Abstract
Both occupational exposure to the leukemogen benzene and in vitro exposure to its metabolite hydroquinone (HQ) lead to the induction of numerical and structural chromosome changes. Several studies have shown that HQ can form DNA adducts, disrupt microtubule assembly and inhibit DNA topoisomerase II (topo II) activity. As these are potential mechanisms underlying endoreduplication (END), a phenomenon that involves DNA amplification without corresponding cell division, we hypothesized that HQ could cause END. We measured END in the human lymphoblastoid cell line, TK6, treated with HQ (0–20 μM) and etoposide (0–0.2 μM) for 48 h. Etoposide was used as a positive control as it is a topo II poison and established human leukemogen that has previously been shown to induce END in Chinese hamster ovary cells. Both HQ and etoposide significantly induced END in a dose-dependent manner (Ptrend < 0.0001 and Ptrend = 0.0003, respectively). Since END may underlie the acquisition of high chromosome numbers by tumour cells, it may play a role in inducing genomic instability and subsequent carcinogenesis from HQ and etoposide. In order to further explore the cytogenetic effects of HQ and etoposide, we also examined specific structural changes. HQ did not induce translocations of chromosome 11 [t(11;?)] but significantly induced translocations of chromosome 21 [t(21;?)] and structural chromosome aberrations (SCA) (Ptrend = 0.0415 and Ptrend < 0.0001, respectively). Etoposide potently induced all these structural changes (Ptrend < 0.0001). The lack of an effect of HQ on t(11;?) and the reduced ability of HQ to induce t(21;?) and SCA, compared with etoposide, further suggests that HQ acts primarily as a topo II catalytic inhibitor rather than as a topo II poison in intact human cells.
Introduction
Benzene, an important industrial chemical and a universal environmental pollutant, is a known human carcinogen (1). Chronic benzene exposure is an established cause of acute myeloid leukaemia (AML) (2,3) and probably causes non-Hodgkin lymphoma (NHL) (4,5). The carcinogenic effects of benzene are not induced directly but are thought to be mediated by a series of phenolic- and quinone-based metabolites, one of which is hydroquinone (HQ) (6). Etoposide, a chemotherapeutic agent employed in the therapy of a wide spectrum of cancers, is also an established human carcinogen (7), causing mainly AML (8).
It is widely accepted that cytogenetic changes may play important roles in the carcinogenicity of benzene and etoposide. Therapy-related AML associated with previous exposure to etoposide is mainly characterized by chromosomal rearrangements involving the mixed lineage leukaemia (MLL) gene on chromosome band 11q23 (9,10). Most MLL rearrangements arise in a region rich in putative DNA topoisomerase II (topo II) cleavage recognition sequences (11). Topo II is an essential nuclear enzyme that catalyzes changes in DNA topology via its cleavage–religation equilibrium (12). Etoposide, a well-established topo II poison, disrupts this equilibrium, and MLL is particularly susceptible to aberrant cleavage and homology-mediated fusion to repetitive elements located on novel chromosome partners (13). As well as translocations of chromosome 11q23, translocations of chromosome 21q22 (10,14) are also commonly observed in etoposide-associated therapy-related AML, and both types of translocation are thought to be causal in the pathogenesis of the disease (15).
Benzene and its metabolites can induce chromosomal damage in vivo and in vitro. We previously reported that benzene causes chromosomal changes commonly associated with AML (16,17) and with NHL (18), including translocations and long-arm deletions, in the blood lymphocytes of exposed workers. We have also demonstrated the induction of non-random aneuploidy in benzene-exposed workers (19). The benzene metabolite HQ has been shown to induce micronuclei in human lymphocytes (20,21) and in a myeloid cell line HL60 (22). We have also demonstrated that HQ can induce chromosomal changes commonly associated with AML (23) and non-random aneuploidy in human lymphocytes (24). The mechanisms underlying the cytogenetic changes induced by benzene are not established but may include the formation of DNA adducts, disruption of microtubule assembly and inhibition of topo II activity, all of which have been shown to be induced by HQ (25–31).
Endoreduplication (END) in eukaryotes is a process that involves DNA amplification without corresponding cell division. In END cell cycles, or endocyles, S phases alternate with distinct gap phases that lack DNA replication, but there is no cell division. Many endocycling cells go directly to the next G1/S phase lacking all vestiges of mitosis, but some retain hallmarks of mitosis (32). The visible mitotic manifestation of the phenomenon of END is diplochromosomes, consisting of four chromatids held together lying side by side, instead of the normal two, as a result of the occurrence of two successive cycles of DNA replication without intervening mitosis, i.e. segregation of daughter chromatids (33). Several mechanisms have been proposed for the induction of END, including DNA damage/modification, cytoskeleton disturbance and topo II inhibition (34), which as discussed above, are potential mechanisms of action of HQ. Etoposide has been shown to induce END in Chinese hamster ovary cells (35). Therefore, we hypothesized that both HQ and etoposide could induce END in human cells and examined END in the human lymphoblastoid cell line, TK6, treated with HQ and etoposide. Our previous human studies showed that benzene exposure leads to the induction of translocations of chromosome 21 [t(21;?)] (16) but not translocations of chromosome 11 [t(11;?)] (18), the major alteration associated with etoposide. In order to confirm the in vivo findings, we further examined t(11;?), t(21;?) and structural chromosome aberrations (SCA) in TK6 cells treated with HQ and etoposide.
Materials and methods
Cell culture, chemical treatment and cytotoxicity
The human lymphoblastoid cell line, TK6, was maintained in RPMI 1640 medium (GIBCO, San Diego, CA) containing 10% foetal bovine serum (Omega Scientific, Tarzana, CA) and 1% penicillin and streptomycin (Omega Scientific) at 37°C in a 5% CO2 moist atmosphere. HQ (≥99%; Aldrich, Milwaukee, WI) was dissolved in sterile 1× PBS immediately prior to treatment for all experiments. Dimethyl sulfoxide was used as the vehicle for etoposide (≥98%, Sigma–Aldrich, St Louis, MO) and was present in cell cultures at a final concentration of 0.1%. TK6 cells were treated with 0, 5, 10, 15 or 20 μM HQ and 0, 0.05, 0.1 or 0.2 μM etoposide for 48 h. Cytotoxicity was measured by the trypan blue exclusion assay.
Metaphase preparation
In order to obtain a sufficient number of metaphase spreads, colcemid (0.1 μg/ml, Gibco BRL, Gaithersburg, MD) was added to each culture 2 h before harvesting. After hypotonic treatment (0.075 M KCl) for 30 min at 37°C, the cells were fixed three times with freshly prepared Carnoy's fixative (methanol:glacial acetic acid = 3:1). The fixed cells were dropped onto glass slides, allowed to air dry and stored at −20°C under a nitrogen atmosphere until use.
END detection
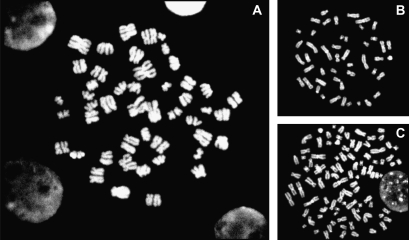
The cells on the slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) and metaphase spreads were scanned and localized automatically using Metafer software (MetaSystems, Altlussheim, Germany). Metaphases were scored at ×1000 magnification to detect END. Metaphase spreads were considered scorable if the cells appeared intact with the chromosomes well spread and condensed and the centromeres and chromatids readily visible. As shown in Figure 1, diplochromosomes, made up of two parallel chromosomes with four chromatids held together lying side by side, are the visible mitotic manifestation of END.
Fig. 1.
Visible mitotic manifestation (×1000) of END. (A) END, (B) normal metaphase and (C) tetraploidy. Diplochromosomes, made up of two parallel chromosomes with four chromatids held together lying side by side, are the visible mitotic manifestation of END.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) was used to detect t(11;?) and t(21;?). Whole-chromosome painting probes for chromosome 11 (directly labelled with SpectrumGreen) (Vysis Inc., Downers Grove, IL) and for chromosome 21 (labelled with digoxigenin) (Oncor Inc., Gaithersburg, MD) were used. Metaphase spreads on each slide were scanned and localized automatically using Metafer software (MetaSystems) following DAPI staining. The FISH procedure was performed as previously described in detail (16).
SCA detection
The cells on the slides were stained with DAPI and metaphase spreads were scanned and localized automatically using Metafer software (MetaSystems). Metaphases were scored at ×1000 magnification to detect SCA, which includes break, acentric fragment, triradial, quadriradial, complex exchange, dicentric chromosome, ring chromosome and marker chromosome according to An International System for Human Cytogenetic Nomenclature (36).
Statistical analysis
For the rare events, END and t(21;?), data from multiple experiments were pooled. Fisher's exact test was applied to test the difference between each dose and control, and a chi-square trend test was used to calculate the Ptrend. For t(11;?) and SCA, Poisson regression was applied to test the difference between each dose and control and calculate the Ptrend.
Results
HQ and etoposide both significantly induced END
Table I shows the frequency of END in TK6 cells following treatment with HQ and etoposide in four independent experiments. Following HQ treatment, END events were detected at each dose and the END frequencies at 15 and 20 μM HQ were significantly higher than that of the control (P < 0.001 and P < 0.0001, respectively). The trend test showed that the END induction by HQ occurred in a dose-dependent manner (Ptrend < 0.0001). Similarly, etoposide treatment induced END at all doses and the END frequency at 0.2 μM etoposide was significantly higher than that of the vehicle control (P < 0.01). As with HQ, induction of END by etoposide occurred in a dose-dependent manner (Ptrend = 0.0003).
Table I.
END induced by HQ and etoposide in TK6 cells
| Doses (μM) | Cells | END (‰) |
| HQ | ||
| 0 | 17 226 | 0 (0.00) |
| 5 | 17 651 | 1 (0.06) |
| 10 | 18 523 | 5 (0.27) |
| 15 | 17 731 | 13 (0.73)*** |
| 20 | 12 111 | 12 (0.99)**** |
| Ptrend | <0.0001 | |
| Etoposide | ||
| 0 | 17 252 | 0 (0.00) |
| 0.05 | 19 896 | 2 (0.10) |
| 0.1 | 20 605 | 3 (0.15) |
| 0.2 | 20 168 | 11 (0.55)** |
| Ptrend | 0.0003 |
**, *** and **** represent P < 0.01, P < 0.001 and P < 0.0001 compared to control, respectively. Ptrend represents the P value of the trend test.
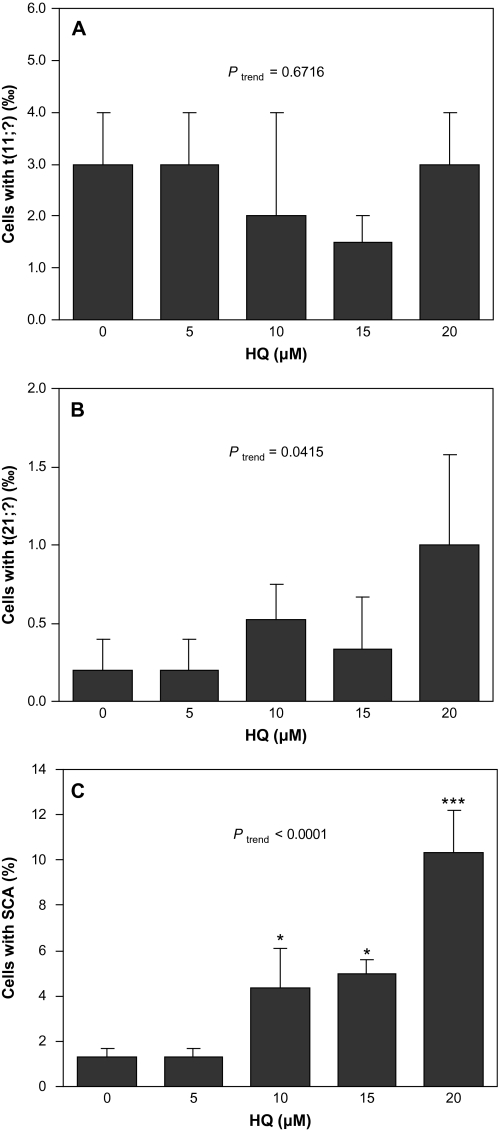
HQ did not induce t(11;?) but significantly induced t(21;?) and SCA
Figure 2 shows the frequency of cells with t(11;?) (A), t(21;?) (B) and SCA (C) in TK6 cells following treatment with HQ. HQ did not induce t(11;?) at any dose examined compared with the control, and the dose–response was not significant (Ptrend = 0.67). The frequency of cells with t(21;?) at 20 μM (1.20‰) was 7-fold higher than that in the control (0.17‰), though the difference was not significant due to the low frequency of events. However, a trend test indicated that HQ treatment significantly induced t(21;?) in a dose-dependent manner (Ptrend = 0.0415). Following HQ treatment, the frequency of cells with SCA at 10, 15 and 20 μM HQ was significantly higher than that of the control (P < 0.05, P < 0.05 and P < 0.001, respectively). The trend test showed that SCA induced by HQ occurred in a dose-dependent manner (Ptrend < 0.0001).
Fig. 2.
t(11;?), t(21;?) and SCA induced by HQ in TK6 cells. Data presented are mean frequency. Error bar represents standard error and * and *** represent P < 0.05 and P < 0.001 compared to control, respectively. Ptrend represents the P value of the trend test. (A) t(11;?) induced by HQ. Two independent experiments were conducted and 2000 metaphases were scored for each dose. (B) t(21;?) induced by HQ. Five independent experiments were conducted and 6015, 6031, 5681, 2997 and 2504 metaphases were scored for 0, 5, 10, 15 and 20 μM HQ, respectively. (C) SCA induced by HQ. Three independent experiments were conducted and 300 metaphases were scored for each dose.
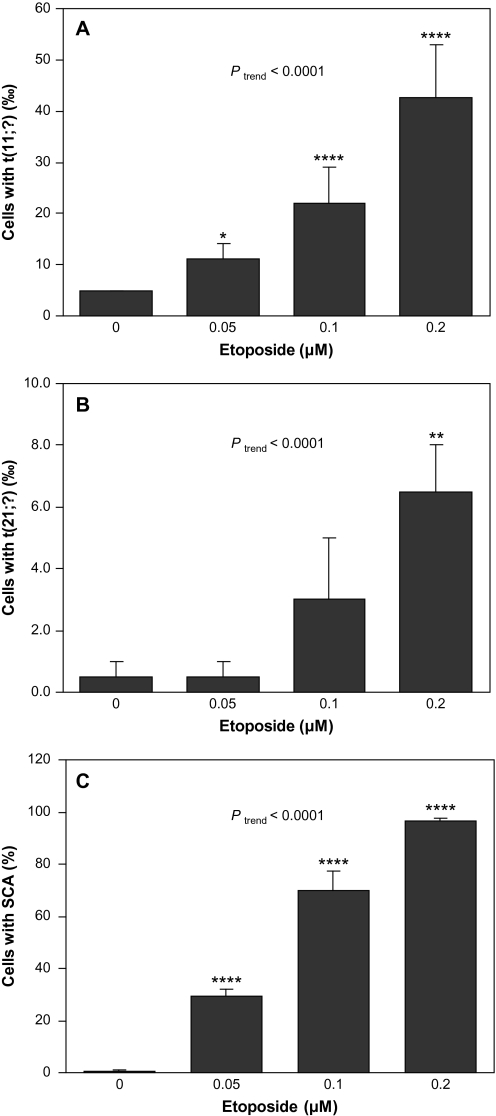
Etoposide significantly induced t(11;?), t(21;?) and SCA
Figure 3 shows the frequency of cells with t(11;?) (A), t(21;?) (B) and SCA (C) in TK6 cells following treatment with etoposide. The frequency of cells with t(11;?) was significantly higher at all doses relative to the control and the induction occurred in a dose-dependent manner (Ptrend < 0.0001). The frequency of cells with t(21;?) at 0.2 μM etoposide was significantly higher than that in the control (P < 0.01) and the induction was dose dependent (Ptrend < 0.0001). Following etoposide treatment, the frequency of cells with SCA was significantly higher at all doses relative to the control and increased in a dose-dependent manner (Ptrend < 0.0001). While HQ and etoposide both significantly induced SCA, etoposide was much more potent than HQ. For example, the percentage of cells with SCA after 0.1 μM etoposide treatment (69.7%) was 16-fold higher than after 10 μM HQ treatment (4.3%), even though the cell viability at 0.1 μM etoposide (85%) was similar to that at 10 μM HQ (81%).
Fig. 3.
t(11;?), t(21;?) and SCA induced by etoposide in TK6 cells. Data presented are mean frequency. Error bar represents standard error and *, ** and **** represent P < 0.05, P < 0.01 and P < 0.0001 compared to control, respectively. Ptrend represents the P value of the trend test. (A) t(11;?) induced by etoposide. Two independent experiments were conducted and 2000 metaphases were scored for each dose. (B) t(21;?) induced by etoposide. Two independent experiments were conducted and 2000 metaphases were scored for each dose. (C) SCA induced by etoposide. Three independent experiments were conducted and 300 metaphases were scored for each dose.
Discussion
END, first defined by Levan and Hauschka (33), has attracted the attention of investigators in cytogenetics and cell cycle biochemistry. It is a process that involves DNA amplification without corresponding cell division. Though it is a common event in plants (37), spontaneous END is rarely observed in mammals, generally as a characteristic feature of specific tissues such as liver (38), tonsils (39) and trophoblast giant cells of the placenta (40). The presence of diplochromosomes, the visible mitotic manifestation of END, indicates incomplete cell division. It has been proposed that END may underlie the acquisition of high chromosome numbers by tumour cells (41), pointing to a possible link between END and carcinogenesis.
In the present study, we found that both HQ and etoposide significantly induced END in human TK6 cells in a dose-dependent manner. To our knowledge, this is the first report that the benzene metabolite, HQ, can induce END in any cell type. Etoposide was previously shown to induce END in Chinese hamster ovary cells (35) and is shown here to do so in human cells.
The three major mechanisms proposed to underlie the induction of END including DNA damage/modification, cytoskeleton disturbance and topo II inhibition have been reviewed (34). Topo II inhibition is one widely accepted pathway since both poisons and catalytic inhibitors of topo II have been consistently shown to induce END in Chinese hamster cells (35,42–46). Etoposide is an established topo II poison, so the inhibition of the enzyme may account for the induction of END in TK6 cells. HQ has been shown to form DNA adducts (25,26), disrupt microtubule assembly (27,28) and inhibit topo II activity (29–31). Any of these mechanisms may cause END induction by HQ in TK6 cells.
In the present study, HQ and etoposide differed in their ability to induce t(11;?) and t(21;?) and SCA in TK6 cells, in agreement with findings in the respective human studies. We found that HQ significantly induced t(21;?) but did not induce t(11;?) even at doses producing high cytotoxicity (data not shown). Similarly, in benzene-exposed workers, t(21;?) (16), but not t(11;?) (18), was elevated by benzene exposure compared to controls. In the present study, etoposide significantly induced t(11;?) and t(21;?) in a dose-dependent manner, findings compatible with those reported in etoposide-treated patients (9,10,14). HQ significantly induced SCA in a dose-dependent manner, a finding consistent with that in benzene-exposed workers (47), though much less potently than etoposide. Although etoposide and HQ exhibited differences in the degree of induction of translocations and SCA, it should be noted that as all analyses were performed at 48 h, an underestimation of the frequency of SCA and a relative over-representation of the more stable translocations is possible.
The lack of an effect of HQ on t(11;?) and the reduced ability of HQ to induce t(21;?) and SCA, compared with etoposide, suggest that HQ acts mainly as a topo II catalytic inhibitor rather than as a poison in intact human cells. HQ can be further converted to p-benzoquinone (BQ) by myeloperoxidase (6) or by autooxidation in absence of the enzyme (48,49). It has been shown that bioactivation of HQ by peroxidase to BQ enhances topo II inhibition (31). Indeed, BQ was shown to be a more potent topo II inhibitor than HQ in a cell-free assay system (29,50). However, it has been unclear whether HQ/BQ acts as a poison or through catalytic inhibition of topo II (29,50,51). Since TK6 cells are myeloperoxidase negative, the cytogenetic changes observed here are likely to be induced mainly by HQ. Further studies comparing the effects of HQ and etoposide in a cell line with myeloperoxidase activity or comparing BQ with etoposide could help to clarify the contribution of topo II inhibition in the mechanism of action of benzene and its metabolites.
The usefulness of TK6 cells to measure genotoxicity has been demonstrated by several groups who have successfully measured instability induced by exposures in these cells (52,53) and found them to have a relatively stable karyotype (54). However, others have found a high degree of genomic instability in TK6 cells (55). While we observed a low background of chromosome aberrations in TK6 cells in the present study, suggesting a low rate of basal instability, the observed effects should be validated in other cell lines or human lymphocytes exposed to HQ and etoposide.
In conclusion, we report that both HQ and etoposide can induce END in human TK6 cells suggesting that END may play a role in inducing genomic instability and subsequent carcinogenesis from HQ and etoposide.
Funding
National Institutes of Health (P42ES004705 and R01ES006721) to M.T.S.
Acknowledgments
We are grateful to Angela Park and Dustin Hang for their laboratory assistance.
Conflict of interest statement: Dr. Smith has received consulting and expert testimony fees from lawyers representing both plaintiffs and defendants in cases involving claims related to exposure to benzene.
References
- 1.IARC. Benzene, IARC Monographs on the Evaluation of Carcinogenic Risks to Man. Vol. 29. Lyon, France: International Agency for Research on Cancer; 1982. pp. 93–148. [Google Scholar]
- 2.Hayes RB, Songnian Y, Dosemeci M, Linet M. Benzene and lymphohematopoietic malignancies in humans. Am. J. Ind. Med. 2001;40:117–126. doi: 10.1002/ajim.1078. [DOI] [PubMed] [Google Scholar]
- 3.Schnatter AR, Rosamilia K, Wojcik NC. Review of the literature on benzene exposure and leukemia subtypes. Chem. Biol. Interact. 2005;153–154:9–21. doi: 10.1016/j.cbi.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 4.Smith MT, Jones RM, Smith AH. Benzene exposure and risk of non-Hodgkin lymphoma. Cancer Epidemiol. Biomarkers Prev. 2007;16:385–391. doi: 10.1158/1055-9965.EPI-06-1057. [DOI] [PubMed] [Google Scholar]
- 5.Steinmaus C, Smith AH, Jones RM, Smith MT. Meta-analysis of benzene exposure and non-Hodgkin lymphoma: biases could mask an important association. Occup. Environ. Med. 2008;65:371–378. doi: 10.1136/oem.2007.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross D. The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. J. Toxicol. Environ. Health A. 2000;61:357–372. doi: 10.1080/00984100050166361. [DOI] [PubMed] [Google Scholar]
- 7.IARC. DNA topoisomerase II inhibitors. IARC Monogr. Eval. Carcinog. Risks Hum. 2000;76:175–344. [PMC free article] [PubMed] [Google Scholar]
- 8.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 9.Bloomfield CD, Archer KJ, Mrozek K, Lillington DM, Kaneko Y, Head DR, Dal Cin P, Raimondi SC. 11q23 balanced chromosome aberrations in treatment-related myelodysplastic syndromes and acute leukemia: report from an international workshop. Genes Chromosomes Cancer. 2002;33:362–378. doi: 10.1002/gcc.10046. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen-Bjergaard J, Philip P. Balanced translocations involving chromosome bands 11q23 and 21q22 are highly characteristic of myelodysplasia and leukemia following therapy with cytostatic agents targeting at DNA-topoisomerase II. Blood. 1991;78:1147–1148. [PubMed] [Google Scholar]
- 11.Mirault ME, Boucher P, Tremblay A. Nucleotide-resolution mapping of topoisomerase-mediated and apoptotic DNA strand scissions at or near an MLL translocation hotspot. Am. J. Hum. Genet. 2006;79:779–791. doi: 10.1086/507791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 13.Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatr. Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 14.Slovak ML, Bedell V, Popplewell L, Arber DA, Schoch C, Slater R. 21q22 balanced chromosome aberrations in therapy-related hematopoietic disorders: report from an international workshop. Genes Chromosomes Cancer. 2002;33:379–394. doi: 10.1002/gcc.10042. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen-Bjergaard J, Christiansen DH, Desta F, Andersen MK. Alternative genetic pathways and cooperating genetic abnormalities in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2006;20:1943–1949. doi: 10.1038/sj.leu.2404381. [DOI] [PubMed] [Google Scholar]
- 16.Smith MT, Zhang L, Wang Y, et al. Increased translocations and aneusomy in chromosomes 8 and 21 among workers exposed to benzene. Cancer Res. 1998;58:2176–2181. [PubMed] [Google Scholar]
- 17.Zhang L, Rothman N, Wang Y, et al. Increased aneusomy and long arm deletion of chromosomes 5 and 7 in the lymphocytes of Chinese workers exposed to benzene. Carcinogenesis. 1998;19:1955–1961. doi: 10.1093/carcin/19.11.1955. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Rothman N, Li G, et al. Aberrations in chromosomes associated with lymphoma and therapy-related leukemia in benzene-exposed workers. Environ. Mol. Mutagen. 2007;48:467–474. doi: 10.1002/em.20306. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Lan Q, Guo W, et al. Use of OctoChrome fluorescence in situ hybridization to detect specific aneuploidy among all 24 chromosomes in benzene-exposed workers. Chem. Biol. Interact. 2005;153–154:117–122. doi: 10.1016/j.cbi.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Yager JW, Eastmond DA, Robertson ML, Paradisin WM, Smith MT. Characterization of micronuclei induced in human lymphocytes by benzene metabolites. Cancer Res. 1990;50:393–399. [PubMed] [Google Scholar]
- 21.Robertson ML, Eastmond DA, Smith MT. Two benzene metabolites, catechol and hydroquinone, produce a synergistic induction of micronuclei and toxicity in cultured human lymphocytes. Mutat. Res. 1991;249:201–209. doi: 10.1016/0027-5107(91)90147-g. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Robertson ML, Kolachana P, Davison AJ, Smith MT. Benzene metabolite, 1,2,4-benzenetriol, induces micronuclei and oxidative DNA damage in human lymphocytes and HL60 cells. Environ. Mol. Mutagen. 1993;21:339–348. doi: 10.1002/em.2850210405. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Wang Y, Shang N, Smith MT. Benzene metabolites induce the loss and long arm deletion of chromosomes 5 and 7 in human lymphocytes. Leuk. Res. 1998;22:105–113. doi: 10.1016/s0145-2126(97)00157-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Yang W, Hubbard AE, Smith MT. Nonrandom aneuploidy of chromosomes 1, 5, 6, 7, 8, 9, 11, 12, and 21 induced by the benzene metabolites hydroquinone and benzenetriol. Environ. Mol. Mutagen. 2005;45:388–396. doi: 10.1002/em.20103. [DOI] [PubMed] [Google Scholar]
- 25.Levay G, Pongracz K, Bodell WJ. Detection of DNA adducts in HL-60 cells treated with hydroquinone and p-benzoquinone by 32P-postlabeling. Carcinogenesis. 1991;12:1181–1186. doi: 10.1093/carcin/12.7.1181. [DOI] [PubMed] [Google Scholar]
- 26.Gaskell M, McLuckie KI, Farmer PB. Genotoxicity of the benzene metabolites para-benzoquinone and hydroquinone. Chem. Biol. Interact. 2005;153–154:267–270. doi: 10.1016/j.cbi.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 27.Irons RD, Neptun DA. Effects of the principal hydroxy-metabolites of benzene on microtubule polymerization. Arch. Toxicol. 1980;45:297–305. doi: 10.1007/BF00293810. [DOI] [PubMed] [Google Scholar]
- 28.Wallin M, Hartley-Asp B. Effects of potential aneuploidy inducing agents on microtubule assembly in vitro. Mutat. Res. 1993;287:17–22. doi: 10.1016/0027-5107(93)90141-2. [DOI] [PubMed] [Google Scholar]
- 29.Hutt AM, Kalf GF. Inhibition of human DNA topoisomerase II by hydroquinone and p-benzoquinone, reactive metabolites of benzene. Environ. Health Perspect. 1996;104(Suppl. 6):1265–1269. doi: 10.1289/ehp.961041265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fung J, Hoffmann MJ, Kim DD, Snyder R. Inhibition of topoisomerase II in 32D.3(G) cells by hydroquinone is associated with cell death. J. Appl. Toxicol. 2004;24:183–188. doi: 10.1002/jat.960. [DOI] [PubMed] [Google Scholar]
- 31.Eastmond DA, Mondrala ST, Hasegawa L. Topoisomerase II inhibition by myeloperoxidase-activated hydroquinone: a potential mechanism underlying the genotoxic and carcinogenic effects of benzene. Chem. Biol. Interact. 2005;153–154:207–216. doi: 10.1016/j.cbi.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 32.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 33.Levan A, Hauschka TS. Endomitotic reduplication mechanisms in ascites tumors of the mouse. J. Natl Cancer Inst. 1953;14:1–43. [PubMed] [Google Scholar]
- 34.Cortes F, Mateos S, Pastor N, Dominguez I. Toward a comprehensive model for induced endoreduplication. Life Sci. 2004;76:121–135. doi: 10.1016/j.lfs.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Sumner AT. Induction of diplochromosomes in mammalian cells by inhibitors of topoisomerase II. Chromosoma. 1998;107:486–490. doi: 10.1007/s004120050333. [DOI] [PubMed] [Google Scholar]
- 36.Shaffer LG, Tommerup N International Standing Committee on Human Cytogenetic Nomenclature. ISCN 2005: An International System for Human Cytogenetic Nomenclature (2005): Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. Basel, Switzerland: Karger; 2005. [Google Scholar]
- 37.Joubes J, Chevalier C. Endoreduplication in higher plants. Plant Mol. Biol. 2000;43:735–745. doi: 10.1023/a:1006446417196. [DOI] [PubMed] [Google Scholar]
- 38.Sigal SH, Rajvanshi P, Gorla GR, Sokhi RP, Saxena R, Gebhard DR, Jr., Reid LM, Gupta S. Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. Am. J. Physiol. 1999;276:G1260–G1272. doi: 10.1152/ajpgi.1999.276.5.G1260. [DOI] [PubMed] [Google Scholar]
- 39.Takanari H, Izutsu K. Studies on endoreduplication. II. Spontaneous occurrence and cellular kinetics of endoreduplication in PHA-stimulated tonsillar lymphocytes. Cytogenet. Cell Genet. 1981;29:77–83. doi: 10.1159/000131554. [DOI] [PubMed] [Google Scholar]
- 40.Zybina EV, Zybina TG, Stein GI. Trophoblast cell invasiveness and capability for the cell and genome reproduction in rat placenta. Early Pregnancy. 2000;4:39–57. [PubMed] [Google Scholar]
- 41.Larizza L, Schirrmacher V. Somatic cell fusion as a source of genetic rearrangement leading to metastatic variants. Cancer Metastasis Rev. 1984;3:193–222. doi: 10.1007/BF00048385. [DOI] [PubMed] [Google Scholar]
- 42.Pastor N, Jose Flores M, Dominguez I, Mateos S, Cortes F. High yield of endoreduplication induced by ICRF-193: a topoisomerase II catalytic inhibitor. Mutat. Res. 2002;516:113–120. doi: 10.1016/s1383-5718(02)00029-3. [DOI] [PubMed] [Google Scholar]
- 43.Pastor N, Cantero G, Campanella C, Cortes F. Endoreduplication induced in cultured Chinese hamster cells by different anti-topoisomerase II chemicals. Evidence for the essential contribution of the enzyme to chromosome segregation. Mutat. Res. 2005;582:11–19. doi: 10.1016/j.mrgentox.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Cantero G, Campanella C, Mateos S, Cortes F. Topoisomerase II inhibition and high yield of endoreduplication induced by the flavonoids luteolin and quercetin. Mutagenesis. 2006;21:321–325. doi: 10.1093/mutage/gel033. [DOI] [PubMed] [Google Scholar]
- 45.Cantero G, Pastor N, Mateos S, Campanella C, Cortes F. Cisplatin-induced endoreduplication in CHO cells: DNA damage and inhibition of topoisomerase II. Mutat. Res. 2006;599:160–166. doi: 10.1016/j.mrfmmm.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Neukam K, Pastor N, Cortes F. Tea flavanols inhibit cell growth and DNA topoisomerase II activity and induce endoreduplication in cultured Chinese hamster cells. Mutat. Res. 2008;654:8–12. doi: 10.1016/j.mrgentox.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L, Eastmond DA, Smith MT. The nature of chromosomal aberrations detected in humans exposed to benzene. Crit. Rev. Toxicol. 2002;32:1–42. doi: 10.1080/20024091064165. [DOI] [PubMed] [Google Scholar]
- 48.Pellack-Walker P, Walker JK, Evans HH, Blumer JL. Relationship between the oxidation potential of benzene metabolites and their inhibitory effect on DNA synthesis in L5178YS cells. Mol. Pharmacol. 1985;28:560–566. [PubMed] [Google Scholar]
- 49.Hiraku Y, Kawanishi S. Oxidative DNA damage and apoptosis induced by benzene metabolites. Cancer Res. 1996;56:5172–5178. [PubMed] [Google Scholar]
- 50.Baker RK, Kurz EU, Pyatt DW, Irons RD, Kroll DJ. Benzene metabolites antagonize etoposide-stabilized cleavable complexes of DNA topoisomerase IIalpha. Blood. 2001;98:830–833. doi: 10.1182/blood.v98.3.830. [DOI] [PubMed] [Google Scholar]
- 51.Lindsey RH, Jr, Bender RP, Osheroff N. Effects of benzene metabolites on DNA cleavage mediated by human topoisomerase II alpha: 1,4-hydroquinone is a topoisomerase II poison. Chem. Res. Toxicol. 2005;18:761–770. doi: 10.1021/tx049659z. [DOI] [PubMed] [Google Scholar]
- 52.Kimmel RR, Agnani S, Yang Y, Jordan R, Schwartz JL. DNA copy-number instability in low-dose gamma-irradiated TK6 lymphoblastoid clones. Radiat. Res. 2008;169:259–269. doi: 10.1667/RR1096.1. [DOI] [PubMed] [Google Scholar]
- 53.Moore SR, Papworth D, Grosovsky AJ. Non-random distribution of instability-associated chromosomal rearrangement breakpoints in human lymphoblastoid cells. Mutat. Res. 2006;600:113–124. doi: 10.1016/j.mrfmmm.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Moore SR, Ritter LE, Gibbons CF, Grosovsky AJ. Spontaneous and radiation-induced genomic instability in human cell lines differing in cellular TP53 status. Radiat. Res. 2005;164:357–368. doi: 10.1667/rr3422.1. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz JL, Jordan R, Evans HH, Lenarczyk M, Liber HL. Baseline levels of chromosome instability in the human lymphoblastoid cell TK6. Mutagenesis. 2004;19:477–482. doi: 10.1093/mutage/geh060. [DOI] [PubMed] [Google Scholar]