Abstract
The XPC protein (encoded by the xeroderma pigmentosum Xpc gene) is a key DNA damage recognition factor that is required for global genomic nucleotide excision repair (G-NER). In contrast to transcription-coupled nucleotide excision repair (TC-NER), XPC and G-NER have been reported to contribute only modestly to cell survival after DNA damage. Previous studies were conducted using fibroblasts of human or mouse origin. Since the advent of Xpc−/− mice, no study has focused on the bone marrow of these mice. We used carboplatin to induce DNA damage in Xpc−/− and strain-matched wild-type mice. Using several independent methods, Xpc−/− bone marrow was ∼10-fold more sensitive to carboplatin than the wild type. Importantly, 12/20 Xpc−/− mice died while 0/20 wild-type mice died. We conclude that G-NER, and XPC specifically, can contribute substantially to cell survival. The data are important in the context of cancer chemotherapy, where Xpc gene status and G-NER may be determinants of response to DNA-damaging agents including carboplatin. Additionally, altered cell cycles and altered DNA damage signalling may contribute to the cell survival end point.
Introduction
The xeroderma pigmentosum Xpc gene is defective in a subset of human patients exhibiting the cancer-prone disease xeroderma pigmentosum which results from defective nucleotide excision DNA repair (NER). XPC patients are sensitive to sunlight and ultraviolet (UV) radiation-induced DNA damage and skin cancers. They also exhibit internal cancers with advanced age. The gene products encoded by Xpc and other XP genes A-G have been characterized biochemically (1). Specifically, the XPC protein (encoded by the xeroderma pigmentosum Xpc gene) is required and is rate limiting for global genomic nucleotide excision repair (G-NER). XPC is neither required nor apparently involved in transcription-coupled nucleotide excision repair (TC-NER). Thus, studies of Xpc are good models for G-NER separate from TC-NER (1).
Mice lacking Xpc genes (Xpc−/− mice) were generated some 10 years ago (2). A number of studies of carcinogenesis and mutagenesis have been conducted, consistent with the cancer-prone human genetic disease discussed above (2–4). This is the first study to examine bone marrow in Xpc−/− mice.
Bone marrow is often dose limiting in response to cancer chemotherapy drugs including carboplatin. While cell survival is a complex end point, the goal of chemotherapy is to sensitize cancer cells while retaining bone marrow cellularity. Agents that can protect bone marrow are important adjuncts to chemotherapy. Presently, the cytokine granulocyte macrophage colony stimulating factor is administered which causes proliferation of bone marrow myeloid stem/progenitor cells and thus repopulates bone marrow after chemotherapy (5). The idea of G-NER as a protective mechanism in bone marrow is novel. Xpc−/− mouse bone marrow was highly sensitive to carboplatin. Patients carrying mutant XPC genes may exhibit an adverse bone marrow response to carboplatin.
Materials and methods
Mice
Mice originated from Sands et al. (2) and were purchased from Taconic Farms and bred at Indiana University under licence agreement as B6;129s7-XPCtml/Brd mice. Female mice were 10 weeks old at the time of initiating the experiments. Bone marrow was directly harvested and cultured for 24–72 h for indicated in vitro experiments (Figures 1–3). For in vivo experiments (Figures 4 and 5), carboplatin (Sigma, St Louis, MI, USA) was dissolved in sterile saline and administered intra-peritoneally at weekly intervals over 6 weeks as indicated in the figure (0.5-ml injections, 60 or 100 mg/kg body weight). White blood cell (WBC) counts were by weekly tail vein blood collection in ethylenediaminetetraacetic acid-treated haematocrit tubes (6). Counting was done using a Hemavet 950 (Drew Scientific, Dallas, TX). Statistical analysis was conducted using a t-test using Microsoft Office 2003 or GraphPad Prism software. All mouse treatments were in accord with protocols approved by the Institutional Animal Care and Use Committee at Indiana University. Mice were sacrificed after 41 days using approved methods. Mice that appeared moribund before 41 days were sacrificed by veterinary staff.
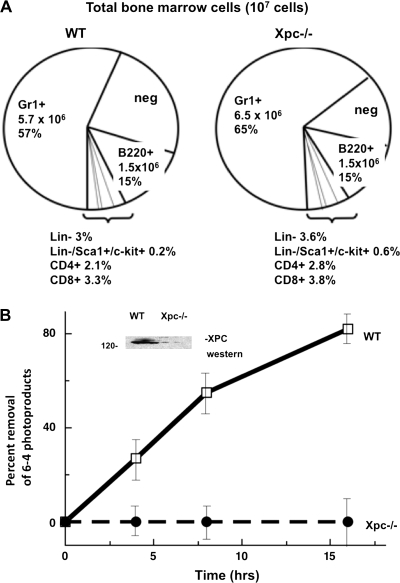
Fig. 1.
G-NER defect in cultured Xpc−/− bone marrow. (A) For cell culture experiments (Figures 1–3) bone marrow was harvested from untreated wild-type or Xpc−/− mice. Cell yields were lower in Xpc−/− compared to wild type; therefore, cell numbers were normalized to 107 viable cells per genotype. Flow cytometry was conducted using six different lineage markers. No significant differences were observed in the lineage markers in comparing 107 cells of each genotype. As shown in the pie graph, lineage markers tested were Gr1+, B220+, Lin−, CD4+, CD8+ and the small subpopulation Lin−/Sca1+/c-kit+. The pie segment labelled ‘neg’ was unreactive for any of the lineage markers tested. (B) XPC protein was undetectable in Xpc−/− bone marrow (inset). Removal of DNA lesions was markedly slow in Xpc−/− bone marrow, consistent with the known rate-limiting role of XPC in G-NER. Pooled bone marrow of three or more untreated 10-week-old mice of each genotype was used. Cells were cultured in cytokine-containing medium for 15 h, irradiated with 20 J/m2 254 nm UV radiation in phosphate-buffered saline and returned to tissue culture. Time points were taken at 0, 4, 8 and 16 h and assayed using an antibody to 6-4 photoproducts (P < 0.02 by t-test).
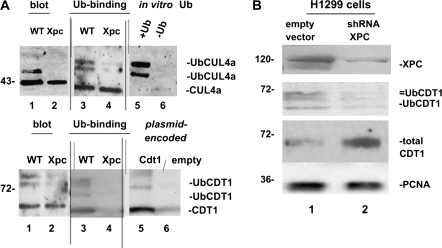
Fig. 3.
(A) CUL4A and CDT1 cell cycle checkpoint proteins in cultured wild-type and Xpc−/− bone marrow. We wanted to determine if the presence or absence of XPC would alter DNA damage signalling. Immunoblots were conducted using 50 μg of total cell lysates (lanes 1 and 2, upper and lower panels). Equal amounts of cellular proteins, 5 mg, were then affinity purified on a ubiquitin-binding resin and immunoblotting of bound proteins was conducted. The higher molecular weight ubiquitinated forms of CUL4A (upper panel) and CDT1 (lower panel) clearly differ between the two genotypes (lanes 3 and 4 of upper and lower panels). The data suggest a defect in CUL4A and CDT1 ubiquitin modification in Xpc−/− mice. To help identify ubiquitinated CUL4A and CDT1, we used in vitro ubiquitin-conjugated proteins. Omission of ubiquitin from the reaction shows that mainly ubiquitinated CUL4A is detected by the CUL4A antibody (lanes 5 and 6, upper panel). Plasmid-encoded CDT1 protein was also used as a marker (lanes 5 and 6, lower panel). (B) Knocking down XPC in H1299 cells alters the ubiquitin modification of CDT1. We used transiently transfected H1299 cells expressing an shRNA to XPC to test if the results shown in panel (A) were due directly to loss of XPC. Although Xpc was not completely silenced, it was clearly decreased by the shRNA. Higher molecular weight ubiquitinated forms of CDT1 were largely absent where XPC was knocked down, consistent with a role for XPC in DNA damage signalling to the CDT1 cell cycle checkpoint protein.
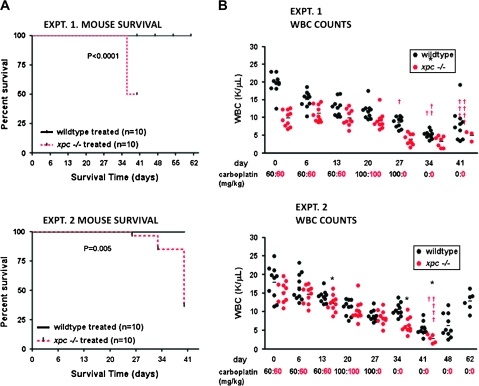
Fig. 4.
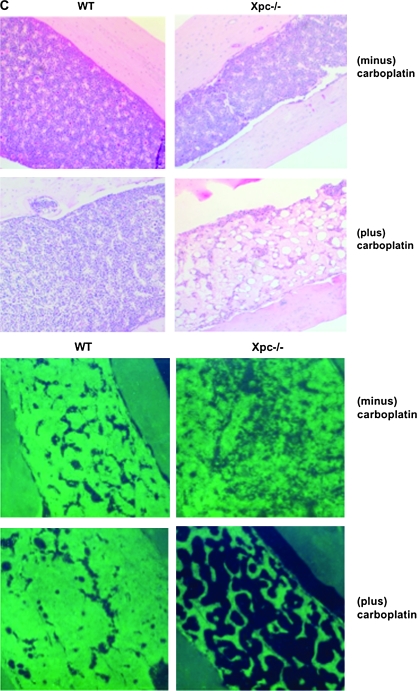
Effect of carboplatin administration in vivo in bone marrow of wild-type and Xpc−/− mice. (A) Kaplan–Meier plots of mouse survival. Twenty mice of each genotype received carboplatin or saline only as controls. Saline-only did not affect mouse survival nor alter WBC counts; saline-only control groups are not shown in order to simplify the figure. Mice received carboplatin at weekly intervals up until day 27. By day 41, 12/20 Xpc−/− mice died, while 0/10 wild-type mice died. (B) WBC counts were monitored during the course of the experiments. WBC counts were not significantly different prior to day 27, but were significantly different after day 27 (P < 0.05, lower panel). Each circle represents an individual mouse, black, wild type; red, Xpc−/−. The horizontal bars represent the mean of each group. By day 41, the few surviving Xpc−/− mice had significantly lower WBC counts (P < 0.05 by t-test). The carboplatin dosing schedule is shown in black, wild type; red, Xpc−/−. Note that wild-type mice received an additional dose on day 27 that was not administered to Xpc−/− mice. (C) Histological evaluation of wild-type and xpc−/− mice in carboplatin-treated and saline-control groups, day 41. Haematoxylin–eosin staining of formalin-fixed femur sections. Marked hypocellularity was observed in Xpc−/− mice receiving carboplatin (upper panel); 4′,6-diamidino-2-phenylindole staining (lower panel). The data are representative of at least three mice of each treatment group and genotype.
Fig. 5.
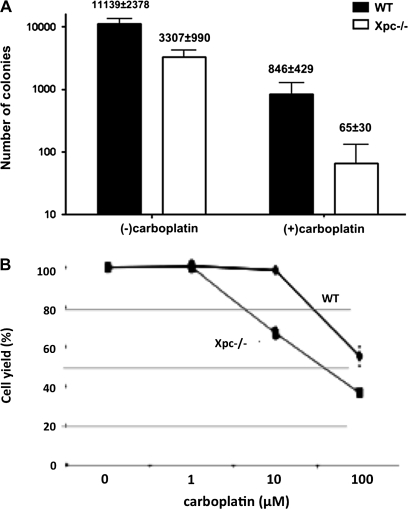
Quantification of bone marrow hypocellularity in Xpc−/− mice compared to wild type. (A) Colony-forming assays of bone marrow harvested from the respective genotypes and carboplatin treatment groups shown in Figure 4. Bone marrow was harvested and grown in complete methylcellulose medium containing interleukin-6 and stem cell factor for 10 days. Total colonies per femur are shown. Xpc−/− bone marrow in the carboplatin-treated group was decreased 10- to 12-fold compared to wild type (P < 0.008 by t-test). Xpc−/− bone marrow in the untreated group was decreased 3-fold compared to wild type (P < 0.02 by t-test). Each set of experiments utilized bone marrow from three or more mice of each genotype. (B) Assay of cell yield in bone marrow cells treated with carboplatin in vitro. Bone marrow of untreated 10-week-old wild-type and Xpc−/− mice was cultured for 24 h and then treated with indicated concentrations of carboplatin for 2 h. Cell yield after 72 h in culture is shown (P < 0.006 by t-test). The data shown were averaged from three experiments. The dose modification factor for 50% cell survival in the presence of carboplatin is ∼10-fold, consistent with the other data.
DNA repair assay
To confirm the DNA repair defect in Xpc−/− bone marrow, we used an immunoassay in which genomic DNA from UV-irradiated bone marrow cells, at 0, 4, 8 or 16 h after UV irradiation, was fixed to 96-well microtiter plates at 10 ng per well. Cells were washed and re-suspended in 2-ml phosphate-buffered saline and irradiated as a thin monolayer in 60-cm2 dishes, then placed back in culture medium. An antibody to 6-4 photoproducts (Trevigen Inc., Gaithersburg, MD) was used to detect removal of lesions. Immunoassays were developed with peroxidase secondary antibody and 2,2′-azino-bis 3-ethylbenzthiazoline-6-sulphonic acid chromogenic substrate and read at 405 nm in a Tecan Spectra plate reader. Immunoblotting of Xpc was with a rabbit polyclonal antibody (Santa Cruz Biotech, Santa Curz, CA).
Cell cycle analysis
Bone marrow of wild-type and Xpc−/− genotypes were cultured in Iscove's modified Dulbecco medium (IMDM)-containing cytokines as above for 48 h to stimulate proliferation and then treated with carboplatin for an additional 15 h. Untreated cultures served as controls for each genotype. Cells were fixed in 70% ethanol and then analysed on a Becton-Dickinson FacsScan for propidium iodide (PI) content. To analyse only proliferating cells, 10-μM bromodeoxyuridine (BrdU) was added to the cultures during the 48-h growth period. Cycling cells were first gated for BrdU using a fluorescein isothiocyanate-conjugated mouse monoclonal antibody to BrdU (Becton-Dickinson, Franklin Lakes, NJ, USA) and then with PI. The data represent ≥15 000 cells per data point.
For lineage marker analysis, antibodies to CD4, CD8, B220, Gr-1, lin, c-kit and Sca-1 were used in accord with manufacturer protocols (Becton-Dickinson). Samples were analysed on a Becton-Dickinson FACScalibur using CellQuest software, scoring at least 10 000 events per sample.
Immunoblotting of cell cycle proteins
Initially whole-cell lysates were used to probe for differences in cell cycle proteins between wild-type and Xpc−/− bone marrow. Given apparent differences in Cdt1 and Cul4a, we purified total ubiquitinated proteins on a ubiquitin-binding resin (Pierce Chemical, Rockford, IL). Cell lysates corresponding to 107 viable cells per sample were prepared in RIPA lysis buffer to which 50 μl of resin was added and rotated overnight at 4°C. The resin was collected by centrifugation and then boiled 15 min in sodium dodecyl sulphate/gel-loading buffer and subject to electrophoresis in 4–20% polyacrylamide gels (Invitrogen, Carlsbad, CA). Immunodetection was on nitrocellulose membranes using rabbit anti-Cul4a (34897; Abcam, Cambridge, MA) or rabbit anti-Cdt1 (sc-28262; Santa Cruz Biotech).
Human H1299 lung cancer cell line was transfected using a FuGene (Boehringer-Mannheim, Indianapolis, IN) and a short hairpin RNA (shRNA) plasmid to partially silence XPC (Origene, Gaithersburg, MD). The empty plasmid served as a control (Origene). Cells were analysed by immunoblotting after 72 h.
Histochemical staining
Femurs were fixed in 10% buffered formalin overnight, then paraffin embedded and thin sectioned for histological examination by a veterinary pathologist. Slides were stained with haematoxylin and eosin for morphological analysis or 4′,6-diamidino-2-phenylindole to visualize intact nuclear DNA. Slides were visualized by a Nikon HB-10101AF fluorescence microscope using a ×100 objective. Digital photographs were captured.
Colony-forming assays
Bone marrow was collected from femurs, fibulas and iliac crests. Total bone marrow cells were then added to complete methylcellulose medium consisting of IMDM liquid medium (Sigma), 20% foetal bovine serum, interleukin-6 (200 U/ml) and stem-cell factor 100 ng/ml (Stem Cell Technologies, Vancouver, Canada). Triplicate 35-mm culture dishes were seeded with 105 cells and placed in a humidified incubator at 37°C for 10 days at which time colonies were counted manually as in ref. (7).
MTS assay of cell viability
Bone marrow of wild-type and Xpc−/− genotypes was cultured in 96-well plates in IMDM medium with cytokines as above and then treated in vitro with increasing concentrations of carboplatin for 2 h. Cell yield was determined after 72 h in culture by adding MTS (Promega, Madison, WI) to the wells and reading the absorbance at 490 nm in a Tecan Spectra plate reader. Cells that did not receive carboplatin served as controls for each genotype representing 100% cell yield.
Results
The data show a pronounced effect on mouse bone marrow owing to the presence or absence of a functional XPC gene product. The first observation was that the number of viable cells recovered from untreated Xpc−/− bone marrow was always 2- to 3-fold lower than wild type. To remedy this problem, we normalized the number of viable cells for both genotypes for the cell culture experiments. Cell numbers were normalized typically to 107 cells representing pooled cells of three mice of each genotype. The normalized cell populations then were used as a source of cells for tissue culture experiments. Flow cytometric analysis was conducted both immediately after harvest and after tissue culture experiments, to ascertain if any one cell lineage was affected in the Xpc−/− mice. As shown in Figure 1A, no single lineage was uniquely affected in Xpc−/− mice. Populations representing lineage markers Gr-1+, B220+, Lin−, CD4+, CD8+ and the triply gated Lin−/Sca1+/c-kit+ population were not significantly different between wild-type and Xpc−/− mice when comparing equal numbers of cells of both genotypes (Figure 1A). The relative distribution of the lineage markers did not change over the course of cell culture experiments (results not shown), although the cell culture experiments were typically conducted within 48 h and were therefore short-term cultures.
Mice of wild type and Xpc−/− genotypes were assayed for XPC expression and G-NER activity. Bone marrow was used directly for western blotting using an XPC antibody. Bone marrow was additionally cultured 24 h, UV irradiated and then subjected to an immunoassay for removal of DNA damage. Removal of DNA lesions was 80% complete in wild-type mice after 16 h, while Xpc−/− mice were defective in removal of the lesions (Figure 1B). Thus, the Xpc−/− mice exhibit the expected defective NER phenotype.
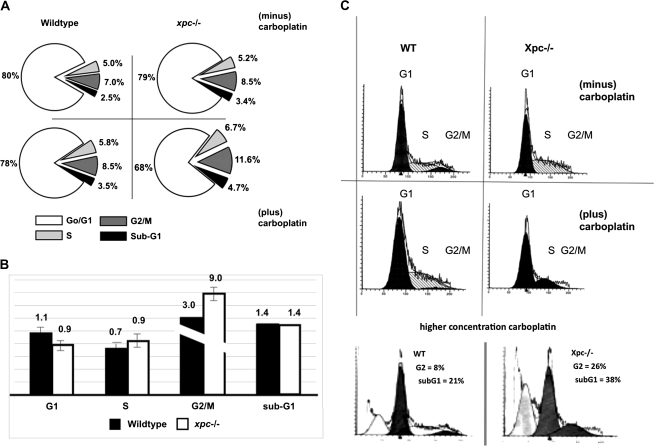
We reasoned that the Xpc−/− DNA repair defect might affect the cell cycle as a secondary consequence of lack of lesion removal. A comparison was made between wild type and Xpc−/− for cell cycle distribution in the presence or absence of carboplatin. The majority (80%) of bone marrow cells when freshly harvested were in the G1 phase of the cell cycle irrespective of genotype (Figure 2A). By PI staining alone, no cell cycle defect of Xpc−/− bone marrow or difference from wild type was observed in any cell cycle phase (Figure 2A). To further analyse the cell cycle, we grew the cells for 48 h in the presence of BrdU, thereby labelling proliferating cells. Cells were then treated with 10-μM carboplatin for 15 h, fixed and stained for PI. Cells were gated for BrdU positivity using a fluorescein-labelled antibody and then for PI staining. A modest but significant decrease in the G1 phase was observed in Xpc−/− compared to wild type (Figure 2B). A G2 arrest by carboplatin was observed in both genotypes, which was more pronounced in Xpc−/− than in wild type (Figure 2B). The flow cytometric profiles corresponding to the bar graph in Figure 2B are shown in Figure 2C. The intent of the cell cycle experiments was to reveal cell cycle differences between wild-type and Xpc−/− bone marrow, while minimally causing apoptosis or other cell death. However, use of a higher concentration of carboplatin revealed a G2 population of 8% in the wild type and 26% in the Xpc−/−. Moreover, the sub-G1 apoptotic populations were 21% for the wild type and 38% for Xpc−/−. Thus, G2 arrest may precede apoptosis, both of which were more pronounced in the Xpc−/− mutant bone marrow compared to wild type (Figure 2C).
Fig. 2.
Evidence of G1 and G2 cell cycle checkpoint alterations in cultured Xpc−/− bone marrow. (A) Bone marrow harvested from untreated 10-week-old mice was cultured in cytokine-containing medium for 15 h and then treated with 10-μM carboplatin for an additional 24 h. Cells were fixed in 70% ethanol and analysed by PI staining. By PI staining alone, cell cycle differences were not significant and 68–80% of cells were in G1. (B) Bone marrow harvested from untreated 10-week-old mice was stimulated with cytokines and cultured for 48 h in the presence of 10-μM BrdU to label proliferating cells, then treated with 10-μM carboplatin for 15 h and then fixed and stained with PI. A fluorescein isothiocyanate-conjugated antibody to BrdU was used to gate the BrdU-labelled population, which were then assayed for PI content. Values for carboplatin-treated bone marrow were divided by values for untreated bone marrow conducted side by side. The data represent three pooled mice of each genotype. Two separate experiments yielded similar results. A modest but significant decrease in G1 population was observed in Xpc−/− mice compared to wild-type mice (P < 0.05 by t-test). A significant increase was observed in the G2 population in Xpc−/− mice compared to wild-type mice (P < 0.02 by t-test). The plot shows relative cell cycle distribution after carboplatin treatment. (C) Raw flow cytometric data corresponding to the bar graph shown in panel (B). Cell number is plotted on the y-axis, at least 15 000 events per sample; PI staining is shown on the x-axis. Use of a higher concentration of carboplatin (40 μM) revealed a sub-G1 apoptotic population which was more pronounced in the Xpc−/− mutant bone marrow. The G2 population was also more pronounced in the mutant at the higher carboplatin concentration.
We examined several key cell cycle regulatory proteins for differences between Xpc−/− and wild type, including Rb, cyclin D1 and cyclin E, which were unremarkable (results not shown). Our analyses by western blotting would reveal not only protein steady-state levels but also phosphorylation and/or other modifications that might be affected as a secondary consequence of lack of lesion removal in the Xpc−/− mutant. Interesting results were obtained for CUL4A and CDT1 cell cycle checkpoint proteins, which are involved in DNA damage signalling. The rationale for studying CUL4A and CDT1 was that XPC is a key substrate for CUL4A after DNA damage; thus, in the absence of XPC, other downstream targets including CDT1 might be either exaggerated or attenuated. Higher molecular weight forms of CUL4A and CDT1 were largely absent in Xpc−/− bone marrow suggesting that Xpc may play a role in the DNA damage signalling mechanism (Figure 3A). The differences in higher molecular weight CUL4a and CDT1 were observed in untreated, freshly harvested bone marrow. Both CUL4A and its downstream substrate CDT1 are ubiquitinated as evidenced by binding to a ubiquitin-binding resin (Figure 3A), although we do not exclude other possible post-translational modifications that may occur as well. The differences are not merely reflective of cell cycle differences, as cell cycle differences in untreated cells are not significant (Figure 2A). The differences are not merely reflective of bone marrow cell subpopulations, which could theoretically differ between wild type and Xpc−/−. No significant differences were observed between wild type and Xpc−/− for any of the six lineage markers shown in Figure 1A. Thus, the apparent defective ubiquitination of CUL4A and its downstream substrate CDT1 appears to be a characteristic of Xpc−/− bone marrow. We used transiently transfected H1299 cells to test if knocking down Xpc would mirror the mouse bone marrow findings. H1299 cells in which Xpc was knocked down by an shRNA plasmid showed a decrease in ubiquitinated CDT1, similar to that observed in the bone marrow (Figure 3B). XPC may therefore affect DNA damage signalling to the cell cycle checkpoint protein CDT1, although further studies are needed.
Having conducted cell culture experiments to characterize Xpc−/− bone marrow (Figures 1–3), we conducted experiments to address carboplatin sensitivity in vivo. Twenty mice of each genotype were divided into four treatment groups and administered carboplatin at weekly intervals. Mice were 10 weeks old at the time of initiating the experiment. By day 41 of carboplatin regimen, 12/20 Xpc−/− mice had died unexpectedly. No deaths were observed in wild-type mice receiving carboplatin (Figure 4A). No deaths were observed in saline-only control groups of either genotype (results not shown). During the course of carboplatin treatments, peripheral WBC counts were measured weekly (6). Peripheral WBC counts in Xpc−/− mice were significantly lower than in wild-type mice (Figure 4B). Remaining mice were sacrificed on or after day 41 using approved protocols. Mice were evaluated by a veterinary pathologist. The most significant pathology was marked hypocellularity in bone marrow in 10/10 Xpc−/− mice examined by the pathologist (Figure 4C). Brain lesions were observed in ∼50% of the mice which, however, occurred with equal frequency in wild type and Xpc−/− (results not shown). No significant lesions were found in cecum, colon, duodenum, heart, ileum, jejunum, kidney, liver, lymph nodes, spleen, stomach or thymus. Perivascular lymphoid infiltrates were observed in lung in both genotypes (results not shown).
To quantify and further characterize the hypocellularity in Xpc−/− bone marrow, we conducted colony-forming assays and cell viability assays using a vital dye. A hallmark of myeloid stem/progenitor cell populations is their colony-forming ability when placed in culture and stimulated with cytokines. Colony-forming assays of bone marrow were conducted as in ref. (7). The data were plotted as total colonies per femur. In the absence of carboplatin, Xpc−/− mice exhibited a 3-fold decrease in colony-forming units compared to wild type (Figure 5A). With carboplatin, the difference was 10- to 12-fold (Figure 5A). We also used the vital stain thiazolyl blue to measure cell viability after 4 days in culture. Using this method, carboplatin concentrations corresponding to 50% cell yield (known as IC50 dose) was approximately 10-fold lower in Xpc−/− compared to wild type (Figure 5B). It is likely that a higher fraction of apoptosis contributed to decreased cell yield in Xpc−/− bone marrow, evidenced by the sub-G1 (apoptotic) fractions shown in Figure 2C. We also conducted experiments utilizing annexin V, an indicator of apoptosis that is assayed by flow cytometry. The Xpc−/− bone marrow showed a higher apoptotic fraction by annexin V staining (results not shown), although neither of these findings of apoptosis can exclude other mechanisms such as irreversible cell cycle arrests that may also contribute to overall cell yield.
Untreated Xpc−/− mice always yielded 2- to 3-fold fewer cells per femur compared to wild type. We used equal numbers of viable cells for cell culture experiments shown in Figures 1–3, while Figures 4 and 5 reflect the difference in overall bone marrow yield between wild type and Xpc−/−. All six lineage markers were decreased in proportion to total cell yield in Xpc−/− as compared to wild type, i.e. no single lineage was uniquely affected. Table I shows raw cell numbers of distinct lineages, per femur, recovered from bone marrow of wild type and Xpc−/− genotypes after 41 days receiving either saline only or carboplatin. The Gr-1+, B220+, Lin−, CD4+ and CD8+ lineages differed significantly between wild type and Xpc−/−, as indicated by P values shown in Table I.
Table I.
Lineage marker distribution after six weeks carboplatin administration
| Genotype | WT |
Xpc−/− |
|||
| Treatment | Saline | Carboplatin | Saline | Carboplatin | |
| Gr-1+ | P < 0.005a | 5.7 (±0.3) × 106 | 1.6 (±0.1) × 106 | 3.5 (±0.5) × 106 | 0.3 (±0.1) × 106 |
| B220+ | P < 0.005a | 1.5 (±0.2) × 106 | 6.0 (±1.5) × 105 | 7.4 (±0.3) × 105 | 1.5 (±0.1) × 105 |
| Lin− | P < 0.008a | 3.0 (±0.5) × 105 | 0.8 (±0.2) × 105 | 1.8 (±0.3) × 105 | 0.3 (±0.1) × 105 |
| CD4+ | P < 0.05a | 2.1 (±0.4) × 105 | 1.6 (±0.2) × 105 | 1.4 (±0.2) × 105 | 1.2 (±0.7) × 105 |
| CD8+ | P < 0.05a | 3.3 (±0.5) × 105 | 1.3 (±0.1) × 105 | 1.9 (±0.2) × 105 | 1.0 (±0.1) × 105 |
| Lin−/Sca1+/c-kit+ | Not significant | 2.0 (±0.4) × 104 | 2.1 (±0.5) × 104 | 3.0 (±0.5) × 104 | 0.3 (±0.1) × 104 |
Indicates statistical significance for the comparison of wild type and Xpc−/− for carboplatin treatment groups; values represent the mean and standard deviation of three mice per group.
Discussion
It has long been thought that, in contrast to TC-NER, G-NER mediated by XPC contributes only modestly to cell survival after DNA damage (8). Essentially all studies were conducted in human or mouse fibroblasts which showed 2- to 3-fold differences in cell survival after DNA damage (8–12). The present study is the first to examine bone marrow in Xpc−/− mice. Xpc−/− bone marrow exhibited 10-fold greater sensitivity to carboplatin compared to strain-matched wild-type bone marrow (Figures 4 and 5 and Table I). The data indicate that the Xpc gene can contribute substantially (10-fold) to cell survival in bone marrow. The cell survival end point was biologically relevant as bone marrow myelosuppression was dose limiting for carboplatin, consistent with other studies (13) and the majority of Xpc−/− mice died during the course of the experiments (Figures 4 and 5). No deaths were observed in wild-type mice irrespective of the carboplatin regimen and/or duration (Figure 4 and results not shown).
Carboplatin and cisplatin produce a similar DNA damage spectrum, mainly 1,2 and 1,3 platinum diadducts that are substrates for NER in vitro (14,15). However, our data do not exclude that minor lesions such as platinum monoadducts, interstrand cross-links and/or base damage by reactive oxygen species may also contribute to the cell survival end point. Recent studies point to a role for XPC in repair of oxidative DNA damage (12). A catalytic role for XPC in base-excision DNA repair (BER) was suggested by the finding that XPC enhanced the activity of the DNA glycosylase 8-oxoguanine DNA glycosylate (OGG1) (12). Colony-forming ability of Xpc−/− bone marrow was decreased 3-fold compared to wild type even in the absence of carboplatin (Figure 5A). As endogenous base damage is removed by the separate BER pathway, the decreased cell yield in Xpc−/− mice may be due to the XPC role in BER (12). Interestingly, Xpc−/− fibroblasts exhibited sensitivity to a 20% oxygen environment compared to wild-type fibroblasts consistent with a role in base damage repair (11).
Cell survival though is a complex end point that involves cell cycle effects. We found that Xpc−/− bone marrow cells were defective in a G1 checkpoints and showed increased accumulation in G2 compared to wild type (Figure 2). One possibility is that Xpc−/− bone marrow cells exit the cell cycle after a prolonged G2 or may be irreversibly arrested in G2 (Figure 2). We show that CUL4A and CDT1 cell cycle proteins exhibit defective ubiquitination even in untreated Xpc−/− mice. The data suggest an alteration in DNA damage signalling in Xpc−/− bone marrow (Figure 3), although further studies are warranted. Given that these data were obtained in the absence of carboplatin, it is possible that XPC signalling of oxidative base damage involves the CUL4A/CDT1 mechanism (16–18). Thus, XPC may contribute to DNA damage sensitivity including base damage by (i) controlling the BER protein OGG (12), (ii) altering cell cycles and (iii) affecting DNA damage signalling to downstream effectors including CUL4A and CDT1. Our data pertain to bone marrow; thus, it is not known if absence of functional XPC would exhibit these same features in other cell types.
Human patients with xeroderma pigmentosum XPC mutant alleles typically develop skin cancers well prior to age 20 and develop internal cancers with ageing. Similarly, Xpc−/− mice develop spontaneous lung cancers at ∼1 year of age (4). We used mice as young as possible for the current study. Perhaps human XPC patients would exhibit myelosuppression were they to live to advanced age. On the other hand, the mice are completely null for Xpc genes, while the human mutant XPC alleles probably exhibit variable penetrance. One study did show myelosuppression in an XPC patient (19). Thus, cancer therapeutic regimens may require modification in the case of XPC patients.
In summary, the Xpc gene product protects mouse bone marrow from exogenous and probably endogenous DNA damage with cell survival as an end point. The biological relevance is indicated by the fact that 12/20 Xpc−/− mice receiving carboplatin died while 0/20 wild-type mice died. No deaths were observed in saline-only controls of either genotype. The proximal cause of death appeared to be bone marrow failure, although other tissues may have contributed. The XPC protein may be linked to the CUL4A/CDT1 cell cycle checkpoint, probably relevant to DNA damage signalling and may contribute to the cell death mechanism.
Funding
National Institutes of Health (R01 HL086978) to M.L.S.; American Institute for Cancer Research (04B010) to M.L.S.; US Department of Defense (BC051172) to J.L.F. T.M.H. was supported by 1R25 GM079657 training grant to Dr. Hal E. Broxmeyer.
Acknowledgments
We thank Mark R. Kelley, Laura Haneline and D. Wade Clapp for comments on the figures. We thank four anonymous reviewers for their helpful comments.
Conflict of interest statement: None declared.
References
- 1.Friedberg EC, Aguilera AM, Gellert M, et al. DNA repair, from molecular mechanism to human disease. DNA Repair (Amst.) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Sands AT, Abuin A, Sanchez A, Conti CJ, Bradley A. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature. 1995;377:162–165. doi: 10.1038/377162a0. [DOI] [PubMed] [Google Scholar]
- 3.Berg RJ, Ruven HJ, Sands AT, deGruiji FR, Mullenders LH. Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J. Invest. Dermatol. 1998;110:405–409. doi: 10.1111/j.1523-1747.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 4.Hollander MC, Philburn RT, Patterson AD, Velasco-Miguel S, Friedberg EC, Linnoila RI, Fornace AJ., Jr Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc. Natl Acad. Sci USA. 2006;102:13200–13205. doi: 10.1073/pnas.0503133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf T, Densmore JJ. Pegfilgrastim use during chemotherapy: current and future applications. Curr. Hematol. Rep. 2004;3:419–423. [PubMed] [Google Scholar]
- 6.Cai S, Ernstberger A, Wang H, et al. In vivo selection of hematopoietic stem cells transduced at a low multiplicity-of-infection with a foamy viral MGMT(P140K) vector. Exp. Hematol. 2008;36:283–292. doi: 10.1016/j.exphem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haneline LS, Broxmeyer HE, Cooper S, Hangoc G, Carreau M, Buchwald M, Clapp DW. Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from Fac−/− mice. Blood. 1998;91:4092–4098. [PubMed] [Google Scholar]
- 8.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 9.Muotri AR, Marchetto MC, Zerbini LF, Libermann TA, Ventura AM, Sarasin A, Menck CF. Complementation of the DNA repair deficiency in human xeroderma pigmentosum group A and C cells by recombinant adenovirus-mediated gene transfer. Hum. Gene Ther. 2002;13:1833–1844. doi: 10.1089/104303402760372936. [DOI] [PubMed] [Google Scholar]
- 10.Anaudeau-Begard C, Brellier F, Chevallier-Lagente O, Hoeijmakers J, Bernerd F, Sarasin A, Magnaldo T. Genetic correction of DNA repair-deficient/cancer-prone xeroderma pigmentosum group C keratinocytes. Hum. Gene Ther. 2003;14:983–996. doi: 10.1089/104303403766682241. [DOI] [PubMed] [Google Scholar]
- 11.Melis JP, Wijnhoven SWW, Beems RB, et al. Mouse models for xeroderma pigmentosum group A and group C show divergent cancer phenotypes. Cancer Res. 2008;68:1347–1353. doi: 10.1158/0008-5472.CAN-07-6067. [DOI] [PubMed] [Google Scholar]
- 12.D'Errico M, Parlanti E, Teson M, Bernardes de Jesus BM, et al. New functions of Xpc in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colby C, Koziol S, McAfee SL, Yeap B, Spitzer TR. High-dose carboplatin and regimen-related toxicity following autologous bone marrow transplant. Bone Marrow Transplant. 2002;29:467–472. doi: 10.1038/sj.bmt.1703417. [DOI] [PubMed] [Google Scholar]
- 14.Shivji MK, Moggs JG, Kuraoka I, Wood RD. Assaying for the dual incisions of nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 2006;314:435–456. doi: 10.1385/1-59259-973-7:435. [DOI] [PubMed] [Google Scholar]
- 15.Trego KS, Turchi JJ. Pre-steady state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45:1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Xiong Y. Targeted ubiquitination of Cdt1 by the DDB1-Cul4a-Roc1 ligase in response to DNA damage. J. Biol. Chem. 2006;281:3753–3756. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 17.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 18.Sugasawa K, Okuda Y, Saijo M, et al. UV-induced ubiquitinylation of Xpc protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 19.Salob SP, Webb DKH, Atherton DJ. A child with xeroderma pigmentosum and bone marrow failure. Br. J. Dermatol. 1992;126:372–374. doi: 10.1111/j.1365-2133.1992.tb00681.x. [DOI] [PubMed] [Google Scholar]