Abstract
Influenza A virus (IAV) is one of the most common infectious pathogens in humans and causes considerable morbidity and mortality. The recent spread of highly-pathogenic avian IAV H5N1 viruses has reinforced the importance of pandemic preparedness. In the pathogenesis of IAV infection, cellular proteases play critical roles in the process of viral entry into cells that subsequently leads to tissue damage in the infected organs. Since there are no processing protease for the viral membrane fusion glycoprotein hemagglutinin precursor (HA0) in IAV, entry of the virus into cells is determined primarily by the host cellular HA0 processing proteases that proteolytically activate membrane fusion activity. HA0 of seasonal human IAV has the consensus cleavage site motif Q(E)-T/X-R and is selectively processed by at least seven different trypsin-type processing proteases identified to-date in animal model experiments using mouse-adapted IAV or gene expression system in MDCK cells. As is the case for the highly pathogenic avian influenza (HPAI) A virus, endoproteolytic processing of the HA0 occurs through ubiquitous cellular processing proteases, which selectively recognize the multi-basic consensus cleavage site motifs, such as R-X-K/R-R, and K-X-K/R-R. The cleavage enzymes for the R-X-K/R-R motif, but not K-X-K/R-R motif, have been reported to be furin and pro-protein convertase (PC)5/6 in the trans-Golgi network. Here we report new members of type II transmembrane serine proteases of the cell membrane, mosaic serine protease large form (MSPL) and its splice variant TMPRSS13, which recognize and cleave both R-X-K/R-R and K-X-K/R-R motifs without calcium. Furthermore, IAV infection significantly up-regulates a latent ectopic pancreatic trypsin, one of the potent HA processing proteases, and pro-matrix metalloprotease-9, in various organs. These proteases may synergistically damage the blood-brain barrier in the brain and basement membrane of blood vessels in various organs, resulting in severe edema and multiple organ failure. In this review, we discuss these proteases as new drug target molecules for IAV treatment acting by inhibition of IAV multiplication and prevention of multiple organ failure, other than anti-viral agents, viral neuraminidase inhibitors.
Keywords: Influenza virus, highly pathogenic avian influenza virus, hemagglutinin processing protease, tryptase Clara, mini-plasmin, trypsin, TMPRSS-13
INTRODUCTION
Post-translational proteolytic cleavage of precursors of membrane fusion glycoprotein hemagglutinin (HA0) of the enveloped influenza A virus (IAV) is a prerequisite for viral membrane fusion activity and virus entry into the cell (Homma and Ohuchi, 1973; Scheid and Choppin, 1974; Klenk and Rott, 1988; Kido et al, 1999). However, IAV has no HA0 processing protease in its genes. Thus, cellular proteolytic conversion of HA0 to HA1 and HA2 subunits is indispensable for viral entry and multiplication within the infected host, and is a major determinant of IAV pathogenicity.
To-date, two main HA0 processing motifs have been described in IAV. One is the single basic HA0 processing motif Q(E)-T/X-R sequence of human seasonal IAVs (Klenk and Garten, 1994; Kido et al, 1999), and the R-E-T-R and P-K-X-R motifs of avirulent avian IAV H5 and H7, respectively (Tsukamoto et al, 2008), which are readily cleaved by several extracellular trypsin-type proteases. Among many of the cellular trypsin-type serine proteases, only few enzymes selectively convert HA0 to HA1 and HA2. In addition, the processing proteases for the single basic motif are predominantly distributed in the airway and enteric duct, and most of the epidemic human IAV known to date are pneumotropic in which the virus proliferates in these organs, except for a few neurotropic strains, such as IAV WSN/33(H1N1) strain (Ward, 1996). The other HA0 processing motif is a multiple-basic-residues motif of the highly pathogenic avian influenza (HPAI) viruses. In the multiple basic residues motifs, the R-K/R-K/R/X-R motif with R at position P4 is readily cleaved by ubiquitously present intracellular processing proteases, such as furin and proprotein convertases (PCs)5/6, in the presence of calcium (Klenk and Garten, 1994; Horimoto et al, 1994; Remacle et al, 2008). However, replacement of P4 R by K and a non-basic amino acid in the multi-basic motif significantly suppresses the processing activities of furin and PCs (Barr, 1991; Thomas, 2002; Remacle et al, 2008). We recently reported the identification of novel type II membrane processing proteases of the cell surface, which preferentially cleaved after the carboxy-terminal R residue in the multiple basic residues motifs of both R-K/R-K/R/X-R and K-K/R-K/R/X-R sequences without calcium.
In the progress of IAV pathogenicity, infection results in up-regulation of host cellular proteases, such as latent ectopic pancreatic trypsin and matrix metalloprotease-9 (MMP-9) in various organs, potentially causing damage of connective tissues and destruction of tight junctions of endothelial cells followed by severe edema.
HA0 PROCESSING PROTEASES IN THE AIRWAY FOR HUMAN SEASONAL INFLUENZA VIRUSES
Figure 1 shows the detection of mouse-adapted IAV Aich/2/68(H3N2) antigen in airway epithelial cells of mice at 2 and 6 days after intranasal infection. The viral antigen was initially detected on the cilia of airway epithelial cells at day 2 after infection (typically >90% of infected cells were ciliated airway epithelial cells in the mouse bronchus) (Figure 1A). Along with the spread of virus infection at day 6, the virus antigen spread from the ciliated epithelial cells to neighboring non-ciliated secretory epithelial cells in the bronchus and was detectable in the whole cell but sparing the nucleus (Figure 1B). The comb-like structure of epithelial cells cilia became swollen, forming fused-structures at day 6 after viral infection (Figure 1C and 1D), which subsequently fell off.
Figure 1.

Immunofluorescence detection of mouse-adapted human IAV Aichi/2/68(H3N2) in vivo after infection of mice for 2 days (A) and 6 days (B). C57BL/6 female mice weighing 10-12 g were instilled intranasally with 6.6 × 104 PFU of mouse-adapted human IAV. Two and 6 days after infection, mouse bronchi were isolated, fixed and immunostained for virus HA antigen (green). C and D: Scanning electron micrographs of the surface of bronchus of mice before (C) and (D) 6 days after IAV infection. Cilia of airway epithelial cells of female mouse with a comb-like structure before virus infection became swollen, forming fused-structures at 6 days after intranasal IAV infection and many of such cilia subsequently fell off. Bar = 1 μm.
The HA0 processing proteases reported for human IAV in animals and humans that recognize single basic motif are pancreatic trypsin (Klenk et al, 1975), plasmin from calf and chicken serum (Lazarowitz et al, 1973), blood clotting factor Xa from chick embryo (Gotoh et al, 1990), tryptase Clara from rat lungs (Kido et al, 1992), mini-plasmin from rat lungs (Murakami et al, 2001), ectopic anionic trypsin from rat lungs (Towatari et al, 2002), porcine mast cell tryptase (Chen et al, 2000), tryptase TC30 from porcine lungs (Sato et al, 2003) and transmembrane protease serine (TMPRSS) 2 and type II membrane protein human airway trypsin-like protease (HAT) (Böttcher et al, 2006) (Table 1). Why various trypsin-type HA0 processing proteases capable of potentiating IAV infections exist in the airways? We found different distribution for these cellular proteases in the airways (Kido et al, 2007) as well as different proteolytic potentiation of various strains of the viruses (Murakami et al, 2001). Among the proteases we have examined, trypsin efficiently activated the infectivity of all strains. Mini- and micro-plasmin activated almost all strains, though less efficiently than trypsin. Different HA cleavability of some of the processing proteases was found among species: mast cell tryptase from porcine lungs processed HA0, whereas human and rat mast cell tryptase did not (Chen et al, 2000). However, among the reported proteases, blood clotting factor Xa from human serum, not from chick embryo, hardly cleaved HA0 of IAV Aich/2/68(H3N2) without loss of their amidolytic activities against synthetic peptide substrates (Kido H et al, unpublished data). In addition to the host cellular proteases, microbial proteases also proteolytically activate influenza virus HA0 in bacterial infection of the airways and may play a role in the spread of the virus (Tashiro et al, 1987; Akaike and Maeda, 2000). Mutational evolution of IAV HA0 to adapt to these host trypsin-type processing proteases in the airway allows efficient multiplication of the virus in vivo which ultimately increase to becomes epidemic.
Table 1.
Comparison of viral activating enzymes
| Enzyme | MW (kDa) SDS-PAGE | MW (kDa) Non-reducing condition (composition) | Optimal substrate | Inhibitor | Localization | Reference |
|---|---|---|---|---|---|---|
| Tryptase Clara | 30 | 180 (hexamer) | QAR | aprotinin, leupeptin, antipain, KSTI | bronchiolar epithelial Clara cells | Kido et al., 1992 |
| Mini-plasmin | 28 + 12 | 38 (heterodimer) | QAR | aprotinin, KSTI, BBSTI, leupeptin | folded epithelial cells in bronchiolar divisions | Murakami et al., 2001 |
| Ectopic anionic trypsin I | 22 | 31 (monomer) | EGR QGR | aprotinin, soybean trypsin inhibitor, leupeptin | stromal cells in peri-bronchiolar region | Towatari et al., 2002 |
| Mast cell tryptase | 32-35 | 120 (tetramer) | QAR | antipain, leupeptin | mast cells | Chen et al., 2000 |
| Tryptase TC30 | 30 | 30 (monomer) | SIQSR | aprotinin, benzamidine, leupeptin | ND | Sato et al., 2003 |
| HAT | 28 | 28 (monomer) | FSR | aprotinin, leupeptin, antipain, KSTI | ciliated epithelial cells | Yasuoka et al., 1997 |
| TMPRSS2 | 70 (32) | 70 (32 kDa: released form) (monomer) | GGR | Benzyloxycarbonyl lysine dephenylphosphonate | Epithelial cells of respiratory, gastrointestinal, and urogenital tracts | Wilson et al., 2005 |
HAT: type II membrane protein human airway trypsin-like protease; TMPRSS2: transmembrane protease serine 2; M.W.: molecular weight, KSTI; Kunitz-type soybean trypsin inhibitor; BBSTI: Bowman-Birk soybean trypsin inhibitor; ND: Not determined.
HA0 PROCESSING PROTEASES FOR THE HIGHLY PATHOGENIC AVIAN INFLUENZA VIRUSES
To-date, all HPAI viruses of H5 and H7 subtypes have cleavage site motifs with multiple basic amino acids (Zambon, 2001; Tsukamoto, 2008) and HA0 of HPAI virus is postulated to be cleaved by ubiquitous proteases, such as subtilisin-like proteases, furin and PC5/6 (Klenk and Garten, 1994). In this review, we add a new member of type II membrane serine proteases on the cell surface, other than the subtilisin-like proteases in the trans-Golgi network, which recognize and cleave HA0 of HPAI viruses with the multiple basic motifs of both R-K/R-(K/R/X)-R sequence with R at position P4 and K-K/R-(K/R/X)-R sequence with K at position P4.
The cleavage preferences of subtilisin-like proteases, furin, PC4, PC5/6, PC7, PACE4, are R at P1 and P4 positions in the cleavage motif R-X-(R/K/X)-R and replacement of P4 R by K and a nonbasic amino acid significantly suppresses their processing activity (Bently et al, 1986; Barr 1991; Thomas 2002; Remacle et al, 2008). These subtilisin-like proteases are calcium-dependent with a pH optimum ranging from 6.5 to 7.5 (Stieneke-Gröber et al, 1992) and are mainly localized intracellularly in the trans-Golgi network and some cycle between the trans-Golgi and the cell surface membrane (Thomas, 2002). The relationship between the pathogenesis of HPAI viruses and proteolytic HA0 cleavage of the multiple basic residue motif with R at P1 and P4 positions has been studied extensively (see reviews by Zambon, 2001; Stieneke-Gröber et al, 1992; Walker et al, 1994; Shiryaev et al, 2007). In the HPAI viruses, however, there is another type of HA0 cleavage sequence with multi-basic cleavage motif with R at P1 and K at P4, which is not a preferable or not a substrate of furin.
Type II membrane serine proteases (TTSP) of mosaic serine protease large form (MSPL) and its splice variant transmembrane protease serine 13 (TMPRSS13) with a pH optimum ranging from 7.0 to 8.5 and no calcium requirement, are identified as new candidate HA0 processing proteases for HAPI virus on the cell surface. MSPL and TMPRSS13 were recently cloned from human lungs (Kim et al, 2001; Kido and Okumura, 2008). Although furin selectively cleaves the multiple basic residues motif with R at P1 and P4, these novel membrane proteases preferentially recognize the double basic residues at the cleavage site, and R or K at P4 position in the multiple basic residues motifs greatly enhances the efficiency of processing.
Figure 2 shows alignment of the amino acid sequences of MSPL with 581 residues and TMPRSS13 with 567 residues and schematic representation of the domain structure of MSPL. The serine protease domains at residues 321–581 for MSPL and 326–567 for TMPRSS13 are identical except for the C-terminal ends, i.e., 555–581 for MSPL and 560–567 for TMPRSS13 (Figure 2A). They have a typical catalytic triad (His, Asp and Ser) of serine proteases, and a putative proteolytic activation site (R) in the R320-I-V-G-G324 in MSPL and R325-I-V-G-G329 in TMPRSS13. The consensus protease domain of MSPL/TMPRSS13 exhibits 42, 39 and 43% identity with those of human plasma kallikrein (Beaubien et al, 1991), hepsin (Tsuji et al, 1991), and transmembrane protease serine 2 (TMPRSS2) (Paoloni-Giacobino et al, 1997), respectively, but poor identity with subtilisin family protease furin. MSPL and TMPRSS13 have putative transmembrane domains (P162–W184 and P167–W189, respectively) at the N-terminal sides. The cytoplasmic domain of these variants has tandem repeat motifs corresponding to the phosphorylation sites of cyclin-dependent kinase Cdc2, CaM kinase II and protein kinase C (Kennelly and Krebs, 1991; Wang and Roach, 1993), and is the longest among the TTSPs known to date (Figure 2B).
Figure 2.
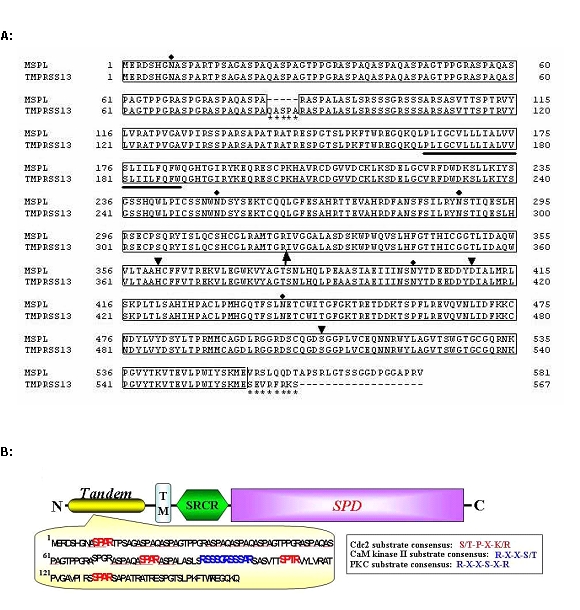
Alignment of amino acid sequences of hMSPL and hTMPRSS13 and schematic representation of the domain structures of hMSPL. (A) Alignment of the amino acid sequences of hMSPL and hTMPRSS13. Numbering starts with Met1 of the first amino acid. The putative transmembrane domain is bold-underlined. Arrow points to the putative cleavage site for active serine protease. The catalytic triad (His, Asp and Ser) are indicated by arrowheads. Potential N-glycosylation sites are indicated by closed diamonds. hTMPRSS13 differs from hMSPL by the 5-amino acid insertion in the tandem repeat sequences (from Gln83 to Ala87) and the 8-amino acid extension at the C-terminus (from Ser560 to Ser567) (asterisks). (B) Schematic representation of predicted domain structure of hMSPL. TM, transmembrane domain; SRCR, scavenger receptor cysteine-rich domain; SPD, serine protease domain. The putative kinase phosphorylation sites found in the N-terminal side tandem repeat sequence. The sequence motifs matching the consensus sequences, such as S/T-P-X-K/R of Cdc2, R-X-X-S/T of CaM kinase II and R-X-X-S-X-R of protein kinase C, are in boldface. Homologous sequences, ASPA, are underlined.
The expression profiles of MSPL and TMPRSS13 in various human tissues and cells assessed by reverse transcription-polymerase chain reaction (RT-PCR) with specific primer pairs for each C-terminal of the serine protease domains (Figure 3A and B), show that both genes are predominantly expressed in human lung, placenta, pancreas and prostate, although TMPRSS13 is almost ubiquitously expressed in various organs except a limited expression in skeletal muscle. TMPRSS13/MSPL are expressed at high levels in blood vessels, human choriocarcinoma (BeWo) cells and human umbilical vein endothelial cells (HUVEC) in primary cultures, but not detectable in human endothelial cell line ECV-304, although furin is detectable in almost all human organs and the cells examined, including ECV-304 (data not shown). The high expression of chicken TMPRSS13 (cTMPRSS13) is also detected in blood vessels, lungs, trachea, colon, small intestines and kidneys (Figure 3C).
Figure 3.

Gene expression profiles of hMSPL and hTMPRSS13 in various human tissues (A), cells (B) and those of cTMPRSS13 (C). RT-PCR analysis was carried out using MSPL- and TMPRSS13-specific primers in the indicated tissues and cells. As a template, Human Multiple Tissue cDNA Panels were used. The expression level of GAPDH mRNA in each sample was used as an internal control. HUVEC; human umbilical vein endothelial cells.
The recombinant extracellular regions of human MSPL (hMSPL) and hTMPRSS13 (i.e., SRCR domain and serine protease domain) after transfection of the constructs into HEK 293T cells were purified from the serum-free culture medium and then characterized. The substrate specificities of the recombinant hMSPL are shown in Table 2. Among the synthetic substrates tested, hMSPL preferentially hydrolyzes dibasic substrates (R/K at P1 and P2 positions) and furin substrate L-pyroglutamyl-R-T-K-R-4-methylcoumaryl-7-amide (MCA) with R at P1 and P4 being the best. Peptide substrate of HPAI virus A/chick/Penn/1370/83 (H5N2), M-R-N-V-P-Q-K-K-K-R↓-G-L-F-G with K at position P4 is also cleaved by hMSPL efficiently, the activity being equivalent to the furin substrate. However, substrates with Lys at P1 position are less hydrolyzed. Unlike trypsin and plasmin, Bz-R-MCA and Boc-V-L-K-MCA are not hydrolyzed by MSPL and no activity is observed on the substrates for elastase and chymotrypsin-type proteases. Recombinant hTMPRSS13 shows similar substrate specificity to that of hMSPL. The active site mutants, hMSPLS506A and hTMPRSS13S511A, do not hydrolyze any substrates at all. The activity of hMSPL is inhibited by trypsin inhibitors, such as aprotinin, benzamidine and Bowman-Birk trypsin inhibitor, but, unlike trypsin, α1-antitrypsin does not inhibit the enzyme activity (Kido and Okumura 2008). In the furin inhibitors tested, decanoyl-RVKR-chloromethylketone, but not α1-antitrypsin Portland, inhibits the enzyme activity with a low Ki value of 2.9 nM (Okumura et al, submitted). The inhibitor specificity of recombinant hTMPRSS13 is similar to that of hMSPL.
Table 2.
Substrate specificities of recombinant soluble hMSPL
| Substrate (100 μM) | Relative activity (%) |
|---|---|
| Boc-Q-A-R-MCA | 100.0±1.8 |
| Pyr-R-T-K-R-MCA | 3620.7±21.6 |
| M-R-N-V-P-Q-K-K-K-R↓-G-L-F-G | 3312.1±43.2 |
| Boc-Q-R-R-MCA | 707.7±5.9 |
| Boc-R-V-R-R-MCA | 667.4±5.2 |
| Boc-L-R-R-MCA | 350.0±2.3 |
| Boc-L-K-R-MCA | 192.3±1.9 |
| Boc-E-K-K-MCA | 57.8±0.8 |
| Boc-E(OBzl)-A-R-MCA | 66.7±1.1 |
| Boc-L-S-T-R-MCA | 46.7±0.3 |
| Boc-F-S-R-MCA | 33.3±0.2 |
| Boc-Q-G-R-MCA | 20.0±0.2 |
| Boc-L-T-R-MCA | 18.9±0.1 |
| P-F-R-MCA | 12.2±0.1 |
| Boc-E(OBzl)-G-R-MCA | 11.1±0.1 |
| Boc-V-L-K-MCA | 0.0±0.0 |
| Bz-R-MCA | 0.0±0.0 |
| Suc-A-P-A-MCA | 0.0±0.0 |
| Suc-A-A-P-F-MCA | 0.0±0.0 |
The amidolytic activities of the purified hMSPL were analyzed toward various synthetic peptides in 0.1 M Tris-HCl (pH 8.0) at 37°C. The amount of 7-amino-4-methylcoumarin and the C-terminal fragment peptide liberated from the substrate were determined. One unit of enzyme activity was defined as the amount that degraded 1 μmol of substrate per min. Data are mean ± SD of three separate experiments and are expressed as percentages of activity toward Boc-Q-A-R-MCA.
PROTEOLYTIC ACTIVATION OF HA0 OF HIGHLY PATHOGENIC AVIAN INFLUENZA VIRUS BY hMSPL AND hTMPRSS13
A large number of HPAI (H5 and H7) viruses has the HA cleavage site sequence of R-K/R-(K/R/X)-R, which is cleavable by furin, PC5/6, hMSPL and hTMPRSS13. On the other hand, part of HPAI viruses has the other cleavage site sequence of K-K/R-(K/R/X)-R with K at P4 position. To analyze the cleavage specificities of hMSPL and hTMPRSS13 against the motifs of HPAI, we performed mass spectrometry analyses of the synthetic HA peptides containing the cleavage motif sequence and part of membrane fusion domain G-L-F-G sequence at the C-terminal side, M-R-N-V-P-Q-K-K-K-R-G-L-F-G (A/chicken/Pennsylvania/1370/83(H5N2)), and P-E-P-S-K-K-R-K-K-R-G-L-F-G (A/FPV/Rostock/34(H7N1)), after incubation with these enzymes for 1 h. Both enzymes preferentially cleaved the C-terminal side of R adjacent to the G-L-F-G sequence. There was no significant difference in cleavage efficiency by hMSPL and hTMPRSS13 between the peptide M-R-N-V-P-Q-K-K-K-R-G-L-F-G with K at P4 position and P-E-P-S-K-K-R-K-K-R-G-L-F-G with R at P4 position (Okumura et al, submitted). Similar results were obtained in the experiments of eight different synthetic peptides with K/R at P4 from HPAI viruses.
For exo vivo experiments, we established full-length human MSPL and TMPRSS13 stable transfectant ECV-304 cells, ECV304-MSPL and ECV304-TMPRSS13, respectively. Since recombinant HA expression system of HPAI virus with the K-K-K-R cleavage site is not currently available in spite of various trials, we used in the following experiments the expression of highly virulent influenza A/chick/Penn/1/83 (H5N2) variant 47 (Ohuchi et al, 1989) as an example of poor-susceptible strain to furin. This variant has R-K-K-R cleavage motif but poor cleavage susceptibility to furin. After infection of ECV304-MSPL, ECV304-TMPRSS13 cells and wild-type ECV304 (ECV304-WT) cells with the A/chicken/ Penn/1/83 (H5N2) variant 47 HA/SV40 recombinant virus for 48 h, HA cleavage efficiencies and cell fusion activity monitored by giant cell formation were analyzed. The recombinant HA0 in ECV304-WT cells was converted to HA1 subunit with low efficiency of 9-14%, although higher conversion rates of 37-42% were observed in both ECV304-MSPL and ECV304-TMPRSS13 cells (Okumura et al, submitted). On the other hand, furin-susceptible recombinant H7 HA0 with R-K-K-R motif showed 40-45% conversion in WT and MSPL/TMPRSS13-transfectant cells after infection with the A/FPV/Rostock/34 H7 HA/SV40 recombinant virus (Ohuchi et al. 1994) under the same experimental conditions. The proteolytically activated recombinant HA0 of HPAI viruses facilitated cell fusion. These results suggest that MSPL and TMPRSS13 are novel candidate HA0 processing proteases for HPAI viruses, in addition to furin and PC5/6.
UPREGULATION BY INFECTION OF LATENT PANCREATIC TRYPSIN IN VARIOUS ORGANS
Most of the epidemic human IAV are pneumotropic but a few neurotropic strains can multiply both in the airway and brain. In addition, a rare but often fatal encephalopathy caused by infection with a pneumotropic IAV has been reported in children with Reye's syndrome and influenza-associated encephalopathy (Delorme and Middleton 1979; Fujimoto et al, 1998). Recently, we reported that IAV infection can markedly upregulate latent pancreatic trypsin in the brain (Le et al, 2006) and various organs that may have not only a HA0 processing activity but also a maturase activity of upregulated proMMP-9 (Yamada et al, 2006).
Figure 4 shows the localization of brain trypsin I determined by in situ hybridization. The results showed that this enzyme clusters in the hippocampal pyramidal and dentate gyrus neuronal cells and also distributes in endothelial cells of the brain capillaries, particularly in the allocortex. The enzyme distribution pattern supports the accumulation of neurovirulent IAV WSN(H1N1) antigen in hippocampal neuronal cells (Takahashi et al, 1995) and that of pneumotropic IAV Aichi/2/68(H3N2) antigen in the brain capillaries (Yao et al, 2006). Upregulation of proMMP-9 and its convertase trypsin seems to synergistically enhance the destruction of basement membrane component type IV collagen and endothelial cell tight junction components, resulting in sever edema and connective tissue damage in various organs.
Figure 4.
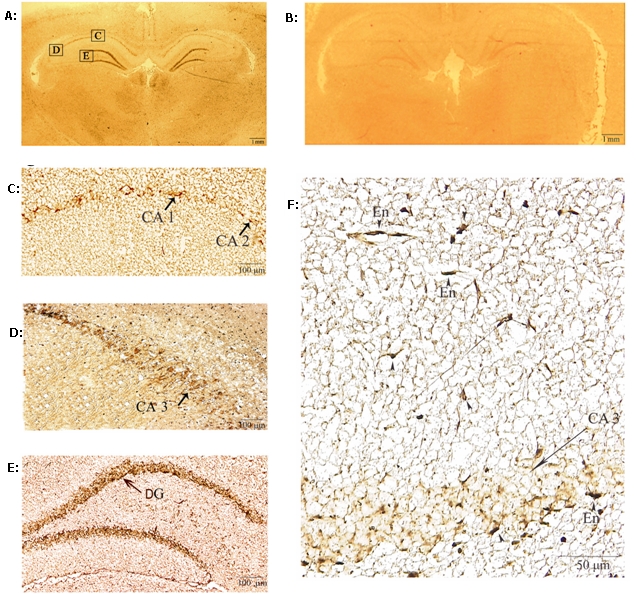
In situ hybridization of trypsin I in rat brain. Low-power photomicrographs of rat brain section, hybridized with rat trypsin I antisense probe (A) and sense probe (B). Bars in A and B represent 1 mm. High power photographs of A show that the positive staining is localized in pyramidal neurons of the cornuammonis (CA)1-2 (C) and CA-3 (D), and in the granular cells of the dentate gyrus (DG) (E). Capillary endothelial cells (En) were also heavily stained (D and F). Bars, C, D and E = 100 μm, F = 50 μm.
To provide evidence for the roles of cellular proteases in influenza virus infection, phenotype analyses of trypsin I knockout mouse and TMPRSS13 knockout mouse after influenza virus infection are now under investigations.
CONCLUSIONS
Cellular HA0 processing enzymes are prerequisite for the infectivity and pathogenicity of human IAV and HPAI viruses. In addition, IAV-induced trypsin and proMMP-9 causes further tissue damage. Anti-influenza virus drugs, inhibitors of viral neuraminidase, are going to be stockpiled in many countries as part of their anti-pandemic planning. However, adverse effects have recently been observed from its therapeutic or prophylactic administration (Nicholson et al, 2000), including reports of increasingly frequent neuraminidase inhibitor resistant virus (Kiso et al, 2004). To prevent the emergence of drug-resistant mutants and to improve the therapeutic potential of these drugs, the combined use of several anti-viral drugs with different modes of action seems preferable. One promising candidate anti-influenza drug could be inhibitors of cellular processing proteases of viral envelope fusion glycoprotein precursor HA0, which inhibit viral entry into cells. The other strategy is inhibition of up-regulation of trypsin and proMMP-9, which might be useful to prevent tissue damage in various organs.
ACKNOWLEDGEMENTS
Our studies were supported in part by Grants-in-Aid for The Special Coordination Funds for Promoting Science and Technology of Ministry of Education, Culture, Science and Technology and Promotion of Fundamental Studies in Health Science of the National Institute of Biomedical Innovation of Japan.
LIST OF ABBREVIATIONS
- Boc
t-Butyloxycarbonyl
- Bz
Benzoyl
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HUVEC
Human umbilical vein endothelial cell
- HA0
Hemagglutinin precursor
- HAT
Human airway trypsin-like protease
- HPAI
Highly pathogenic avian influenza A virus
- IAV
Influenza A virus
- MCA
Methylcoumaryl-7-amide
- MMP-9
Matrix metalloprotease-9
- MSPL
Mosaic serine protease large form
- PC
Proprotein convertase
- TMPRSS
Transmembrane protease serine
- TTSP
Type II membrane serine protease
COMPETING INTERESTS
None declared.
REFERENCES
- Akaike T, Maeda H. Nitric oxide and virus infection. Immunology. 2000;101:300–308. doi: 10.1046/j.1365-2567.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr PJ. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Beaubien GI, Rosinki-Chupin MG, Mattei M, Mbikay M, Chrestien M, Seidah NG. Gene structure and chromosomal localization of plasma kallikrein. Biochemistry. 1991;30:1628–1635. doi: 10.1021/bi00220a027. [DOI] [PubMed] [Google Scholar]
- Bently AK, Rees DJG, Rizza C, Brownlee GG. Defective propeptide processing of blood clotting factor IX caused by mutation of arginine to glutamine at position-4. Cell. 1986;45:343–348. doi: 10.1016/0092-8674(86)90319-3. [DOI] [PubMed] [Google Scholar]
- Böttcher E, Matrosovich T, Beyerle M, et al. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shiota M, Ohuchi M, et al. Mast cell tryptase from pig lungs triggers infection by pneumotropic Sendai and influenza viruses. Purification and characterization. Eur J Biochem. 2000;267:3189–3197. doi: 10.1046/j.1432-1327.2000.01346.x. [DOI] [PubMed] [Google Scholar]
- Delorme L, Middleton PJ. Influenza A virus associated with acute encephalopathy. Am J Dis Child. 1979;133:822–824. doi: 10.1001/archpedi.1979.02130080062011. [DOI] [PubMed] [Google Scholar]
- Fujimoto S, Kobayashi M, Uemura O, et al. PCR on cerebrospinal fluid to show influenza-associated acute encephalopathy or encephalitis. Lancet. 1998;352:873–875. doi: 10.1016/S0140-6736(98)12449-2. [DOI] [PubMed] [Google Scholar]
- Gotoh B, Ogasawara T, Toyoda T, Inocencio NM, Hamaguchi M, Nagai Y. An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J. 1990;9:4189–4195. doi: 10.1002/j.1460-2075.1990.tb07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Ohuchi M. Trypsin action on the growth of Sendai virus in tissue culture cell. J Virol. 1973;12:1457–1465. doi: 10.1128/jvi.12.6.1457-1465.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Nakayama K, Smeekens SP, Kawaoka Y. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol. 1994;68:6074–6078. doi: 10.1128/jvi.68.9.6074-6078.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- Kido H, Murakami M, Oba K, Chen Y, Towatari T. Cellular proteinases trigger the infectivity of influenza A and Sendai viruses. Mol Cells. 1999;9:235–244. [PubMed] [Google Scholar]
- Kido H, Okumura Y. MSPL/TMPRSS13. Front Biosci. 2008;13:754–758. doi: 10.2741/2717. [DOI] [PubMed] [Google Scholar]
- Kido H, Okumura Y, Yamada H, Le QT, Yano M. Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr Pharma Des. 2007;13:405–414. doi: 10.2174/138161207780162971. [DOI] [PubMed] [Google Scholar]
- Kido H, Yokogoshi Y, Sakai K, et al. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem. 1992;267:13573–13579. [PubMed] [Google Scholar]
- Kim DRS, Sharmin M, Inoue M, Kido H. Cloning and expression of novel mosaic serine proteases with and without a transmembrane domain from human lung. Biochem Biophys Acta. 2001;1518:204–209. doi: 10.1016/s0167-4781(01)00184-1. [DOI] [PubMed] [Google Scholar]
- Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- Klenk H-D, Rott R, Orlich M, Blödom J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- Klenk H-D, Garten W. Host cell processing proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–42. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Klenk H-D, Rott R. The molecular biology of influenza virus pathogenicity. Adv Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Goldberg AR, Choppin PW. Proteolytic cleavage by plasmin of the HA polypeptide of influenza virus: host cell activation of serum plasminogen. Virology. 1973;56:172–180. doi: 10.1016/0042-6822(73)90296-1. [DOI] [PubMed] [Google Scholar]
- Le TQ, Kawachi M, Yamada H, Shiota M, Okumura Y, Kido H. Identification of trypsin I as a candidate for influenza A virus and Sendai virus envelope glycoprotein processing protease in rat brain. Biol Chem. 2006;387:467–475. doi: 10.1515/BC.2006.062. [DOI] [PubMed] [Google Scholar]
- Menninger KA. Influenza and schizophrenia. An analysis of post-influenza “dementia precox” as of 1918 and five years later. Am J Psychiatr. 1926;5:469–529. doi: 10.1176/ajp.151.6.182. [DOI] [PubMed] [Google Scholar]
- Murakami M, Towatari T, Ohuchi M, et al. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur J Biochem. 2001;268:2847–2855. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- Nicholson KG, Aoki FY, Osterhaus AD, et al. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomized controlled trial. Neuraminidase inhibitor Flu treatment investigator group. Lancet. 2000;355:1845–1850. doi: 10.1016/s0140-6736(00)02288-1. [DOI] [PubMed] [Google Scholar]
- Ohuchi M, Orlich M, Ohuchi R, Simpson BE, Garten W, Klenk HD, Rott R. Mutations at the cleavage site of the hemagglutinin alter the pathogenicity of influenza virus A/chik/Penn/83 (H5N2) Virology. 1989;168:274–280. doi: 10.1016/0042-6822(89)90267-5. [DOI] [PubMed] [Google Scholar]
- Ohuchi M, Cramer A, Vey M, Ohuchi R, Garten W, Klenk HD. Rescue of vector-expresses fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J Virol. 1994;68:920–926. doi: 10.1128/jvi.68.2.920-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoloni-Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRSR domains and maps to 21q22.3. Genomics. 1997;44:309–320. doi: 10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- Remacle AG, Shiryaev SA, Oh E-S, et al. Substrate cleavage analysis of furin and related proprotein convertases. J Biol Chem. 2008;283:20897–20906. doi: 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Yoshida S, Iida K, Tomozawa T, Kido H, Yamashita M. A novel influenza A virus activating enzyme from porcine lung: purification and characterization. Biol Chem. 2003;384:219–227. doi: 10.1515/BC.2003.024. [DOI] [PubMed] [Google Scholar]
- Scheid A, Choppin PW. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- Shiryaev SA, Remacle AG, Ratnikov BI, et al. Targeting host cell furin proprotein convertases as a therapeutic strategy against bacterial toxins and viral pathogens. J Biol Chem. 2007;282:20847–20853. doi: 10.1074/jbc.M703847200. [DOI] [PubMed] [Google Scholar]
- Stieneke-Gröber A, Vey M, Angliker H, et al. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtillisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yamada T, Nakaima S, Yamamoto T. The substantia is a major target for neurovirulent influenza A virus. J Exp Med. 1995;181:2161–2169. doi: 10.1084/jem.181.6.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M, Ciborowski P, Klenk H-D, Pulverer G, Rott R. Role of Staphylococcus protease in the development of influenza pneumonia. Nature. 1987;352:536–537. doi: 10.1038/325536a0. [DOI] [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towatari T, Ide M, Ohba K, et al. Identification of ectopic anionic trypsin I in rat lungs potentiating pneumotropic virus infectivity and increased enzyme level after virus infection. Eur J Biochem. 2002;269:1–9. doi: 10.1046/j.1432-1033.2002.02937.x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Ashizawa H, Nakanishi K, et al. Subtyping of H1 to H5 hemagglutinin genes of avian influenza virus by RT-PCR assay and molecular determination of the pathogenic potential. J Clin Microbiol. 2008 doi: 10.1128/JCM.02386-07. doi: 10.1128/JCM.02386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A, Torres-Rosado A, Arai T, et al. Hepsin, a cell membrane-associated protease. Characterization, tissue distribution, and gene localization. J Biol Chem. 1991;266:16948–16953. [PubMed] [Google Scholar]
- Walker JA, Molloy SS, Thomas G, et al. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. J Virol. 1994;68:1213–1218. doi: 10.1128/jvi.68.2.1213-1218.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Roach PJ. Purification and assay of mammalian protein (serine/threonine) kinases. In: Hardie DG, editor. Protein phosphorylation: A practical approach. Oxford, NY: Oxford University Press; 1993. pp. 121–144. [Google Scholar]
- Ward AC. Neurovirulence of influenza A virus. J Neurovirol. 1996;2:139–151. doi: 10.3109/13550289609146876. [DOI] [PubMed] [Google Scholar]
- Yamada H, Le QT, Kousaka A, Higashi Y, Tsukane M, Kido H. Sendai virus infection up-regulates trypsin I and matrix metalloprotease-9, triggering viral multiplication and matrix degradation in rat lungs and lung L2 cells. Arch Virol. 2006;151:2529–2537. doi: 10.1007/s00705-006-0807-y. [DOI] [PubMed] [Google Scholar]
- Yao D, Kuwajima M, Chen Y, Shiota M, et al. Impaired long-chain fatty acid metabolism in mitochondria causes brain vascular invasion by a non-neurotropic epidemic influenza A virus in the newborn/suckling period: implications for influenza-associated encephalopathy. Mol Cell Biochem. 2007;299:85–92. doi: 10.1007/s11010-005-9046-x. [DOI] [PubMed] [Google Scholar]
- Zambon MC. The pathogenesis of influenza in humans. Rev Med Virol. 2001;11:227–241. doi: 10.1002/rmv.319. [DOI] [PubMed] [Google Scholar]


