Abstract
The Y-chromosome is responsible for sex determination in mammals, which is triggered by the expression of the SRY gene, a testis-determining factor. This particular gene, as well as other genes related to male fertility, are located in the non-recombining portion of the Y (NRY), a specific region that encompasses 95% of the human Y-chromosome. The other 5% is composed of the pseudo-autosomal regions (PARs) at the tips of Yp and Yq, a X-chromosome homologous region used during male meiosis for the correct pairing of sexual chromosomes. Despite of the large size of the human NRY (about 60 Mb), only a few active genes are found in this region, most of which are related to fertility. Recently, several male fertility dysfunctions were associated to microdeletions by STS mapping. Now that the complete genetic map of the human Y-chromosome is available, the role of particular NRY genes in fertility dysfunctions is being investigated. Besides, along with the description of several nucleotide and structural variations in the Y-chromosome, the association between phenotype and genotype is being addressed more precisely. Particularly, several research groups are investigating the association between Y-chromosome types and susceptibility to certain male dysfunctions in different population backgrounds. New insights on the role of the Y-chromosome and maleness are being envisaged by this approach.
Keywords: Y-chromosome, male dysfunction, infertility, pseudo-autosomal region, non-recombining Y, population structure
INTRODUCTION
The mammalian Y-chromosome is a degenerated X chromosome (both derived from an ancestral autosome pair), which has lost several housekeeping genes becoming specialized in male sex determination. A number of studies have suggested, erroneously, that some traits such as violent character, hairy ear, etc., could be mapped on the Y-chromosome. More recently, it has been demonstrated that some of the Y-chromosome genes are implicated in dysfunctions, such as infertility, gonadal sex reversion, gonadoblastoma and non-syndromic hearing impairment. Furthermore, a number of studies also claim that other Y-chromosome genes are associated with high blood pressure and prostate cancer. However, this requires further investigation. This review intends to provide an overview of new evidences of the Y function and of the probable genes involved in male phenotypes.
THE HUMAN Y-CHROMOSOME
The sex chromosomes, X and Y, emerged from an autosome pair around 300 million years ago among the first mammals (Lahn et al, 2001). Since then, they have been diverging because of their different functional roles. The Y-chromosome has become specialized in the male sex determination, progressively decreasing its gene content by losing and gaining sequences through the action of events, such as, deletions, mutations, insertions, recombination and transposition, etc., (reviewed in Charlesworth et al, 2005). The divergence between X- and Y-chromosomes has broken down their homology, generating obstacles for recombination between the two sex chromosomes. This evolutionary process has also produced Y-chromosomes with remarkable differences, including between closely-related species as human and chimpanzee (Hughes et al, 2005). In the human Y-chromosome, meiotic recombination takes place in two relatively short regions (approximately 3 Mb of its ∼60 Mb length), namely Yp- and Yq-PAR (pseudo-autosomal region) that pair with the X-chromosome. They are placed at the tips of the X- and Y-chromosomes (Figure 1) and the high homology between them is kept by the obligatory crossing-over during male meiosis, which assures the correct pairing and segregation of X and Y. Despite of the occurrence of many X-homologous regions distributed along the Y-chromosome, most of these do not recombine and are designated as NRY (Non-Recombining Y). Skaletsky et al (2003) renamed NRY region to MSY (Figure 1) or “male-specific region of the Y” because they observed that the designation “non-recombining region” did not reflect the dynamic evolutionary events happening in this part of the Y-chromosome.
Figure 1.
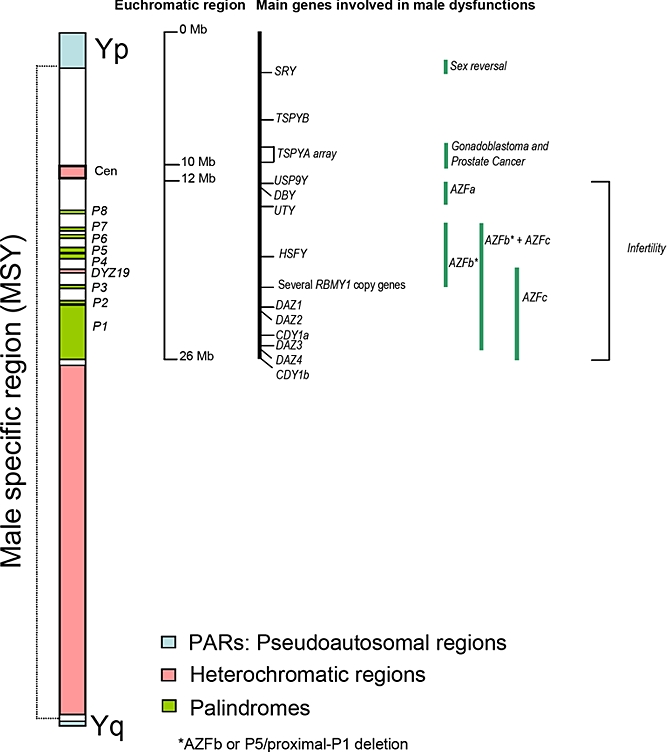
Schematic display of the human Y-chromosome showing pseudo-autosomal regions (PARs), the male specific region (MSY or NRY), the heterochromatic regions, the main genes on the euchromatic region and the regions associated with male dysfunctions (adapted from Skaletsky et al, 2003, and Jobling and Tyler-Smith, 2003).
The Y-chromosome forms 2-3% of the haploid genome, most of which is heterochromatic, around ∼40 Mb in size (Figure 1). The heterochromatic region is made up of three blocks (Figure 1); the first (1 Mb) is placed at the centromere (cen), the second one (DYZ19) in the middle of Yq (∼400 Kb) and the third one comprise most of distal Yq. All three blocks consist of massively amplified tandem repeats of low sequence complexity.
The long arm heterochromatin presents such high length variability among individuals that it can be used to differentiate two male siblings (Rahman et al, 2004). The centromeric heterochromatic region is composed of alphoid satellite repeats and also shows significant variability in length and sequence among male lineages. Furthermore, this region also contains several Y-chromosome genetic markers commonly used in population studies (Santos et al, 1995).
The euchromatic region of MSY encompasses 23 Mb and is made up of three main groups of genes (Skaletsky et al, 2003). The first group is composed of X-transposed genes, represented by two coding units with approximately 99% identity to the corresponding X copies. The second group consists of 16 coding genes that are generally single-copy and expressed widely; however, some of these are also pseudogenes. The genes in this group are likely relics of the ancient autosome pair from which X and Y evolved, but most of them have X similarity, coding similar but non-identical protein isoforms. The third group is formed of genes derived from three converging process; amplification of X-degenerated genes, transposition and duplication or retroposition and duplication of autosomal genes. This is a multicopy class that encompasses approximately 10.2 Mb distributed in Yp and Yq, which consists of large regions where gene copies show ∼99.9% identity, usually maintained by gene-conversion. There are eight palindromic regions, six of which possess genes that are expressed only in testis. The second and third group of genes diverge in the expression patterns: the second group is involved in cellular housekeeping activities that are important for both males and females and the third group is exclusively composed of genes related to male functions, mainly spermatogenesis (Skaletsky et al, 2003).
Recently, Kirsch et al (2005) found eight putative genes spread over the pericentromeric Yq11 region. These potential genes were revealed by sequencing of a 554 kb genomic segment containing a 450 kb euchromatic island. Four tandem copies of a homeobox-containing DUX gene family were apparently functional. These finding open new possibilities to search for candidate genes responsible for Y chromosome dysfunctions, such as gonadoblastoma (GBY, see below).
Y-CHROMOSOME VARIATION AS A POPULATION MARKER
The Y-linked loci in the MSY region are haploid, paternally inherited and devoid of recombination with the X-chromosome. Because of these particular characteristics, variation results in the accumulation of mutations along generations, called as paternal or male lineages. Population geneticists have extensively studied human male lineages to trace migrations and reconstruct human history (reviewed by Jobling and Tyler-Smith, 2003). Many Y-chromosome polymorphisms, also called unique event polymorphisms (UEPs), are biallelic markers, and can be combined in haplogroups that define Y lineages with specific geographic distributions around the world (The Y Chromosome Consortium - YCC 2002). Figure 2 shows a phylogenetic tree as proposed by the Y-chromosome Consortium (YCC 2002; Jobling and Tyler-Smith, 2003) with the main haplogroups defined by the UEPs. The haplogroup distributions reflect past demographic events and human migration routes. The worldwide pattern of distribution of the haplogroups suggests a recent origin (between 60 and 150 thousands years ago) in Africa for all present-day human Y-chromosomes (Underhill et al, 2000; Wilder et al, 2004).
Figure 2.
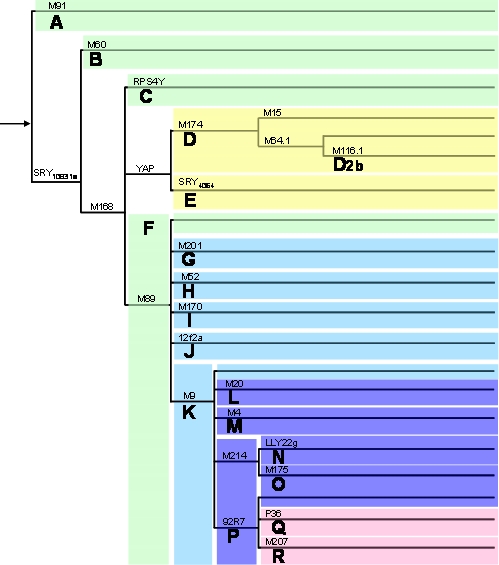
Phylogenetic tree of the main Y-chromosome lineages or haplogroups (based on YCC, 2002; Jobling and Tyler-Smith, 2003). Above the branches are shown loci with derived mutations characterizing each major haplogroup (haplogroup names are below the branches). The subhaplogroup named D2b is also depicted, originating from haplogroup D.
The worldwide distribution of Y lineages is thought to be a consequence of random evolutionary forces such as genetic drift, population expansion and migration. The influence of natural selection on these processes is unknown, but is usually regarded to be of little significance. However, one cannot dismiss natural selection so easily because the Y-chromosome carries important genes involved in spermatogenesis, which can be a target for adaptive processes. Two possible selection mechanisms could conceivably have had strong effect in the population genetic variation of Y-chromosomes: hitchhiking and background selection. The hitchhiking effect occurs when an allele increases in frequency because it is linked to a beneficial allele at a neighbouring locus subjected to positive selection, even if it does not play itself any direct role in fitness (Otto, 2000). The background selection process is the opposite; in this process an allele is less likely to persist in a population and may be eliminated because it is linked to deleterious alleles. The consequence of these two processes can thus be the spread and eventual fixation, or the decrease and eventual extinction of a particular Y haplogroup or lineage.
Y-CHROMOSOME GENES AND MALE DYSFUNCTION
Skaletsky et al (2003) predicted at least 27 distinct proteins or protein families conferred by the MSY region, most of them involved in male specific functions (spermatogenesis) and in sex determination. Ginalski et al (2004) used distinct softwares to detect domains in Y-chromosome proteins and found several DNA-binding motifs. These findings suggest the existence of genes whose proteins act as transcriptional repressors, such as SRY (sex determining region of chromosome Y), HSFY (heat shock transcription factor Y), ZFY (zinc-finger Y) and SMCY (selected mouse cDNA Y). Other identified domains are probably involved in protein-protein interactions, including complex assembly (UTY – ubiquitous tetracopeptide repeat motif Y), histone binding, histone acetylation and chromatin structure changing (CDY – chromodomain Y), RNA recognition (RBMY – RNA-binding motif Y, DAZ – deleted in Azoospermia), etc. The emerging picture is of a Y-chromosome protein “package” involved in regulation of gene expression on different levels such as transcription, pre-mRNA processing and translation.
Y-chromosome and sex reversion
The first and essential step in normal sexual differentiation takes place during the 5th-6th week of gestation. During organogenesis the process of cell proliferation, differentiation, migration and death are precisely regulated by complex signaling networks. The Y-chromosome specific SRY gene, initially named TDF (testis determining factor), is one of the key genes involved in human sex determination: It directs an undifferentiated gonad into a testis, which secretes hormones responsible for normal male development (reviewed by Fleming and Vilain, 2004). After this event, male development unfolds automatically, unless a number of genetic accidents interrupt the male-determining pathway, generating a female baby. Many other genes are required to produce a male (Chaboissier et al, 2004), which are being identified through the study of a variety of sex reversal patients (Vernole et al, 2000; Saavedra-Castillo et al, 2005).
Defects or deletions of the SRY gene are assigned as the likely cause of 15% of XY females (reviewed by Knower et al, 2003). In contrast, males with an XX karyotype usually present a tiny piece of the Y-chromosome with the SRY gene added to the X chromosome. Frequently, these reversal events are also associated with other developmental malformations. The SRY gene, which is located close to the Yp-PAR, is responsible for triggering the testis development during gonadogenesis. It is a testis-specific transcription factor that plays key role in sexual differentiation and development in males. SRY is an intronless gene that spans 3.8 kb, with an open reading frame encoding a 204 amino acids protein with an estimated molecular mass of 24 kDa. The SRY protein displays a DNA-binding HMG (High-Mobility-Group) box, a motif that characterizes a class of non-histone proteins that binds to a consensus target DNA sequence. The regulation of SRY activity has been suggested to proceed by acetylation/deacetylation (Thevenet et al, 2004). Houssain and Saunders (2001) reported that the product of the wt1 (Wilms' tumor) gene – essential for male sex determination and differentiation in mammals – acts as a transcriptional activator of SRY. Despite of being discovered 14 years ago, no target gene for SRY has been identified so far. The autosomal gene, Sox9, a candidate target, encodes a related HMG box-containing factor, is up-regulated specifically in XY gonads shortly after the onset of SRY transcription and is thought to be essential for the differentiation of Sertoli cells (Kent et al, 1996; Morais da Silva et al, 1996). Sekido et al (2004) found that Sox9 expression is altered in SRY mutants and suggested that Sox9 action is downstream of SRY. DAX1, a gene placed at the X chromosome, is an orphan nuclear receptor originally proposed to act as an “anti-testis” factor, but Meeks et al (2003) showed that it is required at several points of embryonic development. They suggested that SRY and DAX1 are both required for testis development in humans.
Gonadal dysgenesis, gonadoblastoma and prostate cancer
Mutations in SRY gene have been reported in approximately 10-15% of 46,XY males with gonadal dysgenesis. The majority of the remaining cases may have mutations in the SRY regulatory elements or other genes involved in the sex differentiation pathway. Recent reports support the relationship between SRY alterations, gonadal dysgenesis and/or primary infertility (Shahid et al, 2004).
Uehara et al (1999; 2002) found mutations in the SRY gene of patients with XY gonadal dysgenesis and gonadal tumor formation. Patients with gonadal dysgenesis and Y-chromosome are at high risk of developing gonadoblastoma, a rare benign tumor with potential for malignant transformation, which affects a subset of patients with intersex disorders. Patients with intersex syndromes presenting gonadal dysgenesis (45,X/46,XY) and a subset of patients with Turner syndrome (45,XO) are particularly subjected to developing gonadoblastoma (Gravholt et al, 2000; Mancilla et al, 2003; Mazzanti et al, 2005). Some studies have reported an association between the occurrence of gonadoblastoma and either macroscopic or molecular evidence of presence of a Y-chromosome (Uehara et al, 1999 and 2002). Even though the gonadoblastoma diagnosis is not completely reliable and the malignant potential of the pathogenesis is still rather obscure, the presence of Y-chromosome material can be definitive for prognostic. Despite the controversial studies, in Y-chromosome positive cases the gonadectomy is generally recommended (Kondi-Pafiti et al, 2005; Pena-Alonso et al, 2005). The frequency of intersex patients with fragments of Y-chromosome and presenting gonadoblastoma may vary from 7-10% (Gravholt et al, 2000) to 30% (Verp and Simpson, 1987).
It has been hypothesized that the expression of a gene (or genes) on the Y-chromosome is directly involved in the etiology of gonadoblastoma. Page (1987) postulated the existence of a Y-chromosome gene (GBY) with an undefined physiologic function in normal males, which would have the ability to predispose dysgenetic gonads to develop malignancy when present in females. Women 46,XY without SRY but carrying GBY locus developed gonadoblastoma in their dysgenic ovaries. Using an approach based on STS mapping, Tsuchya et al (1995) identified a critical region where gonadoblastoma genes could be near the Y-chromosome centromere. Their analysis indicated two candidate genes – both dispersed Y-linked gene families: TSPY (testis-specific protein Y-encoded) and YRRM (Y-chromosome RNA recognition motif) that were present in all gonadoblastoma patients.
YRRM or RBMY gene encodes a nuclear protein containing a RNA recognition motif, whose expression is restricted to the testis and it is one of the candidate genes related to fertility. It is now considered an unlikely candidate for GBY (Lau, 1999), but Tsuei et al (2004) detected RBMY transcripts in testis and liver cancer tissues. Besides, they were able to transform rodent fibroblast, suggesting RBMY as a candidate oncogene. TSPY gene is present in 20-40 copies on the human Y-chromosome (or 50 copies according to Shubert et al, 2003) and falls dispersed within two GBY critical regions: interval 3E-3G and 4B in the short arm according to Tsuchiya et al (1995) and intervals 4B and 5E in the long arm according to Salo et al (1995). TSPY is the main candidate for GBY and there is some evidence supporting its role as an oncogene. Tsuchiya et al (1995) detected TSPY expression of this gene in tumor samples. Recently, Delbridge et al (2004) mapped its homolog in the X chromosome and observed that its cDNA was involved in cell cycle regulation, suggesting that TSPY is required for germ cell proliferation. Comparison between TSPY and TSPX sequences revealed that TSPY evolved much more rapidly in sequence structure and gene copy numbers. TSPY has six exons and occurs in multiple copies but does not form palindromic repeats like other male specific genes (YRRM and DAZ). Like other testis-specific genes, the multiple copies of TSPY have been suggested to be resulted from a requirement for critical doses during spermatogenesis (Graves et al, 1998).
In addition to the evidence of TSPY involvement in gonadoblastoma, aberrant TSPY expression was also observed in prostate cancer tissues and cell lines (Dasari et al, 2001), and at elevated levels in tumor cells of prostate cancers at various degrees of malignancy (Lau et al, 2003). Vijayakumar et al (2005) transferred a human Y chromosome to a human prostate cancer cell lineage and found a different result. They observed that the Y- chromosome suppressed tumor formation in athymic nude mice. Paracchini et al (2003) performed a case-control study using 118 Y markers in 930 prostate cancer cases and observed a statistical association between a common Japanese Y-chromosome lineage, haplogroup O3 (derived from haplogroup O – Figure 2) and a fourfold increased risk of severe prostate cancer. It would be interesting to verify whether TSPY presents alteration in gene copy number or mRNA transcription and protein expression in this Japanese male lineage, which could be linked to prostate cancer. Taken together, these results suggest that the likely association between TSPY or another Y chromosome gene and cancer can be much more complex than previously understood.
Y-chromosome genes and non-syndromic hearing impairment
Recently, Wang et al (2004) reported a Chinese family with non-syndromic deafness, whose clinical phenotype was only present in the paternal lineage along seven generations. Wang et al (2004) suggested a likely candidate in Yp11, PCDH11, which codes for a protocadherin protein. However, another gene located in Yp, TBL1Y, can be likely associated to this dysfunction. This gene, Transducin beta-like 1Y, has a X chromosome homologue related to a X-linked late-onset sensorineural deafness (Yan et al, 2005). Interestingly, two recent reports pointed out the occurrence of deletions or Yp translocation associated to deafness (Graham and Bacino, 2003; Klein et al, 2005).
Y-chromosome genes and male infertility
In 1976, Tiepolo and Zuffardi observed for the first time the involvement of Yq deletions in male infertility when they were analyzing cells from idiopathic infertile males. Since then, many structural abnormalities in the Y-chromosome have been observed, including microdeletions detectable only by molecular methods. Molecular studies have shown that microdeletions at Yq11 may represent the etiological factor in as many as 10-15% of cases with idiopathic azoospermia or severe oligozoospermia (Reijo et al, 1995; Vogt et al, 1996). The microdeletion events appear in three critical regions of the long arm of the Y-chromosome, initially considered non-overlapping, called Azoospermia factor (AZFa, AZFb and AZFc) (Vogt et al, 1996 – Figure 1). Around 71% of men with Y chromosomal microdeletions and severe oligozoospermia or idiopathic azoospermia were found to have AZFc deletions compared with 13% with AZFa and 31% with AZFb deletions (Ferlin et al, 1999; Reynolds and Cooke, 2005). Challenges for current research include the elucidation of the genomic mechanisms that generate such recurrent deletions and also the identification of the genes that cause infertility when deleted or damaged. Recombination between repetitive regions is believed to be the cause of the high incidence of de novo microdeletions in the Y-chromosome long arm. For instance, Kuroda-Kawaguchi et al (2001) demonstrated that 47 out of 48 men with AZFc deletions had the same proximal and distal breakpoints in 229 kb direct repeats flanking AZFc. Furthermore, Repping et al (2002) demonstrated that recombination between repetitive regions in Yq can explain the majority of AZFb and AZFb + AZFc deletions. Recently, it was shown that homologous recombination events are highly recurrent in the MSY region, especially at the AZFc locus (Skaletsky et al, 2003; Repping et al, 2003; Machev et al, 2004).
The spermatogenic impairment caused by AZFb and AZFc deletions can be actually caused by genes mapped at these regions as proposed by several studies (Mahadevaiah et al, 1998; Brown et al, 1998). Since the first deletion mapping studies until the most recent and detailed physical map of the human Y chromosome (Skaletsky et al, 2003) several genes related to spermatogenesis were discovered, which are described below.
AZF deletions causing male fertility
AZFa
AZFa deletions are rarely found among infertile males but usually present more severe consequences than AZFc deletions. Complete deletion of the region is associated with the Sertoli-Cell-Only (SCO) syndrome, observed in testicular biopsies (Kamp et al, 2001). This region spans approximately 800 kb (Sun et al, 1999) of Yq and is mapped proximal to the centromere (Figure 1). AZFa contains two main candidate genes: USP9Y (ubiquitin-specific protease 9, also known as DFFRY) and DBY/DDX3Y (DEAD/H box polypeptide). Foresta et al (2000) reported well-defined spermatogenic alterations carrying deletions of both genes, USP9Y and DBY. They suggested that DBY probably is the main AZFa candidate because it is more frequently deleted and its absence produces a significant reduction of germ cells or even their complete absence. The DBY gene has a structural homologue on the short arm of the X-chromosome, DBX (Xp11.4). DBY and DBX are transcribed in several tissues, however, translation of DBY is detected only in the male germ line, predominantly in spermatogonia (Ditton et al, 2004).
The USP9Y gene encodes a specific protease that contains an ubiquitin C-terminal hydrolase domain (Brown et al, 1998). Studying the USP9Y gene, Sun et al (1999) identified the first point mutation causing azoospemia, confirming its role in the phenotype and suggesting the requirement for the ubiquitin system in spermatogenesis. Despite USP9Y seems to play a critical role in human spermatogenesis, the chimpanzee Y chromosome encodes a smaller USP9Y ORF, without the catalytic domain (Hughes et al, 2005). It is an open question whether the chimpanzee USP9Y counterpart is inactivated or is still functionally active.
Two paralogous sequences of human endogenous retrovirus (HERV) flanking the AZFa locus are diagnosed as the main cause of recurrent deletion events and, sometimes duplications, in this region (Sun et al 2000). HERVs account for about 1% of the human genome and are widely distributed throughout the chromosomes. The two AZFa HERV copies, each one consisting of 10-12 kb in length, are located ∼700 kb apart and present ∼94% similarity (Sun et al, 2000). Because of the long similar sequences between them, they may work as substrate for non-homologous recombination, producing chromosomal rearrangements. Despite that AZFa deletions differ among them, most of breakpoints fall within the two HERV sequences (Sun et al, 2000).
AZFb or P5/proximal-P1 deletion
Until recently, AZFb was considered a distinct Yq deletion interval, responsible for male infertility in a fraction of idiopathic infertile males. However, Repping et al (2002) demonstrated that the previously considered non-overlapping AZFb deletions actually encompass 1.5 Mb of AZFc, including two copies of the main AZFc gene, DAZ. They named this recurrent deletion as P5/proximal-P1, because most breakpoints in this region localized inside of P5 and P1 proximal palindromes (Repping et al, 2002) (Figure 3). It was also observed that different length deletions have breakpoints localized in other palindromes, such as P5/distal-P1 and P4/distal-P1, which also delete part of the AZFc region. Because the usual markers employed to detect alterations provided a low resolution mapping, the actual deletion lengths were not known. Total P5/proximal-P1 deletion encompasses 6.2 Mb and removes 32 genes and transcripts. The associated infertile phenotype could be caused by the absence of candidate genes in AZFb, or also by AZFc candidate genes, or by both.
Figure 3.
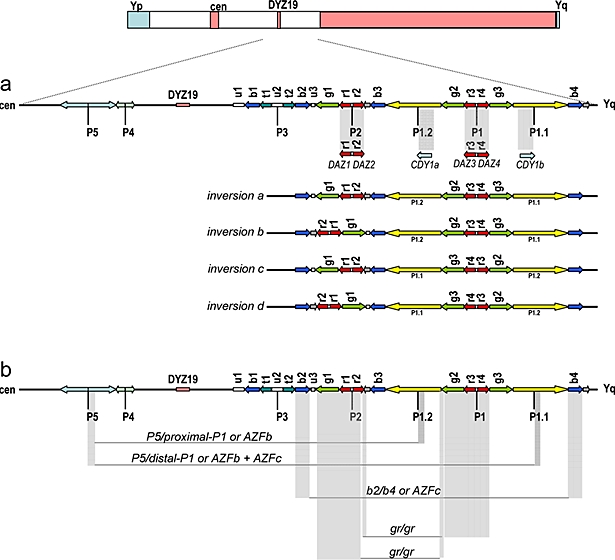
Gene structure of AZFb and AZFc regions with massive presence of repeats and palindromes (adapted from Kuroda-Kawaguchi et al, 2001; Repping et al, 2002; Skaletsky et al, 2003; Machev et al, 2004). Arrows show copies of the various repeats, gene families and palindromes P1 to P5. Cen refers to centromere. (a) The gray bar highlights the main AZFc candidate genes related to infertility. The palindromic copies of DAZ and CDY1 genes are shown in detail. Four polymorphic Y chromosome structures (inversions a to d) with different gene orders are shown below the gene map. These structures are present in distinct Y haplogroup backgrounds (Machev et al, 2004). (b) The gray bars delimit the regions frequently deleted in AZFb/c infertile males: P5/P1.2, P5/P1.1, b2/b4 and gr/gr. The outcome of each deletion is dependent on the gene order depicted by some chromosome structure polymorphisms as the inversions depicted here.
The main candidate for causing infertility in AZFb is the RBMY gene family, which encodes a nuclear protein containing a RNA recognition motif and whose expression is restricted to testis. The presence of an RNA binding motif (RBM) and four tandem repeats (SRGY box), similar to segments of several splicing factors, indicates a probable role of the RBMY protein in RNA metabolism (Ma et al, 1993). RBMY consists of approximately 30 copies of genes and pseudogenes, found on both arms of the Y-chromosome, but probably only the subfamily RBMY1 is functional (Chai et al, 1997). RBMY1 is present in AZFb and consists of seven slightly different genes. Delbridge et al (1999) found an active RBMY homologous copy at X-chromosome (Xq26), RBMX, which was also shown to be present in multiple processed copies in several chromosomes (Lingenfelter et al, 2001). According to Lingenfelter et al (2001), RBMY has acquired a testis-specific function, whereas RBMX is ubiquitously expressed and subjected to X inactivation. Friel et al (2002) used reverse transcription-polymerase chain reaction (RT-PCR) to analyze the presence of some infertility AZFb candidate genes in testicular cells of men with idiopathic azoospermia. They found one AZFb deleted patient lacking RBM mRNA, corroborating the hypothesis that RBM expression is exclusive to AZFb and that the lack of testicular RBMY1 mRNA results in suppressed spermatogenesis.
Ferlin et al (2003) identified four infertile males with similar AZFb breakpoints but all of them carrying the candidate RBMY. It seems probable that other genes in that region may be also important in spermatogenesis. The other candidate gene encodes for a heat shock protein, HSFY, which is deleted in an idiopathic azoospermic male (Vinci et al, 2005).
AZFc and b2/b4 deletion
AZFc represents the most frequently deleted region among infertile males. The estimated frequency of AZFc deletion is approximately 1 in 4000 males, accounting for about 12% cases of non-obstructive azoospermia and for about 6% cases of severe oligozoospermia (Kuroda-Kawaguchi et al, 2001). The phenotype observed in AZFc deleted males can vary from azoospermia to oligozoospermia, including sporadic cases of AZFc deleted males who were able to conceive children naturally (Gatta et al, 2002; Kuhnert et al, 2004). Kuroda-Kawaguchi et al (2001) observed that most of deletions share similar breakpoints and resulted from homologous recombination between two direct repeats called b2/b4 (Figure 3). The estimated length of most of AZFc deletions that arises de novo with these characteristics is about 3.5 Mb.
The main candidate gene in AZFc is the DAZ cluster, a set of genes transcribed in the adult testis and expressed exclusively in germ cells, apparently encoding an RNA binding protein (Reijo et al, 1995; Yen et al, 1997). The DAZ gene has been implicated in both the establishment and maintenance of the primordial germ cell (PGC) population as well as in the control of meiotic division (reviewed by Reynolds and Cooke, 2005). Using a single male as Y-reference chromosome, Saxena et al (2000) reported the existence of at least four DAZ copies with different numbers of intragenic tandem repeats, organized in two blocks, each one comprising an inverted pair of DAZ genes, all localized in the AZFc region (Figure 3a). The DAZ has a homologous autosomal copy on chromosome 3, called DAZL. It presents high similarity to the boule gene of Drosophila, which causes spermatogenic failure when mutated. DAZ genes in the Y-chromosome are a recent acquisition in Primates (humans and old world monkeys). Mulhall et al (1997) proposed that the DAZ cluster is preferentially involved in quantitative rather than qualitative production of sperm.
Infertility predisposition and Y-chromosome variation
The Y-chromosome has evolved into a highly specialized chromosome to perform male functions, mainly spermatogenesis. The recurrent AZFc deletions associated to infertility raise the hypothesis that some Y-chromosome types could be more subjected to deletions. Alternatively, they could carry less copies of spermatogenesis related genes, thus, such males would present infertility predisposition. RBMY and DAZ are members of multicopy gene families, which suggest the possibility that expression levels vary according to copy gene number. Vogel et al (2000) investigated the phenotype resulting from modifications introduced in RBMY and DAZ in transgenic mice and observed a variable penetrance, as we observe in human males with Y-chromosome deletions. Possibly, environmental effects and background genetic profile can change gene expression and male phenotype (Mulhal et al, 1997; Foresta et al, 2000). Most genes at MSY present motifs or domains generally involved in transcriptional or translational regulation, supporting the possible correlation between Y-chromosome genes expression and quantitative sperm variation.
Kuroki et al (1999) found that Japanese males with Y-chromosomes belonging to haplogroup D (probably belonging to subhaplogroup D2b – see below) were associated with low sperm count, suggesting that they are more likely subjected to spermatogenic failure. Repping et al (2003) proposed an explanation of Kuroki's results through the observation of a recurrent AZFc deletion of 1.6 Mb, called gr/gr (Figure 3b). This deletion was observed to arise independently in 13 out of 29 lineages of the Y-chromosome genealogical tree. They observed that one specific lineage in Japan, D2b (subtype of D – Figure 2), contains only gr/gr-deleted chromosomes. This deletion does not eliminate all testis-specific genes present in the region, but deletes two of four DAZ copies. According to Repping et al (2003), men carrying Y-chromosomes with gr/gr deletions should be at an increased infertility risk, since the deletion eliminates some transcription units, which would reduce the sperm production. They claimed that the D2b haplogroup, which has fixed the gr/gr deletion and occurs in ∼30% of Japanese males, is subjected to negative selection.
Recent studies conducted in our laboratory found no evidence of significant reduction of fitness related to infertility when comparing Y haplogroup frequencies from infertile and control males using populations from Israel and Japan (Carvalho et al, 2003; 2004). If Japanese males of the haplogroup D2b are subjected to negative selection, thus predisposed to infertility, we should be able to detect significant haplogroup differences between the infertile and control groups. We have recently improved our Y-chromosome analysis using more UEPs to detect several haplogroups, and more STSs from the AZFc region, specifically to detect gr/gr deletions. Using these improved approaches we obtained the same result, i.e., we could not detect any statistical association between Y-chromosome haplogroups and infertility predisposition (unpublished data). Despite we had no sperm count data for a direct comparison with their results, it is intriguing that we did not observe any increase in D2b males (actually, we observed a reduction) in the infertile group (25%) when compared with the control group (35%). We suggested that, probably, Japanese gr/gr chromosomes may have structural or nucleotide differences that compensate the AZFc gene loss, as gain-function alterations. It is also possible that gr/gr deletions could increase the risk of infertility in some genetic backgrounds but not in another ones. Perhaps, different Y-chromosome lineages could have different gene numbers of multicopy families involved in spermatogenesis, increasing or diminishing the effect of gr/gr deletions. Interestingly, recombination between b2/b4 segments from sister chromatids generated duplication of DAZ genes, restoring two copies of DAZ erased after a gr/gr deletion event (Repping et al, 2003). Of course, to properly evaluate the infertility risk we may also have to consider variations in autosomal or Xchromosomal genes interacting in the spermatogenic complex. These epistatic interactions could be responsible for the phenotypic variability in different individuals with a gr/gr deletion in their Y-chromosomes.
Despite the deletion of the whole AZFc region leads to sterility, the direct relationship between partial AZFc deletions and infertility is still very controversial. The recent findings suggest that AZFc presents a high variability in genomic structure among individuals (Figure 3a). Some individuals carrying a gr/gr deletion can lose, at least, nine transcription units and still be fertile. Machev et al (2004) found four different gr/gr deletion types, which remove several genes. They also observed several individuals with inversions and duplications in the AZFc region (figure 3a), but none presented spermatogenic alterations. Those events are highly recurrent, occurring in different Y-chromosome lineages. Indeed, a particular deletion event (b2/b3) loses 1.8 Mb from AZFc (Repping et al, 2004). It was suggested to happen in polymorphic chromosomes with an inversion between gr/rg repeats (see Figure 3a) and subsequent deletion involving repeats b2 and b3. However, there is no evidence of significant reduction in fitness of males from haplogroup N (Figure 2), which have the b2/b3 deletion, fixed, as we also suggested for the D2b lineage from Japan.
Recent studies (Table 1) have analyzed the association between gr/gr deletions and infertility. Many of them found a positive correlation (Repping et al, 2003; Lynch et al, 2005; Ferlin et al, 2005; de Llanos et al, 2005; Giachini et al, 2005), but others not (Hucklenbroich et al, 2004; Machev et al, 2004). There are some possible causes to explain those disparities, one of them is the lack of agreement in parameters, such as, the sperm count (see Table 1). According to WHO (World Health Organization) recommendations, an individual is considered oligozoospermic if presents sperm concentration <20×106 sperm/ml. However, Lynch et al (2005) considered as oligozoospermic those males who presented ∼40×106 sperm/ml (table 1). They found association between gr/gr deleted males and infertility, but if WHO recommendations were used, they would have not detected a significant association with infertility (see table 1): males with >20×106 sperm/ml representing 4.6% (Monash Male Infertility database) or 5.4% (Men referred for Yq testing), similar to azoospermic (3.6% or 5.0%).
Table 1.
Comparative data of recent studies about gr/gr deletions and male infertility
| Reports | N | Spermatogenic failure | gr/gr N (%) | Y haplogroups | Association gr/gr + infertility |
|---|---|---|---|---|---|
| Repping et al 2003 | 471 | 471 Non-obstructive azoospermia or oligozoospermia (<5×106 sperm/ml) | 15 (3.2) | DE* (xD2b) F*(xJK) J K*(xP) P*(xR1a) |
Yes |
| 246 | <10×106 sperm/ml or <20×106 total | 9 (3.7) | DE* (xD2b) P*(xR1a) R1A |
||
| 215 | Unknown fertility | 4 (1.9) | D2b, J, K*(xP) |
||
| 148 | Controls: >40×106 total | 0 | - | ||
| Lynch et al 2005 | 197 | Azoospermia | 7 (3.6) | All gr/gr: E, F*(xJK), J, K*(xP), P*(xR1a) |
Yes |
| Monash Male Infertility (MMI) database | 119 | 0.1–0.5×106 sperm/ml | 4 (3.4) | ||
| 188 | 0.6–5.0×106 sperm/ml | 8 (4.3) | |||
| 122 | 5.0–19×106 sperm/ml | 4 (3.3) | |||
| 56 | 20–40×106 sperm/ml | 1 (1.8) | |||
| 106 | >40×106 sperm/ml | 3 (2.8) | |||
| Men referred for Y chromosome (Yq) testing | 119 | Azoospermia | 6 (5.0) | ||
| 131 | 0.1–0.5×106 sperm/ml | 3 (2.3) | |||
| 198 | 0.6–5.0×106 sperm/ml | 12 (6.0) | |||
| 114 | >5.0–19×106 sperm/ml | 5 (4.4) | |||
| 37 | ≥20×106sperm/ml | 2 (5.4) | |||
| 234 | Controls | 1 (0.4) | |||
| Ferlin et al 2005 | 73 | Azoospermia | 3 (4.1) | Yes | |
| 193 | <5.0×106 sperm/ml | 10 (5.2) | - | ||
| 71 | 5.0–20×106 sperm/ml | 3 (4.2) | |||
| 263 | Controls: >20×106 sperm/ml | 1 (0.4) | |||
| De Llanos et al 2005 | 66 | Azoospermia | 12 (4.2) | Yes | |
| 217 | <5×106 sperm/ml | 11 (5.1) | - | ||
| 232 | Controls | 0 | |||
| Giachini et al 2005 | 89 | Idiopathic infertile males | 4 (4.5) | Yes | |
| 61 | Males with mild abnormal andrological findings | 3 (4.9) | - | ||
| 189 | Controls | 1 (0.5) | |||
| Hucklenbroich et al 2005 | 61 | Azoospermia | 2 (3.3) | All gr/gr: F*(xIJK), R1 |
No |
| 133 | <1.0 ×106 sperm/ml | 6 (4.5) | |||
| 154 | 1.0–20×106 sperm/ml | 6 (3.9) | |||
| 170 | Controls | 3 (1.8) | |||
| Machev et al 2004 | 263 | Azoospermia or Oligozoospermia <5×106 sperm/ml | 15 (5.7) | DE, J, P, Y*(xDEJP) |
No |
| 46 | >5×106 sperm/ml | 3 (6.5) | DE, J, P | ||
| 210 | Unknown fertility | 10 (4.8) | J, P, Y*(xDEJP) |
||
| 189 | Controls | 6 (3.2) | DE, Y*(xDEJP) |
||
Some reports (Machev et al, 2004; Ferlin et al, 2005) have showed that the deletion size and the genes excluded in each gr/gr deletion can be completely different from each other. This raises another problem, since gr/gr deletions mean different Y chromosome structures and rearrangements. Furthermore, haplogroup R (Kuroda-Kawaguchi et al, 2001) presents four copies of the DAZ genes, which may not be true for all Y chromosomes. Recently, Butler and Schoske (2005) studied the DYS464 microsatellite, which is present in four copies and located near to the DAZ genes. Apparently, most of males have four DYS464 copies in their Y chromosomes, but they also reported many males with three to seven copies, which may also reflect the number of DAZ genes. Indeed, Lin et al (2005) have reported the occurrence of six DAZ copies in different Y chromosome lineages (O3, I and R1). Thus, if different Y chromosomes can display two, three, five, six or seven DAZ genes, how can we confidently infer gr/gr deletions using only STSs markers? To answer this question it will be needed to generate detailed genomic maps through sequencing of AZFc regions of different Y chromosome lineages with and without gr/gr deletions.
The true role of genes placed at AZFb/c regions as causative of the infertile phenotype is still a puzzle. Intriguingly, a single gene has shown a point mutation causing infertility (USB9Y, candidate gene of the AZFa region). Genes at AZFb/c are correlated to infertility because they are deleted in some Y-chromosomes of infertile patients and play a probable role in spermatogenesis, inferred by their expression in testis or their X or autosomal homologous counterpart. Recently, Ferlin et al (2005) described a family case whose father, bearing a gr/gr deleted Y chromosome, had five sons. The fertility state of each son, all of them are gr/gr deleted, vary from fertile, oligozoospermia to azoospermia. Interestingly, the fertility level is correlated to the age of each son. The correlations between the sperm production in gr/gr males and age have not been tested yet, but it seems also an important influence to be further evaluated.
This male infertility picture is so complex that the testis biopsies of males with Y-chromosome deletions reveal diverse outcomes, ranging from a complete absence of germ cells (Sertoli-Cell-Only Syndrome) to the presence of cells arrested in different meiotic stages, occasionally producing mature sperm (Brandell et al, 1998). Ferlin et al (1999) suggested that long deletions are associated with the most severe spermatogenic failure, but so far it was not possible to establish a definitive correlation between testis phenotype and infertility genes. Many hypotheses have been postulated to explain such discordant facts. One hypothesis proposes the probable influence of the genetic “background” on fertility among individuals because it is estimated that >4000 genes may be involved in human spermatogenesis (Gianotten et al, 2004). Meschede et al (2000) demonstrated familial clustering associated with male infertility, corroborating this view. Moreover, environmental factors are also likely to influence the phenotype and the advanced paternal age has also been pointed out as a contributing factor.
Nevertheless, much evidence suggests another interesting hypothesis. It may be possible that infertility in AZFb/c deleted males may not be caused by the absence of Y genes itself but due to the impairment of the spermatogenic process either because of the lack of 6.1 Mb AZFb deletion or 3.5 Mb AZFc deletion. Yogev et al (2004) observed a strong impairment in the normal alignment of X and Y pseudo-autosomal regions (PARs) in AZFb-c deleted individuals (∼29%) and a moderated decrease (70-94%) in AZFc deleted males. It has been suggested that the spermatogenic failure could be either caused by the absence of genes necessary to bivalent formation complex in meiosis or by recombination disturbance caused by the absence of a long DNA stretch. If the first possibility were true, we would expect lower bivalent formation involving other chromosome pairs, unless Y infertility genes were specific for bivalent Y-chromosome complex formation. Solari (1999) observed the heterogeneous stages of meiotic cell degeneration in the histopathology of cells carrying chromosomal abnormalities, which could be compared to the spermatogenesis infertile male picture (arrested in different meiotic stages). Nevertheless, in most mammals, the DAZL alone is sufficient for complete gametogenesis (Skaletsky et al, 2003). Vogel et al (2002) used transgenic mice carrying either human DAZL or human DAZ (Y) on a mouse DAZL-knockout and observed that both transgenics enabled partial rescue of the spermatogenesis failure. However, the autosomal gene resulted in a larger amount of early germ cells, but this proposal still needs further investigation.
CONCLUSION
The human Y chromosome has just been completely sequenced (Skaletsky et al, 2003) but a lot of work still needs to be done to reveal its role in maleness. The peculiar nature of its genes, mostly appearing in multiple copies, generates difficulties for understanding its function and the correlation between genotype and phenotype. Besides, the DNA sequence of a single Y-chromosome reveals only a snapshot of a dynamic chromosome, which presents several distinct gene structures among individuals (Figure 3a). It poses a dramatic challenge to molecular biologists and medical geneticists investigating Y-chromosome-mediated dysfunctions, and also reveals the complexity of the functional pathways related to inherited disorders affecting humans.
ACKNOWLEDGEMENTS
This work was supported by the Brazilian agency CNPq. We would like to thank Professor Sérgio D. J. Pena for his extensive contribution to this review.
LIST OF ABBREVIATIONS
- AZF
Azoospermia Factor
- CDY
Chromodomain Y
- DAZ
Deleted in Azoospermia
- DBY
DEAD box Y
- DFFRY
Drosophila Fat Facet related Y
- GBY
Gonadoblastoma locus on the Y Chromosome
- HERV
Human Endogenous Retrovirus
- HMG
High-Mobility-Group
- HSFY
Heat Shock Transcription Factor Y
- MSY
Male-specific region of the Y Chromosome
- NRY
Non-Recombining portion of the Y Chromosome
- PARs
Pseudo-Autosomal Regions
- RBMY
RNA-Binding Motif Y
- RT-PCR
Reverse Transcription-Polymerase Chain Reaction
- SCO
Sertoli-Cell-Only Syndrome
- SMCY
Selected Mouse cDNA Y
- Sox9
SRY-box containing gene 9
- SRY
Sex determining region of Chromosome Y
- STS
Single-Tagged-Sequences
- TDF
Testis Determining Factor
- TSPY
Testis-Specific Protein Y-encoded
- UEPs
Unique Event Polymorphisms
- USP9Y
Ubiquitin-Specific Protease 9
- UTY
Ubiquitous Tetracopeptide repeat motif Y
- YCC
Y-chromosome Consortium
- YRRM
Y-chromosome RNA Recognition Motif
- ZFY
Zinc-Finger Y
STATEMENT OF COMPETING INTERESTS
The authors declared no competing interests.
REFERENCES
- Brandell RA, Mielnik A, Liotta D, Ye Z, Veeck LL, Palermo GD, et al. AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod. 1998;13:2812–2815. doi: 10.1093/humrep/13.10.2812. [DOI] [PubMed] [Google Scholar]
- Brown GM, Furlong RA, Sargent CA, Erickson RP, Longepied G, Mitchell M, et al. Characterization of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum Mol Genet. 1998;7:97–107. doi: 10.1093/hmg/7.1.97. [DOI] [PubMed] [Google Scholar]
- Butler JM, Schoske R. U.S. population data for the multicopy Y-STR locus DYS464. J Forensic Sci. 2005;50:975–977. [PubMed] [Google Scholar]
- Carvalho CMB, Fujisawa M, Shirakawa T, Gotoh A, Kamidono S, Paulo TF, et al. Lack of association between Y chromosome haplogroups and male infertility in Japanese men. Am J Med Genet A. 2003;116:152–158. doi: 10.1002/ajmg.a.10827. [DOI] [PubMed] [Google Scholar]
- Carvalho CMB, Rocha JL, Santos FR, Kleiman SE, Paz G, Yavetz H, et al. Y-chromosome haplotypes in azoospermic Israeli men. Hum Biol. 2004;76:469–478. doi: 10.1353/hub.2004.0042. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- Chai NN, Salido EC, Yen PH. Multiple functional copies of the RBM gene family, a spermatogenesis candidate on the human chromosome. Genomics. 1997;45:355–361. doi: 10.1006/geno.1997.4944. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Dasari VK, Goharderakhshan RZ, Perinchery G, Li LC, Tanaka Y, Alonzo J, et al. Expression analysis of Y chromosome genes in human prostate cancer. Urol. 2001;165:1335–1341. [PubMed] [Google Scholar]
- de Llanos M, Ballesca JL, Gazquez C, Margarit E, Oliva R. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod. 2005;20:216–220. doi: 10.1093/humrep/deh582. [DOI] [PubMed] [Google Scholar]
- Delbridge ML, Lingenfelter PA, Disteche CM, Graves JA. The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome. Nat Genet. 1999;22:223–224. doi: 10.1038/10279. [DOI] [PubMed] [Google Scholar]
- Delbridge ML, Longepied G, Depetris D, Mattei MG, Disteche CM, Graves JA, et al. TSPY, the candidate gonadoblastoma gene on the human Y chromosome, has a widely expressed homologue on the X - implications for Y chromosome evolution. Chromosome Res. 2004;12:345–356. doi: 10.1023/B:CHRO.0000034134.91243.1c. [DOI] [PubMed] [Google Scholar]
- Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E, Vogt PH. The AZFa gene DBY DDX3Y is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Moro E, Garolla A, Foresta C. Human male infertility and Y chromosome deletions: role of the AZF candidate genes DAZ, RBM and DFFRY. Hum Reprod. 1999;14:1710–1716. doi: 10.1093/humrep/14.7.1710. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Moro E, Rossi A, Dallapiccola B, Foresta C. The human Y chromosome's azoospermia factor b AZFb region: sequence, structure and deletion analysis in infertile men. J Med Genet. 2003;40:18–24. doi: 10.1136/jmg.40.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, et al. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42:209–213. doi: 10.1136/jmg.2004.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A, Vilain E. The endless quest for sex determination genes. Clin Genet. 2005;67:15–25. doi: 10.1111/j.1399-0004.2004.00376.x. [DOI] [PubMed] [Google Scholar]
- Foresta C, Ferlin M, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet. 2000;9:1163–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- Friel A, Houghton JA, Glennon M, Lavery R, Smith T, Nolan A, et al. A preliminary report on the implication of RTPCR detection of DAZ, RBMY1, USP9Y and Protamine-2 mRNA in testicular biopsy samples from azoospermic men. Int J Androl. 2002;25:59–64. doi: 10.1046/j.1365-2605.2002.00326.x. [DOI] [PubMed] [Google Scholar]
- Gatta V, Stuppia L, Calabrese G, Morizio E, Guanciali-Franchi P, Palka G. A new case of Yq microdeletion transmitted from a normal father to two infertile sons. J Med Genet. 2002;39(6):e27–e27. doi: 10.1136/jmg.39.6.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachini C, Guarducci E, Longepied G, Degl'Innocenti S, Becherini L, Forti G, et al. The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet. 2005;42:497–502. doi: 10.1136/jmg.2004.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianotten J, Lombardi MP, Zwinderman AH, Lilford RJ, van der Veen F. Idiopathic impaired spermatogenesis: genetic epidemiology is unlikely to provide a short-cut to better understanding. Hum Reprod Update. 2004;10:533–539. doi: 10.1093/humupd/dmh045. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Rychlewski L, Baker D, Grishin NV. Protein structure prediction for the male-specific region of the human Y chromosome. Proc Natl Acad Sci USA. 2004;101:2305–2310. doi: 10.1073/pnas.0306306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BH, Bacino CA. Male patient with non-mosaic deleted Y-chromosome and clinical features of Turner syndrome. Am J Med Genet A. 2003;119:234–237. doi: 10.1002/ajmg.a.10147. [DOI] [PubMed] [Google Scholar]
- Graves JA, Disteche CM, Toder R. Gene dosage in the evolution and function of mammalian sex chromosomes. Cytogenet Cell Genet. 1998;80:94–103. doi: 10.1159/000014963. [DOI] [PubMed] [Google Scholar]
- Gravholt CH, Fedder J, Naeraa RW, Muller J. Occurrence of gonadoblastoma in females with Turner syndrome and Y chromosome material: a population study. J Clin Endocrinol Metab. 2000;85:3199–3202. doi: 10.1210/jcem.85.9.6800. [DOI] [PubMed] [Google Scholar]
- Hossain A, Saunders GF. The human sex-determining gene SRY is a direct target of WT1. J Biol Chem. 2001;276:16817–16823. doi: 10.1074/jbc.M009056200. [DOI] [PubMed] [Google Scholar]
- Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, Simoni M. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod. 2005;20:191–197. doi: 10.1093/humrep/deh558. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, Mielnik A, et al. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol Hum Reprod. 2001;7:987–994. doi: 10.1093/molehr/7.10.987. [DOI] [PubMed] [Google Scholar]
- Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P. A male-specific role for Sox9 in vertebrate sex determination. Development. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- Kirsch S, Weiss B, Miner TL, Waterston RH, Clark RA, et al. Interchromosomal segmental duplications of the pericentromeric region on the human Y chromosome. Genome Res. 2005;15:195–204. doi: 10.1101/gr.3302705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Backstrand K, Cotter PD, Marco E, Sherr E, Slavotinek A. Case report: Y;6 translocation with deletion of 6p. Clin Dysmorphol. 2005;14:93–96. [PubMed] [Google Scholar]
- Knower KC, Kelly S, Harley VR. Turning on the male--SRY, SOX9 and sex determination in mammals. Cytogenet Genome Res. 2003;101:185–198. doi: 10.1159/000074336. [DOI] [PubMed] [Google Scholar]
- Kondi-Pafiti A, Grapsa D, Hasiakos D, Kontorabdis A. Bilateral gonadoblastoma with extended calcification: case report of a tumor developing on dysgenetic gonads. Eur J Gynaecol Oncol. 2005;26:330–332. [PubMed] [Google Scholar]
- Kuhnert B, Gromoll J, Kostova E, Tschanter P, Luetjens CM, Simoni M, et al. Case report: natural transmission of an AZFc Y-chromosomal microdeletion from father to his sons. Hum Reprod. 2004;19:886–888. doi: 10.1093/humrep/deh186. [DOI] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Iwamoto T, Lee J, Yoshiike M, Nozawa S, Nishida T, et al. Spermatogenic ability is different among males in different Y chromosome lineage. J Hum Genet. 1999;44:289–292. doi: 10.1007/s100380050162. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- Lau YF. Sex chromosome genetics'99: Gonadoblastoma, testicular and prostate cancers, and the TSPY gene. Am J Hum Genet. 1999;64:921–927. doi: 10.1086/302353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YF, Lau HW, Komuves LG. Expression pattern of a gonadoblastoma candidate gene suggests a role of the Y chromosome in prostate cancer. Cytogenet Genome Res. 2003;101:250–260. doi: 10.1159/000074345. [DOI] [PubMed] [Google Scholar]
- Lin YW, Thi DA, Kuo PL, Hsu CC, Huang BD, Yu YH, et al. Polymorphisms associated with the DAZ genes on the human Y chromosome. Genomics. 2005;86:431–438. doi: 10.1016/j.ygeno.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Lingenfelter PA, Delbridge ML, Thomas S, Hoekstra HE, Mitchell MJ, Graves JA, et al. Expression and conservation of processed copies of the RBMX gene. Mamm Genome. 2001;12:538–545. doi: 10.1007/s00335001-0003-z. [DOI] [PubMed] [Google Scholar]
- Lynch M, Cram DS, Reilly A, O'bryan MK, Baker HW, de Kretser DM, et al. The Y chromosome gr/gr subdeletion is associated with male infertility. Mol Hum Reprod. 2005;11:507–512. doi: 10.1093/molehr/gah191. [DOI] [PubMed] [Google Scholar]
- Ma K, Inglis JD, Sharkey A, Bickmore WA, Hill RE, Prosser EJ, et al. A Y chromosome gene family with RNA-binding protein homology: candidates for the azoospermia factor AZF controlling human spermatogenesis. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- Machev N, Saut N, Longepied G, Terriou P, Navarro A, Levy N, et al. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet. 2004;41:814–825. doi: 10.1136/jmg.2004.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, et al. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- Mancilla EE, Poggi H, Repetto G, Rumie H, Garcia H, Ugarte F, et al. Y chromosome sequences in Turner's syndrome: association with virilization and gonadoblastoma. J Pediatr Endocrinol Metab. 2003;16:1157–1163. doi: 10.1515/jpem.2003.16.8.1157. [DOI] [PubMed] [Google Scholar]
- Mazzanti L, Cicognani A, Baldazzi L, Bergamaschi R, Scarano E, Strocchi S, et al. Gonadoblastoma in Turner syndrome and Y-chromosome-derived material. Am J Med Genet A. 2005;135:150–154. doi: 10.1002/ajmg.a.30569. [DOI] [PubMed] [Google Scholar]
- Meeks JJ, Weiss J, Jameson JL. Dax1 is required for testis determination. Nat Genet. 2003;34:32–33. doi: 10.1038/ng1141. [DOI] [PubMed] [Google Scholar]
- Meschede D, Lemcke B, Behre HM, De Geyter C, Nieschlag E, Horst J. Clustering of male infertility in the families of couples treated with intracytoplasmic sperm injection. Hum Reprod. 2000;15:1604–1608. doi: 10.1093/humrep/15.7.1604. [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- Mulhall JP, Reijo R, Alagappan R, Brown L, Page D, Carson R, et al. Azoospermic men with deletion of the DAZ gene cluster are capable of completing spermatogenesis: fertilization, normal embryonic development and pregnancy occur when retrieved testicular spermatozoa are used for intracytoplasmic sperm injection. Hum Reprod. 1997;12:503–508. doi: 10.1093/humrep/12.3.503. [DOI] [PubMed] [Google Scholar]
- Otto PS. Detecting the form of selection from DNA sequence data. Trends Genet. 2000;16:526–529. doi: 10.1016/s0168-9525(00)02141-7. [DOI] [PubMed] [Google Scholar]
- Page DC. Hypothesis: a Y-chromosomal gene causes gonadoblastoma in dysgenetic gonads. Development. 1987;101(suppl.):151–155. doi: 10.1242/dev.101.Supplement.151. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Pearce CL, Kolonel LN, Altshuler D, Henderson BE, Tyler-Smith C. A Y chromosomal influence on prostate cancer risk: the multi-ethnic cohort study. J Med Genet. 2003;40:815–819. doi: 10.1136/jmg.40.11.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Alonso R, Nieto K, Alvarez R, Palma I, Najera N, Erana L, et al. Distribution of Y-chromosome-bearing cells in gonadoblastoma and dysgenetic testis in 45,X/46,XY infants. Mod Pathol. 2005;18:439–445. doi: 10.1038/modpathol.3800293. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Bashamboo A, Prasad A, Pathak D, Ali S. Organizational variation of DYZ1 repeat sequences on the human Y chromosome and its diagnostic potentials. DNA Cell Biol. 2004;23:561–571. doi: 10.1089/dna.2004.23.561. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, Gianotten J, et al. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Cooke HJ. Role of the DAZ genes in male fertility. Reprod Biomed Online. 2005;10:72–80. doi: 10.1016/s1472-6483(10)60806-1. [DOI] [PubMed] [Google Scholar]
- Saavedra-Castillo E, Cortes-Gutierrez EI, Davila-Rodriguez MI, Reyes-Martinez ME, Oliveros-Rodriguez A. 47,XXY female with testicular feminization and positive SRY: a case report. J Reprod Med. 2005;50:138–140. [PubMed] [Google Scholar]
- Salo P, Kaariainen H, Petrovic V, Peltomaki P, Page DC, de la Chapelle A, et al. Molecular mapping of the putative gonadoblastoma locus on the Y chromosome. Genes Chrom Cancer. 1995;14:210–214. doi: 10.1002/gcc.2870140309. [DOI] [PubMed] [Google Scholar]
- Santos FR, Pena SD, Tyler-Smith C. PCR haplotypes for the human Y chromosome based on alphoid satellite DNA variants and heteroduplex analysis. Gene. 1995;165:191–198. doi: 10.1016/0378-1119(95)00501-v. [DOI] [PubMed] [Google Scholar]
- Saxena R, Vries JWA, Repping S, Alagappan RK, Skaletski H, Brown LG. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–267. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol. 2004;274:271–279. doi: 10.1016/j.ydbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Shahid M, Dhillion VS, Jain N, Hedau S, Diwakar S, Sachdeva P, et al. Two new novel point mutations localized upstream and downstream of the HMG box region of the SRY gene in three Indian 46,XY females with sex reversal and gonadal tumour formation. Mol Hum Reprod. 2004;10:521–526. doi: 10.1093/molehr/gah071. [DOI] [PubMed] [Google Scholar]
- Schubert S, Skawran B, Dechend F, Nayernia K, Meinhardt A, Nanda I, et al. Generation and characterization of a transgenic mouse with a functional human TSPY. Biol Reprod. 2003;69:968–975. doi: 10.1095/biolreprod.103.016501. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Solari AJ. Synaptonemal complex analysis in human male infertility. Eur J Histochem. 1999;43:265–276. [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, et al. Deletion of azoospermia factor a AZFa region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- The Y Chromosome consortium. A nomenclature for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet L, Mejean C, Moniot B, Bonneaud N, Galeotti N, Aldrian-Herrada G. Regulation of human SRY subcellular distribution by its acetylation/deacetylation. EMBO J. 2004;23:3336–3345. doi: 10.1038/sj.emboj.7600352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescentportion of the human Y chromosome long arm. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Reijo R, Page DC, Disteche CM. Gonadoblastoma: molecular definition of the susceptibility region on the Y chromosome. Am J Hum Genet. 1995;57:1400–1407. [PMC free article] [PubMed] [Google Scholar]
- Tsuei DJ, Hsu HC, Lee PH, Jeng YM, Pu YS, Chen CN, et al. RBMY, a male germ cell-specific RNA-binding protein, activated in human liver cancers and transforms rodent fibro-blasts. Oncogene. 2004;23:5815–5822. doi: 10.1038/sj.onc.1207773. [DOI] [PubMed] [Google Scholar]
- Uehara S, Funato T, Yaegashi N, Suziki H, Sato J, Sasaki T, et al. SRY mutation and tumor formation on the gonads of XP pure gonadal dysgenesis patients. Cancer Genet Cytogenet. 1999;113:78–84. doi: 10.1016/s0165-4608(99)00010-2. [DOI] [PubMed] [Google Scholar]
- Uehara S, Hashiyada M, Sato K, Nata M, Funato T, Okamura K. Complete XY gonadal dysgenesis and aspects of the SRY genotype and gonadal tumor formation. J Hum Genet. 2002;47:279–284. doi: 10.1007/s100380200040. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- Vernole P, Terrinoni A, Didona B, De Laurenzi V, Rossi P, Melino G, et al. An SRY-negative XX male with Huriez syndrome. Clin Genet. 2000;57:61–66. doi: 10.1034/j.1399-0004.2000.570109.x. [DOI] [PubMed] [Google Scholar]
- Verp MS, Simpson JL. Abnormal sexual differentiation and neoplasia. Cancer Genet Cytogenet. 1987;25:191–218. doi: 10.1016/0165-4608(87)90180-4. [DOI] [PubMed] [Google Scholar]
- Vijayakumar S, Garcia D, Hensel CH, Banerjee M, Bracht T, Xiang R, et al. The human Y chromosome suppresses the tumorigenicity of PC-3, a human prostate cancer cell line, in athymic nude mice. Genes Chromosomes Cancer. 2005 doi: 10.1002/gcc.20250. in press. [DOI] [PubMed] [Google Scholar]
- Vinci G, Raicu F, Popa L, Popa O, Cocos R, McElreavey K. A deletion of a novel heat shock gene on the Y chromosome associated with azoospermia. Mol Hum Reprod. 2005;11:295–298. doi: 10.1093/molehr/gah153. [DOI] [PubMed] [Google Scholar]
- Vogel T, Speed RM, Teague P, Cooke HJ. Mice with Y chromosome deletion and reduced Rbm genes on a heterozygous Dazl1 null background mimic a human azoospermic factor phenotype. Hum Reprod. 2000;14:3023–3029. doi: 10.1093/humrep/14.12.3023. [DOI] [PubMed] [Google Scholar]
- Vogel T, Speed RM, Ross A, Cooke HJ. Partial rescue of the Dazl knockout mouse by the human DAZL gene. Mol Hum Reprod. 2002;8:797–804. doi: 10.1093/molehr/8.9.797. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn FM. Human Y chromosome azoospermia factors AZF mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Yan HT, Shinka T, Kinoshita K, Sato Y, Umeno M, Chen G, et al. Molecular analysis of TBL1Y, a Y-linked homologue of TBL1X related with X-linked late-onset sensorineural deafness. J Hum Genet. 2005;50:175–181. doi: 10.1007/s10038-005-0237-9. [DOI] [PubMed] [Google Scholar]
- Yen PH, Chai NN, Salido EC. The human DAZ genes, a putative male infertility factor on the Y chromosome, are highly polymorphic in the DAZ repeat regions. Mamm Genome. 1997;8:756–759. doi: 10.1007/s003359900560. [DOI] [PubMed] [Google Scholar]
- Yogev L, Segal S, Zeharia E, Gamzu R, Maymon BB, et al. Sex chromosome alignment at meiosis of azoospermic men with azoospermia factor microdeletion. J Androl. 2004;25:110–116. doi: 10.1002/j.1939-4640.2004.tb02765.x. [DOI] [PubMed] [Google Scholar]
- Wang QJ, Lu CY, Li N, Rao SQ, Shi YB, Han DY, et al. Y-linked inheritance of non-syndromic hearing impairment in a large Chinese family. J Med Genet. 2004;41:e80. doi: 10.1136/jmg.2003.012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder JA, Mobasher Z, Hammer MF. Genetic Evidence for Unequal Effective Population Sizes of Human Females and Males. Mol Biol Evol. 2004;21:2047–2057. doi: 10.1093/molbev/msh214. [DOI] [PubMed] [Google Scholar]


