Abstract
A comprehensive understanding of the brain requires analysis of individual neurons. Here we describe twin-spot MARCM for high-resolution lineage tracing by independent labeling of paired sister clones. We determine patterns of neurogenesis and the influences of lineage on neuron-type specification. Notably, neural progenitors may yield intermediate precursors that make one, two, or more neurons. Furthermore, neurons acquire stereotyped projections according to their temporal position within various brain sublineages. Twin-spot MARCM also permits birth dating of mutant clones, enabling the detection of a single temporal fate that requires chinmo in a sublineage of six Drosophila central complex neurons. In sum, twin-spot MARCM can reveal the developmental origins of neurons and the mechanisms that underlie cell fate.
A mature brain is, in large part, neurons connected to other neurons in complex patterns. The complexity of the connections depends on diverse intricate neuron morphologies and mapping the brain requires identification of individual neurons based on stereotypical morphologies. To understand how connections form, we must elucidate mechanisms by which neurons arrive at these morphologies.
Neurogenesis in Drosophila involves the sequential derivation of distinct neurons from common progenitors, called neuroblasts (Nbs) 1-4. Clonal analyses reveal that specific neurons are the descendants of specific progenitors and that most progenitors make multiple neuron types in an invariant sequence 5-7. Growing evidence shows a similar process of neuronal diversification in other organisms 8, 9. These results suggest that a neuron's type depends on from which progenitor it was generated and its time of birth. An effective strategy for identifying neuron types and studying development is to label individual neurons according to their lineage of origin and birth order.
MARCM (Mosaic Analysis with a Repressible Cell Marker), a genetic mosaic system, highlights post-mitotic neurons based on lineage and neuronal birth timing 10. With MARCM, neurons of the same lineage can be labeled as a multi-cellular Nb clone via recombination in a dividing Nb. MARCM can also label neurons as two-cell or single-cell clones via recombination in a dividing Nb or ganglion mother cell (GMC), respectively, which allows analysis of morphology and birth timing. Moreover, we can alter genotype specifically in targeted neurons, permitting study of lethal mutations within otherwise unperturbed brains. For example, in the Drosophila olfactory learning and memory center, the mushroom bodies (MBs), analysis of single-cell/two-cell clones reveals the presence of four types of MB neurons that can be distinguished based on their axon projections 5, 11. Notably, the multiple types of MB neurons derive from common progenitors in an invariant sequence. Although all types co-exist in full-sized Nb clones, the neurons born at different developmental stages, shown by inducing clones at different developmental times, develop into different types of MB neurons. Genetic mosaic screens for development-defective MB clones led to the identification of Chinmo, a BTB-zinc finger nuclear protein, which acts autonomously in newborn MB neurons to specify their birth-timing-dependent cell fates 12. Thus, cellular and molecular evidences indicate the specification of MB neuron-types based on the birth order of neurons within the MB lineages.
Despite the power of MARCM, it remains challenging to identify individual neurons in the Drosophila brain where most non-MB neuron types are yielded in a very fast tempo 6, 7. MARCM labels one of two daughter cells, owing to the loss of the chromosome arm that carries a repressor gene (Fig. 1b). Not seeing clones in pairs prevents high-resolution lineage analysis. By labeling both cells (and all their progeny) in different colors, we would gain information essential for comprehensive lineage analysis. We therefore sought to revolutionize MARCM for visualization of twin spots in mosaic brains. Our new system reveals a complete proliferation pattern for any neural progenitor, providing crucial information concerning the identity of mitotic clones.
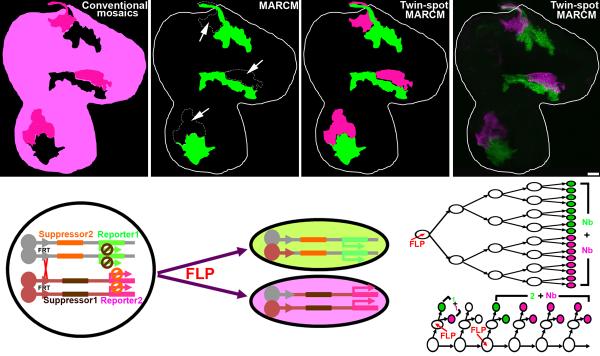
Figure 1. Evolution of mosaic techniques and the twin-spot MARCM design.
Comparison of expected clone patterns from various mitotic recombination-based mosaic systems [conventional mosaics (a), MARCM (b), and twin-spot MARCM (c)] to actual twin-spot MARCM clones in a 3rd instar wing disc (d). (a) In conventional mosaic analysis, clones of interest with the homozygous genotype are invisible (shown in black). The reference clones (dark magenta) are intermingled with the background (light magenta). (b) In MARCM, clones of interest with the homozygous genotype (shown in green) stand out from the unlabeled background (black) and the reference clones (arrows). (c and d) In twin-spot MARCM, clones of interest and the reference clones with homozygous genotypes are visible in green and magenta, respectively, and stand out clearly from the unlabeled background (black). (e) In the twin-spot MARCM system, “suppressor 1” (which inhibits the expression of “reporter 1”) and “reporter 2” are placed distal to one FRT site, while “suppressor 2” (which inhibits the expression of “reporter 2”) and “reporter 1” are placed distal to the other FRT site. After induction of FLP to activate FRT-mediated mitotic recombination, one daughter cell (green) inherits “suppressor 2” and “reporter 1”, while the other (magenta) gets “suppressor 1” and “reporter 2”. A lineage undergoing symmetric (f) or asymmetric (g) proliferation will produce two multi-cellular clones (f) or a two-cell + multi-cellular clone (g). Two single-cell clones (g) appear, one in green and the other magenta, when mitotic recombination occurs during last mitoses (as in a GMC). The scale bar in this and all other figures equals 20 μm.
RESULTS
Design of twin-spot MARCM with reporter-specific silencers
An ideal “twin-spot” MARCM would permit differential expression of markers in paired sister clones and restrict marker expression to the cells homozygous for a particular chromosome arm and its homologous counterpart (Fig. 1c-e). Akin to MARCM, we reasoned it should utilize independent suppressors that reside on opposing homologous chromosome arms and distal to the recombination site whereby loss of heterozygosity would trigger expression of the two distinct markers (Fig. 1e).
For this new MARCM technique, we exploited RNA interference (RNAi) to suppress gene expression in a transcript-dependent manner rather than by targeting promoters or proteins 13-16. Based on our recent success silencing other transcripts with microRNA (miRNA)-encoding transgenes 17, 18, we developed non-interfering UAS-miRNA transgenes that silence UAS-driven mCD8::GFP and UAS-driven rCD2::RFP, respectively (Supplemental Fig. 1b,f). We further ensured the specificity of these two UAS-miRNA transgenes. Co-induction with UAS-GFP-miRNA potently silenced UAS-mCD8::GFP but minimally affected the induction of UAS-rCD2::RFP (Supplemental Fig. 1b,e). Consistently, UAS-rCD2-miRNA only suppressed UAS-rCD2::RFP without compromising the expression of UAS-mCD8::GFP (Supplemental Fig. 1c,f). We can, thus, selectively silence UAS-mCD8::GFP and UAS-rCD2::RFP using UAS-GFP-miRNA and UAS-rCD2-miRNA, respectively. To test the efficacy of the twin-spot MARCM technique, we assembled a pilot system on the 2L chromosome arm, induced clones, and assessed the results in wing discs. Using the ubiquitous tubP-GAL4, we labeled paired clones in different colors in the otherwise unstained wing discs (Fig. 1d), the first evidence that twin-spot MARCM works as we intended.
Twin-spot MARCM differentially labels paired sister clones
We next applied twin-spot MARCM to the MBs to establish its validity for studying the nervous system. Neurogenesis of the MBs has been well studied with the existing MARCM technology and independent methods 5, 19. Past experiments using MARCM, which labels only one side of a twin spot, indicate mutual exclusion of Nb and two-cell clones. However, we cannot ensure the pairing relationship between Nb clones and two-cell clones, tell a two-cell clone from two nearby single-cell clones, or deduce the order of birth from randomly generated clones.
To highlight clones in the MB lineage, we used the strong MB driver GAL4-OK107 in twin-spot MARCM. Of 93 MB Nb clones induced in NHL, we found 91 of them accompanied by exactly two MB γ neurons (Fig. 2a-c). The two exceptions were each paired with a group of three γ neurons (data not shown). The presence of one extra γ neuron on the GMC side of Nb progeny could derive from mitotic recombination within a coexisting GMC of the same or a neighboring lineage. Moreover, the later we induce mitotic recombination, the smaller the size of the resulting Nb clones (Fig. 2d-i; n>10 for each). Irrespective of Nb clone size, we consistently detect a two-cell clone paired with it due to differential labeling by twin-spot MARCM. Notably, MB Nb clones of different sizes are accompanied by two-cell clones of different types, perfectly reflecting the order of birth from γ, α′/β′, to α/β neurons (Fig. 2a-i). These results reinforce the current model of MB neurogenesis and show our twin-spot MARCM system effectively labels paired sister clones in the brain.
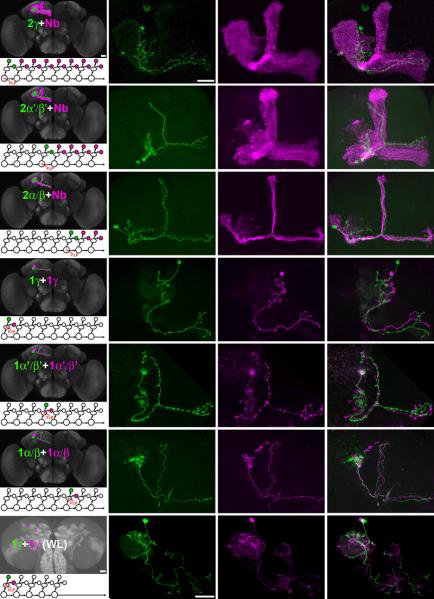
Figure 2. MB lineage analysis with twin-spot MARCM.
Examples of twin-spot MARCM clones used in an analysis of MB lineage are shown: two-cell + multi-cellular Nb clones (a-i) and two single-cell clones (j-u) in adult (a-r) and wandering larval (s-u) brains. Mitotic recombination was induced by FLP activity in MB Nbs (a-i) or GMCs (j-u) at different developmental stages, including 1st instar larval (a-c, j-l and s-u), mid 3rd instar larval (d-f and m-o) and mid-late pupal (g-i and p-r) stages. GAL4-OK107 (a-i and m-r) and MB247-GAL4 (j-l and s-u) were used to visualize twin-spot MARCM clones. In the two-cell + multi-cellular Nb clones: two γ MB neurons (a) are associated with a MB clone (b) highlighting all MB lobes (γ, α, α′, β and β′ lobes); two α′/β′ MB neurons (d) are associated with a MB clone (e) revealing α, α′, β and β′ lobes; two α/β MB neurons (g) are associated with a MB clone (h) containing only α and β lobes. In the two single-cell clones: a green γ MB neuron (j and s) is associated with the other magenta γ MB neuron (k and t); a green α′/β′ MB neuron (m) is associated with the other magenta α′/β′ MB neuron (n); a green α/β MB neurons (p) is associated with the other magenta α/β MB neuron (q). These analyses allow us to distinguish two-cell clones from double inductions in separate GMCs (j-u). The green and magenta merged images are shown in c, f, i, l, o, r and u.
We anticipated pairing of a GFP-labeled neuron with one RFP-labeled neuron in clones produced by mitotic recombination in GMCs. In brains that have multiple GMC-induced clones, it may not be possible to tell which GFP neuron pairs with a particular RFP neuron since sister neurons can reside far apart as shown in our two-cell clones (Fig. 2a). However, the numbers of GFP- and RFP-labeled single-cell clones in each mosaic MB should be constantly equal. While we frequently saw the expected pairing of α′/β′ or α/β neurons (Fig. 2m-r; n>20 for each), this was not the case in clones of γ neurons. First, we seldom obtained γ neurons labeled without Nb clones (only 5 out of 200 mosaic brains derived from recombination in the early 1st instar larval stage). Second, equal numbers of GFP and RFP neurons do not occur in most groups of γ neurons without Nb clones (3 were not paired). Since we never observe a MB Nb clone either alone or accompanied by only one MB neuron, it is unlikely that a significant number of MB γ neurons are negative for GAL4-OK107 or die prematurely. Instead, it is more likely our technique does not faithfully label clones induced in GMCs due to stage-specific differential perdurance of RNAi leading to delayed or incomplete derepression of the UAS-reporters after the loss of their respective UAS-miRNA transgenes. This can also explain why the GMC-induced clones of γ neurons were weakly marked as compared to clones of α′/β′ or α/β neurons (data not shown).
Any perdurance of repression in MARCM derives from expression of repressors prior to mitotic recombination and thus inheritance of repressors directly from heterozygous precursors. In the miRNA-based twin-spot MARCM system, GAL4 drives expression of both the repressors and the reporters; the degree of perdurance should depend on the characteristics of a GAL4 driver. We labeled MB sister clones with MB247-GAL4, a MB driver expressed strongly in post-mitotic neurons but not in precursors 12. We did not detect any delay in the induction of UAS-reporter and we identified γ neuron pairs not only in adult brains (Fig. 2j-l; n=46/48; two cases were one γ neuron paired with two γ neurons) but also in larval brains (Fig. 2s-u; n=12), in which the original MARCM technique cannot label single-cell clones due to tubP-GAL80 perdurance (data not shown).
Twin-spot MARCM reveals different CNS lineage patterns
Visualizing both sides of a twin-spot is essential for depicting a complete proliferation pattern for any progenitor. For example, consistent pairing of a multi-cellular Nb clone and a two-cell clone through development of a lineage would confirm the models that self-renewing Nbs serially produce GMCs and that GMCs produce two neurons. It was long thought that all Drosophila Nbs undergo this simple pattern of neurogenesis until recent identification of PAN (Posterior Asense-Negative) lineages and their transit-amplifying precursors 20, 21. Each transit-amplifying precursor, derived from self-renewing PAN Nbs, may give rise to multiple GMCs through a limited run of self-renewing asymmetric division. In addition, GMCs from many neuronal lineages of the Drosophila ventral nerve cord (VNC) initially produce two daughter cells during larval neurogenesis, but only one of them survives to mature into an adult neuron (Williams D.W. and Truman J.W. personal communication). We imagine twin-spot MARCM easily uncovering diverse unexpected lineage patterns.
We performed a study of neurogenesis patterns in the Drosophila central brain and VNC using twin-spot MARCM with the pan-neuronal GAL4-C155. For mosaic clones in the central brain, 43 of the 108 (40%) Nb clones, including 10 MB Nb clones, are paired with two neurons (Fig. 3a,b,d,e,g,h); 53 of the 108 (49%) Nb clones are accompanied by only one neuron (Fig. 3c,f,i); and 10 of the 108 (9%) Nb clones, located in the dorsomedial region where PAN lineages reside, associate with a cluster of 8 to 12 cells (Fig.3l,o,r). There were two Nb clones without counter pair association (2%; data not shown). In contrast, for mosaic clones in the VNC, only 26 of the 128 (20%) Nb clones are paired with two neurons (Fig. 3j,m,p) while 100 of the 128 (78%) Nb clones are paired with a single neuron (Fig. 3k,n,q) and two Nb clones were not associated with any post-mitotic neurons (2%; data not shown).
Figure 3. Proliferation patterns of Drosophila larval CNS lineages using twin-spot MARCM.

The pan-neuronal driver, GAL4-C155, was used to survey proliferation patterns of Drosophila larval CNS lineages. Twin-spot MARCM clones were induced at a low frequency to simplify the lineage analysis at Drosophila central brain (a-i, l, o and r) and VNC (j, k, m, n, p and q). Like GAL4-OK107, GAL4-C155 can faithfully label two-cell + multi-cellular Nb clones (a, d and g) in MB lineages, which validates the use of GAL4-C155 to probe the overall proliferation patterns of Drosophila CNS lineages. Three types of twin-spot MARCM clones were classified based on the cell number of their associated sister clones. The first class, like MB clones, consists of two-cell + multi-cellular Nb clones (b, e, h, j, m and p) that indicate the familiar production of two progeny by a GMC division. The second class of clones has single-cell + multi-cellular Nb clones (c, f, i, k, n and q) that resemble lineages in which only one daughter cell from the GMC division survives. The lineage identity of [p] and [q] resembles the previously described VNC lineage 13 and lineage 20, respectively 27. The third class is represented by 8-12 cells + large multi-cellular Nb clones (l, o and r) and resemble the recently described PAN lineages in which each transit-amplifying precursor produces multiple GMCs through a limited run of self-renewing asymmetric division.
Identifying all neurons in a central complex sublineage
GAL4-OK107 labels, in addition to MB neurons, a cluster of six neurons in each brain lobe whose projections finally converge in various midline neuropils, collectively called the central complex (Fig. 4a-c). Even in a small lineage such as this, determining the birth order of each neuron requires technology that provides at least the resolution of twin-spot MARCM. By labeling GMC progeny in one color and the remaining lineage in another, we can determine which GMC has given rise to a particular neuron in any protracted neuronal lineage. To show this capability, we characterized the GAL4-OK107 sublineage of the central complex.
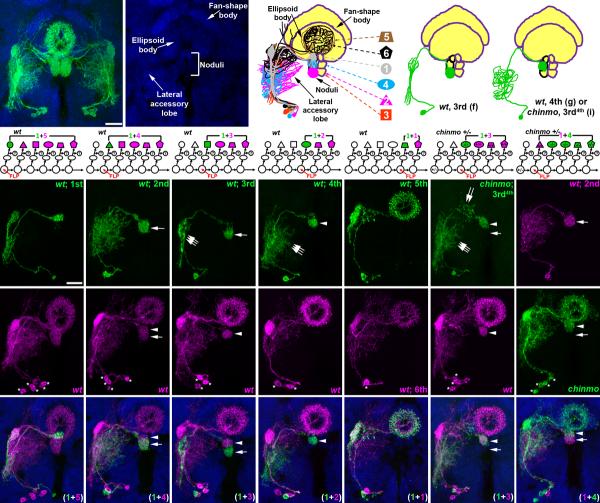
Figure 4. Comprehensive analysis of individual neurons in a central complex sublineage through twin-spot MARCM.
Six central complex neurons (a), nc82 staining for revealing relative positions to these neurons (a-b) and a schematic drawing of these neurons (c). Twin-spot MARCM analyses on this central complex sublineage reveals birth-order of these neurons (d-h and o). The first four neurons (d-g) extend into the noduli with the projections to discrete subcompartments, including the dorsal noduli (d), anterior ventral noduli (e), posterior ventral noduli (f) and middle noduli (g). The last two neurons (h and o) innervate the ellipsoid body. Neuronal morphology alternates between narrow and elaborate neurites (d, f and h vs. e, g and o) in the lateral accessory lobe. A third-born chinmo neuron (3rd4th; i), determined by its reference wild-type Nb clone (p), has the morphology of wild-type fourth-born neuron. This 3rd4th born neuron innervates the middle noduli that normally is innervated by the 4th born neuron (arrowheads in i, p and w). The neurite pattern in the lateral accessory lobe of this 3rd4th born neuron is also transformed from narrow to elaborate (triple arrows in f, g and i). In contrast, the reverse twin-spot MARCM analysis shows that the second-born wild-type single-cell clone is associated with a chinmo Nb clone (j, q and x). Compared to the projection to the posterior ventral noduli in the wild-type Nb clone (arrow in l), the same morphology is not present in the chinmo Nb clone (arrow in q), confirming the transformation of the third-born neuron to the wild-type fourth-born neuron morphology. Asterisk indicates cell body position.
We found that all central complex Nb clones have sister clones that contain only one cell, which suggests we are observing half of the GMC progeny (Fig. 4r-v), and that these six GAL4-OK107-positive central complex neurons acquire different trajectories based on birth order. For example, the first-born neuron, as indicated by five cell bodies in the associated Nb clone, consistently extends into a dorsal sub-compartment of the noduli and extends few processes ventrally (Fig. 4d,k,r and Supplemental Fig. 2a-c; n=10). In contrast, the third-born neuron always innervates a ventral sub-compartment of the noduli (Fig. 4f,m,t and Supplemental Fig. 2d-f; n=12). In addition, each neuron has distinct stereotyped branches off the primary neurite (e.g. triple arrows in Fig. 4f,g). By analyzing many clones produced in twin-spot MARCM, we ultimately identified the unique characteristics and birth order of each GAL4-OK107-positive central complex neuron. The first four neurons each innervate a distinct sub-compartment of the noduli (Fig. 4d-g) and the last two neurons project into the ellipsoid body (Fig. 4h,o). Individual neurons also acquire stereotyped patterns of neurite elaboration in the lateral accessory lobe (Fig. 4d-h,o).
chinmo central complex clones adopt incorrect fates
It is extremely challenging to detect temporal cell fate transformation in a fast-tempo neuronal lineage where neuron fate may change with each self-renewing Nb division. To exemplify how twin-spot MARCM permits better determination of temporal cell fates, we examined whether and how chinmo, which perturbs temporal identity in MB neurons 12, affects temporal fates in the sublineage of six central complex neurons.
Knocking out chinmo from a GMC of the central complex sublineage causes the 3rd sibling to exhibit a temporal cell fate like that of the 4th neuron in the wild-type lineage. Neurons derived from chinmo mutant 3rd GMCs, as determined by associated wild-type Nb clones containing three cells, acquired projections characteristic of wild-type 4th siblings (Fig. 4i,p,w and supplemental Fig. 2g-i; n=8). They innervate the middle sub-compartment of the noduli with the wild-type sibling of the Nb clone (arrow and arrowhead in Fig. 4f,g,i,t,u,w). In addition, mutant 3rd siblings, like wild-type 4th-born neurons, acquire a much more diffuse pattern of neurite elaboration in the lateral accessory lobe than wild-type 3rd-born neurons (triple arrows in Fig. 4f,g,i). As to removal of Chinmo from the central complex Nb, we consistently fail to detect cells with an appropriate 3rd temporal fate in chinmo mutant clones produced prior to birth of the 3rd offspring (n=3). For example, in a chinmo mutant Nb clone induced during production of the second GMC, which leads to a normal 2nd sibling, we do not detect axons of mutant cells in the ventral sub-compartment of the noduli (arrow in Fig. 4e,j,l,q,s,x). We conclude Chinmo is required in only one of the six fates of the GAL4-OK107-positive central complex neurons.
DISCUSSION
Twin-spot MARCM permits the visualization of both sides of a twin spot, enhancing the power of MARCM and supporting creative and informative mosaic studies in the complex nervous system. In particular, labeling GMC clones and their paired Nb clones reveals the developmental origin of neurons. Given the fundamental influence that developmental origin seems to exert on neuron-type specification, single-neuron analysis with twin-spot MARCM promises to identify individual neuron types, resolve their connectivity, and elucidate underlying mechanisms.
As in the original MARCM system, twin-spot MARCM is built on top of GAL4/UAS. Following loss of heterozygosity, only GAL4-positive cells that lack repressors can be labeled. Use of a ubiquitous GAL4 driver is mandatory for revealing mosaic patterns in their entirety. By contrast, one can utilize subtype-specific GAL4 drivers to restrict labeling to subsets of repressor-negative cells for more focused mosaic analysis. Systematic efforts are underway to generate diverse GAL4 drivers that together cover the entire brain 22. Clonal analysis with such GAL4 drivers in twin-spot MARCM should allow us to ultimately identify all neuron types and unambiguously determine their developmental origins. However, most GAL4 drivers label neurons in complex patterns; they mostly label multiple sublineages. We propose combining those subtype-specific GAL4s for comprehensive lineage analysis by twin-spot MARCM.
A power of MARCM is that we can drive additional UAS-transgenes specifically in the MARCM-labeled clones for cell-autonomous rescue of mutant clones or targeted expression of a gain-of-function transgenes. This sort of application is not impossible with the miRNA-based twin-spot MARCM. In theory, and as already demonstrated in other model organisms 23, one can artificially make any gene repressible by a specific miRNA by introducing the miRNA's target sequences into the untranslated regions of the gene of interest. This way, we may express multiple UAS-transgenes differentially between sister clones in mosaic tissues. Twin-spot MARCM clearly preserves all the powers of MARCM and further extends the targeted labeling as well as gene manipulation to both sides of twin spots.
Labeling both sides of a twin spot should allow us to determine a complete proliferation pattern for any neural progenitor which we demonstrated by showing major patterns of post-embryonic neurogenesis with GAL4-C155 in twin-spot MARCM. Several important points are worth discussing here. First, we hypothesize that GAL4-C155 labels every neuron in the larval central brain and VNC. Indeed our observation that 98% of Nb clones have sister clones reinforces this notion. Second, neurons of the same lineage exist as clusters in the Drosophila central brain and VNC, a fact shown without exception in clonal analyses of Drosophila brain. Third, Nb clones generated during early larval life may be accompanied by smaller sister clones of three different sizes. One Nb clone may pair with a single-cell clone, a two-cell clone, or a small multi-cellular clone. The small multi-cellular clones were consistently found in the dorsomedial region of the brain, while no spatial correlation could be detected as to the pairing with single-cell or two-cell clones. Given that neurons and glial cells in the fly central brain derive from distinct post-embryonic precursors 24, 25 and that cell bodies of central neurons of the same lineage are tightly packed together 26, these phenomena indicate unit production of one, two, or a small cluster of mature neurons depending on the lineages. In addition, the recently identified transit-amplifying precursors in PAN lineages probably underlie the production of multiple neurons from some Nb self-renewal 20, 21. Multiple modes of neurogenesis demonstrate the complexity of assembly of the Drosophila CNS, and the use of twin-spot MARCM in lineage and phenotypic analyses should facilitate our understanding on this process.
One major challenge with single-neuron analysis is how difficult it is to determine if we are comparing the same neuron as we look at brains from different organisms. With twin-spot MARCM, we can at least ensure the comparison of neurons with the same developmental origin. Intriguingly, neurons of the same developmental origin consistently acquire the same fate in different organisms. Analysis of single neurons classified based on their developmental origins should permit description of neural circuitry with single-cell resolution. One can further study the genetics of origin-dependent cell fate specification as a step toward elucidating underlying mechanisms. Notably, twin-spot MARCM permits independent assessment of mutant clones' prospective cell fates and has allowed us to demonstrate that Chinmo, a BTB-zinc finger protein, is specifically required for the specification of the third temporal fate in a sublineage of six central complex neurons. In sum, with twin-spot MARCM, we may realistically begin the endeavor to identify every single neuron in the Drosophila brain. Simultaneously, we will come to new understandings about neural development. These together will allow us to build a complete cellular and developmental brain map and lay the foundation for a comprehensive understanding of brain development and function.
Supplementary Material
Acknowledgements
We thank Dr. C. T. Zugates for critical reading of the manuscript, and members of the Lee lab for helpful discussions through the entire project. This work was supported by National Institutes of Health.
Footnotes
Online Methods is available at http://www.nature.com/natureneuroscience/
References
- 1.Hartenstein V, Campos-Ortega JA. Early neurogenesis in wildtype Drosophila melanogaster. Roux's Arch Dev. Biol. 1984;193:308–325. doi: 10.1007/BF00848159. [DOI] [PubMed] [Google Scholar]
- 2.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 3.Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 4.Urbach R, Schnabel R, Technau GM. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–3606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- 5.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 6.Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 7.Lai S-L, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- 8.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 9.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhu S, Chiang A-S, Lee T. Development of the Drosophila mushroom bodies: elaboration, remodeling, and spatial organization of dendrites in the calyx. Development. 2003;130:2603–2610. doi: 10.1242/dev.00466. [DOI] [PubMed] [Google Scholar]
- 12.Zhu S, et al. Gradients of the Drosophila Chinmo BTB-Zinc Finger Protein Govern Neuronal Temporal Identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 13.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 15.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 16.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, Yu HH, Yang JS, Lee T. Specific Drosophila Dscam juxtamembrane variants control dendritic elaboration and axonal arborization. Journal of Neuroscience. 2007;27:6723–6728. doi: 10.1523/JNEUROSCI.1517-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H-H, Yang JS, Wang J, Huang Y, Lee T. Endodoamin diversity in the Drosophila Dscam and its roles in neuronal morphogenesis. J Neurosci. 2009;29:1904–1914. doi: 10.1523/JNEUROSCI.5743-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 20.Bowman SK, et al. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 24.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. Epub 2006 Apr 2002. [DOI] [PubMed] [Google Scholar]
- 25.Awasaki T, Lai S-L, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. Advances in experimental medicine and biology. 2008;628:137–158. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- 27.Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.