 23272966 | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| 2 | 16 | 263.37 | 44 | 13542680 | tubulin, beta, 5 | |
| 3 | 13 | 213.22 | 41 | 135412 | tubulin, alpha 6 | |
| 4 | 7 | 97.29 | 24 | 27370474 | RIKEN cDNA 9630038C02 | |
| 5 | 3 | 58.73 | 9 | 90334 | Ca2+/calmodulin-dependent protein kinase (EC 2.7.1.123) II alpha chain | |
| 6 | 2 | 34.19 | 6 | 26324430 | unnamed protein product | |
| Band 3 | 1 | 11 | 165.47 | 39 | 2118269 | zebrin II |
| 2 | 9 | 154.61 | 35 | 7548322 | aldolase A | |
| 3 | 8 | 147.9 | 31 | 12852054 | creatine kinase, brain | |
| 4 | 9 | 146.56 | 36 | 49868 | put. beta-actin (aa 27–375) | |
| 5 | 9 | 117.73 | 30 | 90313 | aspartate transaminase (EC 2.6.1.1), cytosolic | |
| 6 | 5 | 74.47 | 18 | 20846887 | aspartate aminotransferase precursor | |
| 7 | 4 | 68.36 | 11 | 6679261 | pyruvate dehydrogenase E1 alpha 1 | |
| 8 | 4 | 62.14 | 14 | 21450129 | acetyl-Coenzyme A acetyltransferase 1 precursor | |
| 9 | 3 | 51.62 | 13 | 7305305 | N-myc downstream regulated 2 | |
| 10 | 3 | 49.87 | 11 | 17225449 | SH3 domain protein 2A | |
| 11 | 4 | 49.47 | 14 | 29126784 | Similar to Actin-related protein 14D | |
| 12 | 3 | 38.45 | 11 | 6685763 | Septin 5 (Peanut-like protein 1) (Cell division control related protein 1) (CDCREL-1) | |
| Band 4 | 1 | 5 | 89.12 | 11 | 20857396 | glyceraldehyde-3-phosphate dehydrogenase |
| 2 | 6 | 71.09 | 21 | 18250284 | isocitrate dehydrogenase 3 (NAD+) alpha | |
| 3 | 3 | 40.24 | 9 | 7304909 | ATPase, H+ transporting, V0 subunit D isoform 1 | |
| 4 | 3 | 38.84 | 15 | 26348803 | pyridoxal (pyridoxine, vitamin B6) kinase | |
| 5 | 2 | 35.87 | 8 | 20878370 | similar to glyceraldehyde-3-phosphate dehydrogenase | |
| Band 5 | 1 | 8 | 121.45 | 39 | 10720404 | Voltage-dependent anion-selective channel protein 1 (VDAC-1) (mVDAC1) (mVDAC5) (Outer mitochondrial membrane protein porin 1) (Plasmalemmal porin) |
| 2 | 6 | 107.76 | 24 | 319830 | malate dehydrogenase (EC 1.1.1.37) precursor, mitochondrial | |
| 3 | 5 | 70.52 | 19 | 18152793 | pyruvate dehydrogenase (lipoamide) beta | |
| 4 | 5 | 67.7 | 17 | 6678674 | lactate dehydrogenase 2, B chain; lactate dehydrogenase-B | |
| 5 | 4 | 65.28 | 16 | 6678918 | malate dehydrogenase, soluble | |
| 6 | 4 | 48.78 | 8 | 20888059 | glyceraldehyde-3-phosphate dehydrogenase | |
| 7 | 3 | 40.95 | 11 | 6755965 | voltage-dependent anion channel 2 | |
| 8 | 3 | 39.7 | 15 | 114980 | Beta-soluble NSF attachment protein (SNAP-beta) (N-ethylmaleimide-sensitive factor attachment protein, beta) (Brain protein I47) | |
| 9 | 3 | 37.22 | 12 | 21450053 | RIKEN cDNA 0910001A06 | |
| Band 6 | 1 | 6 | 89.79 | 25 | 6756041 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| 2 | 5 | 56.2 | 31 | 21312520 | quininoid dihydropteridine reductase; DNA segment, Chr 5, ERATO Doi 371, expressed | |
| 3 | 3 | 41.62 | 20 | 21759130 | Rho GDP-dissociation inhibitor 1 (Rho GDI 1) (Rho-GDI alpha) (GDI-1) | |
| 4 | 3 | 38.32 | 14 | 23396786 | NADH-ubiquinone oxidoreductase 30 kDa subunit, mitochondrial precursor (Complex I-30KD) (CI-30KD) | |
| 5 | 2 | 34.41 | 12 | 20909043 | similar to brain-specific protein p25 alpha [Homo sapiens] | |
| 6 | 2 | 33.92 | 12 | 12846508 | triosephosphate isomerase | |
| 7 | 3 | 33.31 | 20 | 20823772 | similar to phophoglycerate mutase (EC 5.4.2.1) B chain - rat |
Identifications based on a unique score of at least 30 and at least 2 unique peptides.
Band number from coomassie stained gel shown in Figure 7
Number of unique tryptic peptides observed by LC/MS/MS
Score based on Spectrum Mill scoring algorithm
Accession number is the gi number from a search of the NCBI nonredundant database limited to mouse species only
Entry name listed is the first name listed within the family of proteins
DISCUSSION
The study of the mechanism of protein radical formation by the wild type SOD1/H2O2 system is important to the understanding of tissue damage in neuroinflammatory pathologies such as fALS. In this report we investigated the H2O2-induced, wild type bovine and human SOD1-driven production of protein radicals in SOD1 itself and the model protein targets HSA and mBH under experimental conditions that clearly separated copper-from CO3 •−-initiated radical chemistry (Scheme 1).
In some studies of the SOD1 and H2O2 reaction, the role of copper- and CO3 •− -triggered oxidations is difficult to distinguish (Scheme I). In the absence of copper chelators [4], Bonini et. al. were the first to detect bovine serum albumin radicals formed by CO3 •− -triggered radical chemistry following the H2O2-induced peroxidative activity of SOD1 in the presence of (bi)carbonate; however, SOD1 radicals were not detected [4, 19]. According to our results, this system would involve the reduction of peroxymonocarbonate to CO3 •− by copper-bound oxidants located at the SOD1 active site and at many other sites in both SOD1 and bovine serum albumin. In the absence of DTPA, the SOD1/HSA/H2O2 reaction in (bi)carbonate buffer produced greater oxidative damage, mainly to side chain residues (Table 1), than that in phosphate buffer, as demonstrated by increases in DMPO-trappable SOD1 radicals and SOD1 inactivation (Figs. 1A and 1B). In the absence of DTPA, reduction of peroxymonocarbonate and formation of CO3 •− will occur at many sites outside the enzyme active site where copper is re-bound. In agreement with these observations, we have previously found that (bi)carbonate protected against site-specific fragmentation, but enhanced side-chain radical formation in HSA [31].
DTPA prevents the re-binding of copper to SOD1 or HSA but, as previously reported, does not prevent oxidative damage to SOD1 at the enzyme active site [12]. In addition, the oxidants formed at the SOD1 active site (reaction 1 and 2) are generated in systems whether or not they contain DTPA due to the fact that this chelating agent does not have access to the SOD1 active site [8, 13]. This can explain the failure of HSA, guaiacol, or ABTS, even at high concentrations, to protect against SOD1’s active site fragmentation (~10 and ~5 kDa fragments) and to completely block SOD1 inactivation.
The copper-bound oxidant (a hydroxyl radical-like species) at the SOD1 active site may well inactivate SOD1 at a different site than CO3 •− does. CO3 •− is the species responsible for the oxidation of peroxidase substrates [11, 13, 21] and for oxidation of a key tryptophan residue and formation of a Trp- Trp cross-linkage in human SOD1 [36]. (Bi)carbonate was required for DMPO-trappable protein radical formation and caused protein aggregation of SOD1, possibly due to the formation of side chain radicals (Table 1). In any case, in our results we have found very little difference between hSOD1 (recombinant or erythrocytic) and bSOD1, although bSOD1 does not contain any Trp. Compare Fig. 1B and Fig. 6A.
Previously, we have shown that carbonic anhydrase enhances the (bi)carbonate-enhanced, H2O2-induced, copper-catalyzed oxidation of a peroxidase substrate, suggesting the involvement of CO2 in the formation of CO3 •− [31]. Indeed, in the present study (Fig. 5C) we show that when carbonic anhydrase is included in the SOD1/H2O2/HSA/DTPA system with physiological concentrations of (bi)carbonate, it produces a dramatic concentration-dependent increase in (bi)carbonate-enhanced, H2O2-induced , SOD1-driven protein radicals. Our results indicate that under our experimental conditions, peroxidase substrates such as guaiacol, ABTS, NADPH, and proteins protect SOD1 against inactivation by 0.1 mM H2O2 in 100 mM (bi)carbonate with 0.1 mM DTPA because they scavenge CO3 •−. Our results also suggest that the diffusible CO3 •− can oxidize critical residues for SOD1 activity or for the transport of the superoxide radical anion through the enzyme cationic channel. In fact, at nonphysiological concentrations above 25 mM, (bi)carbonate increases the rate of inactivation of SOD1 (Fig. 1A) [17]. The peroxidase substrate guaiacol competes with HSA for CO3 •−; thus, guaiacol acts as an alternative target to HSA with a consequent decrease in HSA-centered radical formation (Fig. 5B), but does not completely prevent SOD1 inactivation (data not shown). This could be related to the proximity of SOD1 to CO3 •− as it is formed [15].
In order to explain the pathways of CO3 •−-initiated radical chemistry and the apparent discrepancy between protein radicals and dismutase activity, we propose a novel mechanism involving CO2 based on the enhancing effect of carbonic anhydrase on protein radical formation (Fig. 5C). Indeed, peroxymonocarbonate , which has been proposed to be an important physiological oxidant [4], oxidizes biological targets by a two-electron mechanism, and its oxidizing power is enhanced when reduced to CO3 •− by catalysis with metal centers [4, 31, 37] (Scheme 1). For the formation of CO3 •−, we propose the formation of a free peroxymonocarbonate anion intermediate (HOOCO2 −, see Scheme I); CO2 and the hydrogen peroxide (−OOH/H2O2) are in equilibrium with HOOCO2 − with a Keq = 0.33 M−1 (reaction 3) [19, 38–40]. The addition of the deprotonized water (−OH) to carbon dioxide is catalyzed by carbonic anhydrase. Analogously, we make the novel proposal that addition of deprotonized hydrogen peroxide (−OOH) to CO2 is likewise catalyzed by carbonic anhydrase (reaction 3)
| Reaction 3 |
Accordingly, the equilibrium shown in reaction 3 most likely proceeds via the intermediacy of CO2, where HOOCO2 − is essentially a CO2 adduct of the −OOH [38]. The reduction of HOOCO2 − by H2O2-induced Cu1+-SOD1 (reaction 1) forms CO3 •− directly without any need for the formation of the high-energy, bound oxidant at the enzyme’s active site (reaction 2). It has recently been suggested that the equilibrium constant for Reaction 3 is increased in the presence of biological targets such as proteins, lipids, and carbonic anhydrase mimetics, which may alter the equilibrium of gaseous CO2/dissolved CO2/HCO3 − [19]. Our proposal of a direct role for carbonic anhydrase in peroxymonocarbonate anion formation (reaction 3) is supported by the fact that the established mechanism of carbonic anhydrase for the formation of (bi)carbonate (reaction 4) [41] is very similar for peroxymonocarbonate [38] except the base of hydrogen peroxide (−OOH) nucleophilically attacks CO2 instead of the base of water (−OH) where the Zn2+ of carbonic anhydrase catalyzes the desprotonation of HOOH and H2O, respectively.
| Reaction 4 |
The ease of diffusion of peroxymonocarbonate anion through the anion channel of SOD1 and the substantially lower oxidation potential of CO3 •− (E° ~ 1.78 V) relative to that of the strong oxidant at the active site (E° ~ 2.31 V) should be reflected in a much more kinetically favored formation of protein radicals and would explain the protection afforded by (bi)carbonate against the fragmentation at the SOD1 active site induced by H2O2, which probably is initiated by the abstraction of the hydrogen of the amide bond [42] linking two amino acids near the active site copper of SOD1. In addition, CO3 •− diffuses and produces additional DMPO-trappable radical sites in the side chains of SOD1 itself and HSA (Table 1), but it does not produce protein fragmentation [31]. Our results suggest that CO3 •− can produce protein radicals or oxidize proteins at side chains [43], but it cannot induce backbone cleavages and further protein fragmentation as the copper-bound oxidant does [12, 31]. The enhanced inhibition of the enzyme by (bi)carbonate may be the consequence of the oxidation of residues by the diffusible CO3 •−, which may be important for driving superoxide radical anion through the SOD1 anion channel to its active site.
Our model of hSOD1-driven, CO3 •− -initiated protein oxidation is supported by our finding that the SOD1/H2O2/DTPA/(bi)carbonate system, in which Cu1+ reduces peroxymonocarbonate to form CO3 •−, oxidizes a set of well distinguished bands of proteins in mouse brain homogenate (Fig. 7). Immuno-spin trapping in combination with mass spectrometry technology [30, 44] has allowed the identification of potential targets of CO3 •− -mediated oxidations in mouse brain homogenate. The identification of the protein radicals and their subcellular location using murine models of fALS[45] may be important to understanding the molecular mechanisms of fALS.
We have established experimental conditions that separate copper-from CO3 •−-initiated protein oxidations in the SOD1/H2O2 system, which may help us to develop an experimental model to characterize biological targets in fALS. The identification of these proteins may be a starting point for investigations aimed at identifying critical proteins in neurons overexpressing the hSOD1 mutant proteins that are targets for oxidation and, thus, may have a critical role in this neuroinflammatory disease.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIEHS/NIH. DCR acknowledges the National Institute of Environmental Health Sciences support (Award# R00ES015415) and the start-up funds from the Presbyterian Health Foundation. We thank Ms. Mary Mason and Dr. Ann Motten for helping in the editing of this manuscript.
Footnotes
The abbreviations used are: ABTS, 2,2′-azino-bis-[3-ethylbenzothiazoline]-6-sulfonic acid; CA, carbonic anhydrase; CO3 • −, carbonate radical anion; DMPO, 5,5-dimethyl-1-pyrroline N-oxide; DTPA, diethylenetriamine-pentaacetic acid; ELISA, enzyme-linked immuno-sorbent assay; ESR, electron spin resonance; fALS, familial amyotrophic lateral sclerosis; HSA, human serum albumin; H2O2, hydrogen peroxide; SOD1, Cu,Zn-superoxide dismutase.
REFERENCES
- 1.McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968–1988) Free Radic. Biol. Med. 1988;5:363–369. doi: 10.1016/0891-5849(88)90109-8. [DOI] [PubMed] [Google Scholar]
- 2.Fridovich I. The trail to superoxide dismutase. Protein Sci. 1998;7:2688–2690. doi: 10.1002/pro.5560071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Andrekopoulos C, Joseph J, Chandran K, Karoui H, Crow JP, Kalyanaraman B. Bicarbonate-dependent peroxidase activity of human Cu, Zn-superoxide dismutase induces covalent aggregation of protein- intermediacy of tryptophan-derived oxidation products. J. Biol. Chem. 2003;278:24078–24089. doi: 10.1074/jbc.M302051200. [DOI] [PubMed] [Google Scholar]
- 4.Bonini MG, Fernandes DC, Augusto O. Albumin oxidation to diverse radicals by the peroxidase activity of Cu,Zn-superoxide dismutase in the presence of bicarbonate or nitrite: Diffusible radicals produce cysteinyl and solvent-exposed and -unexposed tyrosyl radicals. Biochemistry. 2004;43:344–351. doi: 10.1021/bi035606p. [DOI] [PubMed] [Google Scholar]
- 5.Valentine JS, Doucette PA, Potter SZ. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Ann. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 6.Hodgson EK, Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: inactivation of the enzyme. Biochemistry. 1975;14:5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- 7.Liochev SI, Fridovich I. Bicarbonate-enhanced peroxidase activity of Cu,Zn SOD: is the distal oxidant bound or diffusible? Arch. Biochem. Biophys. 2004;421:255–259. doi: 10.1016/j.abb.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Uchida K, Kawakishi S. Identification of oxidized histidine generated at the active site of Cu,Zn-superoxide dismutase exposed to H2O2. Selective generation of 2-oxo-histidine at the histidine 118. J. Biol. Chem. 1994;269:2405–2410. [PubMed] [Google Scholar]
- 9.Kurahashi T, Miyazaki A, Suwan S, Isobe M. Extensive investigations on oxidized amino acid residues in H(2)O(2)-treated Cu,Zn-SOD protein with LC-ESI-Q-TOF-MS, MS/MS for the determination of the copper-binding site. J. Am. Chem. Soc. 2001;123:9268–9278. doi: 10.1021/ja015953r. [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Akaike T, Kohno M, Ando M, Maeda H. Hydroxyl radical production by H2O2 plus Cu,Zn-superoxide dismutase reflects the activity of free copper released from the oxidatively damaged enzyme. J. Biol. Chem. 1992;267:25371–25377. [PubMed] [Google Scholar]
- 11.Goss SPA, Singh RJ, Kalyanaraman B. Bicarbonate enhances the peroxidase activity of Cu,Zn-superoxide dismutase. Role of carbonate anion radical. J. Biol. Chem. 1999;274:28233–28239. doi: 10.1074/jbc.274.40.28233. [DOI] [PubMed] [Google Scholar]
- 12.Ramirez DC, Gomez-Mejiba SE, Mason RP. Mechanism of hydrogen peroxide-induced Cu,Zn-superoxide dismutase-centered radical formation as explored by immuno-spin trapping: The role of copper- and carbonate radical anion-mediated oxidations. Free Radic Biol. Med. 2005;38:201–214. doi: 10.1016/j.freeradbiomed.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Sankarapandi S, Zweier JL. Evidence against the generation of free hydroxyl radicals from the interaction of copper,zinc-superoxide dismutase and hydrogen peroxide. J. Biol. Chem. 1999;274:34576–34583. doi: 10.1074/jbc.274.49.34576. [DOI] [PubMed] [Google Scholar]
- 14.Liochev SI, Fridovich I. CO2 enhances peroxidase activity of SOD1: The effect of pH. Free Radic. Biol. Med. 2004;36:1444–1447. doi: 10.1016/j.freeradbiomed.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Liochev SI, Fridovich I. The role of CO2 in metal-catalyzed peoxidations. J. Inorg. Biochem. 2006;100:694–696. doi: 10.1016/j.jinorgbio.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 16.Sankarapandi S, Zweier JL. Bicarbonate is required for the peroxidase function of Cu, Zn-superoxide dismutase at physiological pH. J. Biol. Chem. 1999;274:1226–1232. doi: 10.1074/jbc.274.3.1226. [DOI] [PubMed] [Google Scholar]
- 17.Elam JS, Malek K, Rodriguez JA, Doucette PA, Taylor AB, Hayward LJ, Cabelli DE, Valentine JS, Hart PJ. An alternative mechanism of bicarbonate-mediated peroxidation by copper-zinc superoxide dismutase. J. Biol. Chem. 2003;278:21032–21039. doi: 10.1074/jbc.M300484200. [DOI] [PubMed] [Google Scholar]
- 18.Liochev SI, Fridovich I. CO2, not HCO3−, facilitates oxidations by Cu,Zn superoxide dismutase plus H2O2. Proc. Natl. Acad. Sci. U. S. A. 2004;101:743–744. doi: 10.1073/pnas.0307635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medinas DB, Cerchiaro G, Trindade D, Augusto O. The carbonate radical and related oxidants derived from bicarbonate buffer. I.U.B.M.B. Life. 2007;59:255–262. doi: 10.1080/15216540701230511. [DOI] [PubMed] [Google Scholar]
- 20.Liochev SI, Fridovich I. On the role of bicarbonate in peroxidations catalyzed by Cu,Zn superoxide dismutase. Free Radic. Biol. Med. 1999;27:1444–1447. doi: 10.1016/s0891-5849(99)00190-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Joseph J, Felix C, Kalyanaraman B. Bicarbonate enhances the hydroxylation, nitration, and peroxidation reactions catalyzed by copper, zinc superoxide dismutase. Intermediacy of carbonate anion radical. J. Biol. Chem. 2000;275:14038–14045. doi: 10.1074/jbc.275.19.14038. [DOI] [PubMed] [Google Scholar]
- 22.Alvarez MN, Peluffo G, Folkes L, Wardman P, Radi R. Reaction of the carbonate radical with the spin-trap 5,5-dimethyl-1-pyrroline-N-oxide in chemical and cellular systems: pulse radiolysis, electron paramagnetic resonance, and kinetic-competition studies. Free Radic. Biol. Med. 2007;43:1523–1533. doi: 10.1016/j.freeradbiomed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Liu D, Wen J, Liu J, Li L. The roles of free radicals in amyotrophic lateral sclerosis: reactive oxygen species and elevated oxidation of protein, DNA, and membrane phospholipids. FASEB J. 1999;13:2318–2328. doi: 10.1096/fasebj.13.15.2318. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping analyses of DNA radicals. Nat. Protoc. 2007;2:512–522. doi: 10.1038/nprot.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic. Biol. Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez DC, Chen YR, Mason RP. Immunochemical detection of hemoglobin-derived radicals formed by reaction with hydrogen peroxide: involvement of a protein-tyrosyl radical. Free Radic. Biol. Med. 2003;34:830–839. doi: 10.1016/s0891-5849(02)01437-5. [DOI] [PubMed] [Google Scholar]
- 27.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radic. Biol. Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez DC, Gomez Mejiba SE, Mason RP. Immuno-spin trapping of DNA radicals. Nat. Meth. 2006;3:123–127. doi: 10.1038/nmeth852. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez DC, Mason RP. Immuno-spin trapping: detection of protein-centered radicals. In: Costa LG, Maines MD, Reed DJ, Sassa S, Sipes IG, editors. Curr. Protoc. Toxicol. Hoboken, New Jersey, USA: John Wiley & Sons; 2005. pp. 17.17.11–17.17.16. [DOI] [PubMed] [Google Scholar]
- 30.Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. Identification of free radicals on hemoglobin from its self-peroxidation using mass spectrometry and immuno-spin trapping. J. Biol. Chem. 2004;279:11600–11607. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez DC, Gomez-Mejiba SE, Mason RP. Copper-catalyzed protein oxidation and its modulation by carbon dioxide. Enhancement of protein radicals in cells. J. Biol. Chem. 2005;280:27402–27411. doi: 10.1074/jbc.M504241200. [DOI] [PubMed] [Google Scholar]
- 32.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 33.Gabaldon M. Oxidation of cysteine and homocysteine by bovine albumin. Arch. Biochem. Biophys. 2004;431:178–188. doi: 10.1016/j.abb.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 34.Ivanov AI, Parkinson JA, Cossins E, Woodrow J, Sadler PJ. Bathocuproine-assisted reduction of copper(II) by human albumin. J. Biol. Inorg. Chem. 2000;5:102–109. doi: 10.1007/s007750050013. [DOI] [PubMed] [Google Scholar]
- 35.Cabelli DE, Allen D, Bielski BH, Holcman J. The interaction between Cu(I) superoxide dismutase and hydrogen peroxide. J. Biol. Chem. 1989;264:9967–9971. [PubMed] [Google Scholar]
- 36.Zhang H, Andrekopoulos C, Joseph J, Crow J, Kalyanaraman B. The carbonate radical anion-induced covalent aggregation of human copper, zinc superoxide dismutase, and alpha-synuclein: intermediacy of tryptophan- and tyrosine-derived oxidation products. Free Radic. Biol. Med. 2004;36:1355–1365. doi: 10.1016/j.freeradbiomed.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 37.Bonini MG, Miyamoto S, Di Mascio P, Augusto O. Production of carbonate radical anion during xanthine oxidase turnover in the presence of bicarbonate. J. Biol. Chem. 2004;279:51836–51843. doi: 10.1074/jbc.M406929200. [DOI] [PubMed] [Google Scholar]
- 38.Yao H, Richardson DE. Bicarbonate sufoxidants: Micellar oxidations of aryl sulfides with bicarbonate-activated hydrogen peroxide. J. Am. Chem. Soc. 2003;125:6211–6221. doi: 10.1021/ja0274756. [DOI] [PubMed] [Google Scholar]
- 39.Richardson DE, Regino CAS, Yao H, Johnson JV. Methionine oxidation by peroxymonocarbonate, a reactive oxygen species formed from CO2/bicarbonate and hydrogen peroxide. Free Radic. Biol. Med. 2003;35:1538–1550. doi: 10.1016/j.freeradbiomed.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Richardson DE, Yao H, Frank KM, Bennett DA. Equilibria, kinetics, and mechanism in the bicarbonate activation of hydrogen peroxide: oxidation of sulfides by peroxymonocarbonate. J. Am. Chem. Soc. 2000;122:1729–1739. [Google Scholar]
- 41.Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 42.Hawkins CL, Davies MJ. Generation and propagation of radical reactions on proteins. Biochim. Biophys. Acta. 2001;1504:196–219. doi: 10.1016/s0005-2728(00)00252-8. [DOI] [PubMed] [Google Scholar]
- 43.Augusto O, Bonini MG, Amanso AM, Linares E, Santos CCX, De Menezes SL. Nitrogen dioxide and carbonate radical anion: two emerging radicals in biology. Free Radic. Biol. Med. 2002;32:841–859. doi: 10.1016/s0891-5849(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 44.Deterding LJ, Bhattacharjee S, Ramirez DC, Mason RP, Tomer KB. Top-down and bottom-up mass spectrometric characterization of human myoglobin-centered free radicals induced by oxidative damage. Anal. Chem. 2007;79:6236–6248. doi: 10.1021/ac070935z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cassina P, Cassina A, Pehar M, Castellanos R, Gandelman M, de Leon A, Robinson KM, Mason RP, Beckman JS, Barbeito L, Radi R. Mitochondrial dysfunction in SOD1G93A-bearing astrocytes promotes motor neuron degeneration: prevention by mitochondrial-targeted antioxidants. J. Neurosci. 2008;28:4115–4122. doi: 10.1523/JNEUROSCI.5308-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medinas DB, Cerchiaro G, Trindale DF, Augusto O. The carbonate radical and related oxidants derived from bicarbonate buffer. I.U.B.M.B. Life. 2007;59:1–8. doi: 10.1080/15216540701230511. [DOI] [PubMed] [Google Scholar]