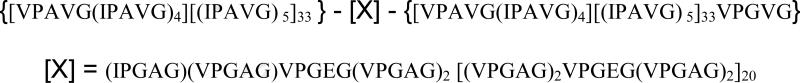Abstract
Recombinant protein polymers were synthesized and examined under various loading conditions in order to assess the mechanical stability and deformation responses of physically crosslinked, hydrated, protein polymer networks designed as triblock copolymers with central elastomeric and flanking plastic-like blocks. Uniaxial stress-strain properties, creep and stress relaxation behavior, as well as the effect of various mechanical preconditioning protocols on these responses were characterized. Significantly, we demonstrate for the first time that ABA triblock copolymers when redesigned with substantially larger endblock segments can withstand significantly greater loads. Furthermore, the presence of three distinct phases of deformation behavior was revealed upon subjecting physically crosslinked protein networks to step and cyclic loading protocols in which the magnitude of the imposed stress was incrementally increased over time. We speculate that these phases correspond to the stretch of polypeptide bonds, the conformational changes of polypeptide chains, and the disruption of physical crosslinks. The capacity to select a genetically engineered protein polymer that is suitable for its intended application requires an appreciation of its viscoelastic characteristics and the capacity of both molecular structure and conditioning protocols to influence these properties.
INTRODUCTION
The emergence of genetically engineered synthetic polypeptides has enabled the design of protein polymers composed of complex peptide sequences in which individual peptide repeat sequences can be selected with distinct mechanical, chemical, or biological properties.1-3 While a large variety of recombinant protein polymers have been reported, those composed of distinct block structures are typically characterized by relatively short block sequences. For example, Cappello and colleagues4-6 have produced a series of silk-elastinlike block copolymers (SELPs) in which silk-like regions, consisting of between 12 and 48 alternating alanine and glycine residues, are found between elastin-mimetic sequences comprised of 8 or 16 repeat sequences of Val-Pro-Gly-Val-Gly. Recently, we have reported the synthesis of high molecular weight recombinant protein block copolymers using an approach, which affords significant flexibility in the selection and assembly of blocks of diverse size and structure.7-11 This has led to the synthesis of a new class of BAB protein triblock copolymer composed of large polypeptide block sequences ranging from 400 to 1200 amino acids in length. This class of protein block copolymers are derived from elastin-mimetic polypeptide sequences in which identical endblocks of a hydrophobic, plastic-like sequence are separated by a central hydrophilic, elastomeric block. The triblock protein copolymer acts as a two-phase network when hydrated, in that the hydrophilic block remains conformationally flexible and elastomeric, while the hydrophobic block forms physical or virtual crosslinks through hydrophobic aggregation.7,8,11
Physically crosslinked protein-based materials possess a number of advantages over their chemically crosslinked counterparts, including ease of processing and the ability to avoid the addition or removal of chemical reagents or unreacted intermediates. Prior studies from our laboratory suggest that the density and strength of the physical crosslinks are important determinants of both mechanical responses and long-term material stability of two-phase protein networks.11 Therefore, in order to enhance the mechanical behavior of these materials a new elastin-mimetic triblock copolymer was synthesized that contains hydrophobic endblocks, which are nearly twice as large than prior versions of this triblock protein polymer. It bears emphasis that most elastin-like protein polymers to date have been less than 100 kD in molecular weight.12-14
Hydrophobic aggregation of the endblocks was examined using differential scanning calorimetry and rheology, and material stability of the physically crosslinked protein networks was accessed through mechanical analysis. In particular, the deformation mechanisms of these protein networks and the conditions under which the physical crosslinks are disrupted were thoroughly examined. In addition to the use of creep and stress relaxation protocols, two new tests, cyclic loading under increasing magnitudes of stress and step loading, were designed to achieve this goal. In the process, three unique phases of deformation-induced mechanical response were defined.
The resilience of a material, which characterizes its capacity for shape and energy recovery under mechanical loading, provides another crucial criterion that dictates the applications for which it may be suitable. Moreover, the magnitude of preconditioning strains and the off-loading period between loading cycles has also been reported to strongly influence the viscoelastic properties of a variety of protein and tissue derived materials.15 Thus, in this investigation we also explored the resilience and viscoelastic behavior of two-phase elastin-mimetic protein polymer networks and the capacity of distinct mechanical preconditioning protocols to affect these properties.
MATERIALS AND METHODS
Synthesis of protein triblock polymer B10
Synthetic methods used to produce the DNA inserts that encode the various elastin-mimetic block copolymers have been described previously.10,11,16 Oligonucleotide cassettes encoding elastic-like (E) and plastic-like (P) repeat units (Table 1) were independently synthesized and inserted into the BamH I and HinD III sites within the polylinker of pZErO-2. Recombinant clones were isolated after propagation in E. coli strain Top10F’, double-stranded DNA sequence analysis verified the identity of the DNA inserts E and P. DNA monomers E and P were liberated from the respective plasmids via sequential restriction digestion with Bbs I and BsmB I, respectively. Self-ligation of each DNA cassette afforded a population of concatemers.
Table 1.
Coding Sequences of Oligonucleotide Cassettes Employed for the Construction of Protein Triblock (PEP) B10
| E Block | ||||||||||||
| Val | Pro | Gly | Ala | Gly | Val | Pro | Gly | Ala | Gly | Val | Pro | Gly |
| GTT | CCA | GGT | GCA | GGC | GTA | CCG | GGT | GCT | GGC | GTT | CCG | GGT |
| CAA | GGT | CCA | CGT | CCG | CAT | GGC | CCA | CGA | CCG | CAA | GGC | CCA |
| Glu | Gly | Val | Pro | Gly | Ala | Gly | Val | Pro | Gly | Ala | Gly | |
| GAA | GGT | GTT | CCA | GGC | GCA | GGT | GTA | CCG | GGT | GCG | GGT | |
| CTT | CCA | CAA | GGT | CCG | CGT | CCA | CAT | GGC | CCA | CGC | CCA | |
| P Block | ||||||||||||
| Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala |
| ATT | CCT | GCT | GTT | GGT | ATT | CCG | GCT | GTT | GGT | ATC | CCA | GCT |
| TAA | GGA | CGA | CAA | CCA | TAA | GGC | CGA | CAA | CCA | TAG | GGA | CGA |
| Val | Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | |
| GTT | GGT | ATC | CCA | GCT | GTT | GGC | ATT | CCG | GCT | GTA | GGT | |
| CAA | CCA | TAG | GGA | GCA | CAA | CCG | TAA | GGC | CGA | CAT | CCA | |
| Modified Polylinker | ||||||||||||
| Met | Val | Pro | Glu | Ser | Ser | Gly | Thr | Glu | Asp | Val | Pro | |
| ATG | GTT | CCA | GAG | TCT | TCA | GGT | ACC | GAA | GAC | GTT | CCA | |
| TAC | CAA | GGT | CTC | AGA | AGT | CCA | TGG | CTT | CTG | CAA | GGT | |
| Gly | Val | Gly | Stop | Stop | ||||||||
| GGT | GTA | GGC | TAA | TAA | ||||||||
| CCA | CAT | CCG | ATT | ATT | ||||||||
Concatemers derived from DNA monomers E and P were inserted into the BsmB I site of their original plasmid containing the monomer cassette. Concatemers encoding 33 repeats of the P monomer and 20 repeats of the E monomer were isolated and identified via restriction cleavage with BamH I and HinD III. Double-stranded DNA sequence analysis confirmed the integrity of the concatemers within the recombinant plasmids, which were labeled pE and pP, respectively. Restriction cleavage of plasmid pE with Bbs I/Xma I and plasmid pP with BsmB I / Xma I afforded two fragments, which were separated via preparative agarose gel electrophoresis. Enzymatic ligation of pE and pP afforded the recombinant plasmid pPE, which encoded the diblock PE as a single contiguous reading frame within plasmid pZErO-2. The diblock, pPE, was used for subsequent construction of the triblock pPEP using the same biosynthetic scheme. Restriction cleavage of plasmid pP with Bbs I/Xma I and plasmid pPE with BsmB I/Xma I afforded two fragments, which were separated via preparative agarose gel electrophoresis. Enzymatic ligation of pP and pPE afforded the recombinant plasmid pPEP, which encoded the triblock PEP as a single contiguous reading frame within plasmid pZErO-2.
The triblock concatemer was liberated from pPEP via restriction cleavage with Bbs I and BsmB I and purified via preparative agarose gel electrophoresis. Enzymatic ligation was used to join the concatemer cassette to the Bbs I sites within the modified polylinker C in plasmid pBAD-A. Double stranded DNA sequence analysis confirmed the integrity of the concatemer within the recombinant plasmid, which was subsequently transferred to the expression plasmid, pET-24 (d) via restriction cleavage with Nco I and HinD III. Double stranded DNA sequence analysis confirmed the integrity of the concatemer within the recombinant plasmid, which was labeled pB10.
Plasmid pB10 encoded the triblock copolymer protein B10 as a single contiguous reading frame within plasmid pET-24 (d) and was used to transform the E. coli expression strain BL21(DE3). This afforded a protein triblock containing flanking endblock sequences [VPAVG(IPAVG)4][(IPAVG) 5]33 and a midblock sequence (IPGAG)(VPGAG)VPGEG(VPGAG)2 [(VPGAG)2VPGEG(VPGAG)2]20 (Scheme 1, Table 2). Large-scale fermentation (4L) was performed at 37°C in Terrific Broth (TB) medium supplemented with kanamycin (50 μg/mL). The fermentation cultures were incubated under antibiotic selection for 48 h at 37 °C with agitation at 225 rpm in an orbital shaker. Cells were harvested via centrifugation at 4 °C and 4,000g for 20 min and the cell pellet was resuspended in lysis buffer (150 mL; 100 mM NaCl, 50 mM Tris-HCl, pH 8.0) and stored at −80°C. The frozen cells were lysed by three freeze/thaw cycles. Lysozyme (1 mg/mL), protease inhibitor cocktail (5 mL), benzonase (25 units/mL), and MgCl2 (1 mM) were added to the lysate and the mixture was incubated at 25 °C for 30 min. The cell lysate was incubated for 12 h at 4°C and was centrifuged at 18,000g for 30 min at 4°C to pellet the cell debris. The target protein was purified from the clarified cell lysate by three to five cycles of temperature-induced precipitation (4°C/37°C) from 5 M NaCl solution. Dialysis and lyophilization afforded protein B10 as a fibrous solid in isolated yields of 250 mg/L of culture. MALDI-TOF mass spectrometry, Obs. (Calc.): B10, 177,608 (176,924.3). Amino acid compositional analysis. B10; Calc. (mol.-%): Ala, 19; Glx, 1; Gly, 25.1; Ile, 14.9; Pro, 20; Val, 20. Obs. (mol.-%): Ala, 18.7; Glx, 1.6; Gly, 25.6; Ile, 12.8; Pro, 19.9; Val, 19.3. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis revealed a single protein band for B10 that migrated higher than its predicted molecular weight (Fig. 1).
Table 2.
Amino acid sequence of B10 and related nucleic acid coding sequence
| Val | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro |
| GTT | CCT | GCT | GTT | GGT | ATT | CCG | GCT | GTT | GGT | ATC | CCA |
| Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala | |
| GCT | GTT | GGT | ATC | CCA | GCT | GTT | GGC | ATT | CCG | GCT | |
| Val | Gly | [Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | |
| GTA | GGT | ATT | CCT | GCT | GTT | GGT | ATT | CCG | GCT | GTT | |
| Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | |
| GGT | ATC | CCA | GCT | GTT | GGT | ATC | CCA | GCT | GTT | GGC | |
| Ile | Pro | Ala | Val | Gly]33 | Ile | Pro | Gly | Ala | Gly | Val | |
| ATT | CCG | GCT | GTA | GGT]33 | ATT | CCA | GGT | GCA | GGC | GTA | |
| Pro | Gly | Ala | Gly | Val | Pro | Gly | Glu | Gly | Val | Pro | |
| CCG | GGT | GCT | GGC | GTT | CCG | GGT | GAA | GGT | GTT | CCA | |
| Gly | Ala | Gly | Val | Pro | Gly | Ala | Gly | [Val | |||
| GGC | GCA | GGT | GTA | CCG | GGT | GCG | GGT | [GGT | |||
| Pro | Gly | Ala | Gly | Val | Pro | Gly | Ala | Gly | Val | Pro | |
| CCA | GGT | GCA | GGC | GTA | CCG | GGT | GCT | GGC | GTT | CCG | |
| Gly | Glu | Gly | Val | Pro | Gly | Ala | Gly | Val | Pro | Gly | |
| GGT | GAA | GGT | GTT | CCA | GGC | GCA | GGT | GTA | CCG | GGT | |
| Ala | Gly]20 | Val | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | |
| GCG | GGT]20 | GTT | CCT | GCT | GTT | GGT | ATT | CCG | GCT | GTT | |
| Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | |
| GGT | ATC | CCA | GCT | GTT | GGT | ATC | CCA | GCT | GTT | GGC | |
| Ile | Pro | Ala | Val | Gly | [Ile | Pro | Ala | Val | Gly | Ile | |
| ATT | CCG | GCT | GTA | GGT | [ATT | CCT | GCT | GTT | GGT | ATT | |
| Pro | Ala | Val | Gly | Ile | Pro | Ala | Val | Gly | Ile | Pro | |
| CCG | GCT | GTT | GGT | ATC | CCA | GCT | GTT | GGT | ATC | CCA | |
| Ala | Val | Gly | Ile | Pro | Ala | Val | Gly]33 | ||||
| GCT | GTT | GGC | ATT | CCG | GCT | GTA | GGT]33 | ||||
| Val | Pro | Gly | Val | Gly | Stop | Stop | |||||
| GTA | CCA | GGT | GTA | GGC | TAA | TAA |
Figure 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of B10 copolymer. B10 was run on a 5% SDS-PAGE and stained with Coomassie G250 (BioRad). Molecular weight markers were Precision Plus Protein Kaleidoscope (BioRad).
Synthesis of triblock protein polymer B9
A recombinant protein that contains flanking hydrophobic endblocks of sequence VPAVG[(IPAVG)4(VPAVG)]16IPAVG separated by a central hydrophilic block VPGVG[(VPGVG)2(VPGEG)(VPGVG)2]48VPGVG was expressed from E. coli and purified, as detailed elsewhere.7 Amino acid compositional analysis. B9; Calc. (mol.-%): Ala, 8.1; Glx, 2.4; Gly, 31.9; Ile, 6.4; Pro, 20.0; Val, 31.2. Obs. (mol.-%): Ala, 10.8; Glx, 2.0; Gly, 28.3; Ile, 7.0; Pro, 22.8; Val, 28.2. MALDI-TOF mass spectrometry, Obs. (Calc.): B9, 165,356 (165,564).
Differential Scanning Microcalorimetry (Micro-DSC)
Differential scanning microcalorimetry was recorded on a Setaram Micro DSC III calorimeter (Setaram Inc., France) at a scan rate of 1 °C/min from 4 to 70 °C. Lyophilized proteins were dissolved at a concentration of 1 mg/mL in distilled, deionized water. MicroDSC data were analyzed using SETSOFT 200 software (Setaram Inc, France).
Rheological analysis of concentrated protein polymer solutions
Rheological data were acquired on an Advanced Rheological Expansion System III rheometer (ARES III, TA instrument, NJ) in parallel plate geometry with a plate diameter of 25 mm. The testing protocol for rheological analysis has been detailed elsewhere.8 In brief, 100 mg/mL protein solutions were prepared by adding distilled, deionized water to lyophilized protein at 4 °C, shaking the solution for 48 h, and then allowing the solution to equilibrate for 72 h. The gap between parallel plates was adjusted between 0.2 − 0.35 mm and dynamic mechanical experiments were performed in shear deformation mode. An initial strain amplitude (γ) sweep was performed at 4°C and 37°C at different frequencies to determine the linear viscoelastic range for the protein polymer.
The gelation temperature was determined by heating samples from 4°C to 40°C at a rate of 1°C per minute. Following temperature equilibration at 37°C, viscoelastic properties were examined by a strain sweep at a fixed frequency of 1Hz and a frequency sweep at fixed strain amplitude of 2%. Experiments were repeated on 5 to 6 samples and representative data presented.
Mechanical analysis of hydrated protein polymer films
For mechanical property analysis, films were cast from protein solutions. In brief, lyophilized proteins were dissolved at a concentration of 100 mg/mL either in 2, 2, 2-trifluoroethanol (TFE) at 23 °C or in water at 4 °C. The protein solution was then poured into Teflon casting molds and solvent evaporation performed either at 23 °C or at 4 °C. Test samples were referred to as TFE-23, water-23, or water-4, indicating the casting solvent and evaporation temperature used for film formation. After complete solvent evaporation, films were hydrated in phosphate buffered saline (PBS) at 37 °C, which contained NaN3 at 0.2 mg/mL to prevent biological contamination. Samples were cut into a dumbbell shape using a stainless steel die with gauge dimensions of 13 mm × 4.75 mm. Hydrated film thickness, as measured by optical microscopy, was typically 0.1 mm for TFE-23 and water-5 films and 0.5 mm for water-23 films.
Mechanical characterization of protein films was performed using a dynamic mechanical thermal analyzer DMTA V (Rheometric Scientific Inc., Newcastle, DE) with a 15 N load cell in the inverted orientation, so that samples could be immersed in a jacketed beaker filled with PBS at 37 °C. The maximum travel distance of the drive shaft of DMTA was 23 mm, which limited maximum strain to 70% of engineering strain. Samples were evaluated by several mechanical test protocols including: (i) Uniaxial tension. Loading and unloading was controlled by displacement at a fixed rate of 5 mm/min. Five to six samples were monotonically stretched to 65% of maximum strain for uniaxial stress-strain analysis. (ii) Creep and stress relaxation. Six to ten samples were prepared for creep analysis. Constant engineering stresses were applied for time periods of up to 30 hours. Four to six samples were prepared for stress-relaxation analysis. Each sample was stretched at 5 mm/min to a fixed strain and the evolution of stress over time was examined. Measurement of stress-relaxation was limited to 30 minutes. (iii) Preconditioning protocols. Five to six samples cast under different conditions were cyclically stretched to 30% strain for 20 cycles with an off-loading period of 5 minutes between cycles. Water-23 films were also stretched to 30% strain for one cycle and then cyclically stretched to 10% strain for 20 cycles; or stretched to 50% strain for one cycle and then cyclically stretched to 30% strain for 20 cycles with an off-loading period of 5 minutes. Resilience was calculated from loading and under the loading curves. (iv) Cyclic loading with increasing stress magnitude. Water-4 samples preconditioned at 30% strain for 20 cycles with an off-loading period of 5 minutes were subjected to cyclic stress of increasing magnitude. Stress was applied for one hour and then removed for one hour followed by reimposition of the load at a higher stress. Reproducibility was examined in three replicate samples. (v) Step loading. Water-4 samples preconditioned at 30% strain for 20 cycles with an off-loading period of 5 minutes were subjected to step loading, in which stress was increased by 50 kPa every two hours. Reproducibility was examined in three replicate samples.
RESULTS AND DISCUSSION
The inverse transition temperature is consistent with protein block structure
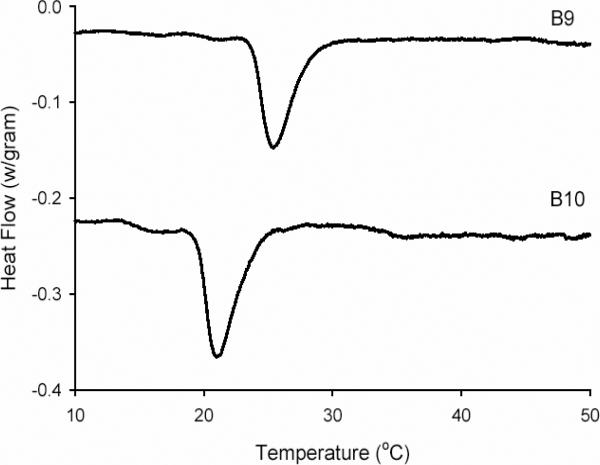
Differential scanning microcalorimetry of dilute aqueous solutions of B10 (1 mg/mL) confirmed the presence of a single endothermic transition at 21°C consistent with coacervation of the hydrophobic endblocks (Fig. 2). The inverse transition temperature of B10 was 4 °C lower than that observed for B9 due to an increase in the size and hydrophobicity of the endblocks. Specifically, the B10 endblocks were nearly twice as large as those of the B9 triblock copolymer and contained a larger mole fraction of isoleucine (20 vs. 16 mol%) in the first position of the pentapeptide repeat sequence IPAVG. Both factors may contribute to a reduction in the transition temperature. Reversibility of the phase transition was confirmed upon repeating microcalorimetry measurements after a 12 h equilibration at 4 °C (data not shown).
Figure 2.
Differential scanning microcalorimetry of B9 and B10. Signals are shifted along Y-axis for clarity.
Rheological analysis confirms formation of a protein gel
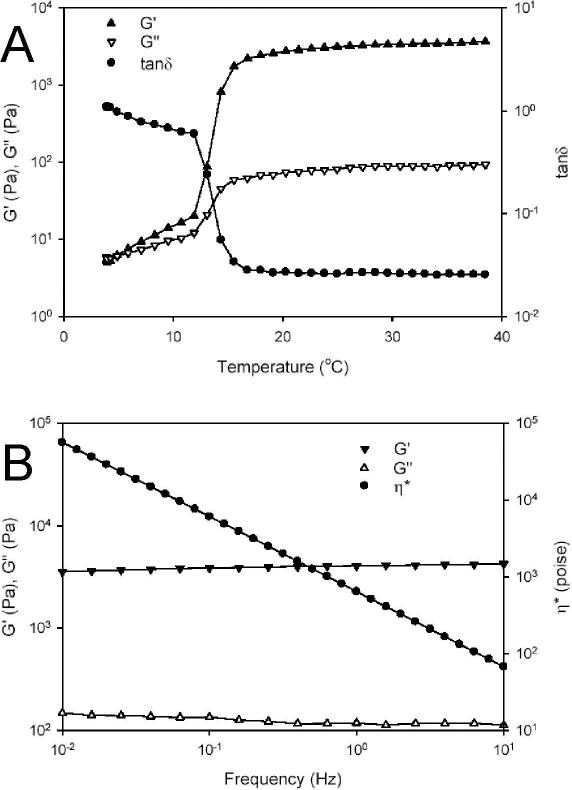
Above 18°C, the shear storage (G’) and loss (G”) modulus of concentrated solutions of B10 increased by a factor of approximately 103 and 10, respectively, while tan δ (G’/G”) decreased, consistent with the formation of a viscoelastic gel (Fig. 3A). Observation of a lower transition temperature for protein gelation than that noted by microcalorimetry studies of dilute protein solutions was likely due to the effect of extensive intermolecular interactions present in the concentrated protein solution used for rheological studies. At 37°C, G’ and G” were independent of frequency between 0.01 to 10 rad/s at a fixed strain amplitude of 2% (Fig. 3B). In addition, the logarithm of complex viscosity (η*) was a linear function of the logarithm of frequency with a slope of −1. All of this suggests that within this frequency range the mechanical response of the protein hydrogel is consistent with a rubbery solid.
Figure 3.
Rheological behavior of B10 in water. (A) Dynamic shear storage (G′), loss modulus (G″), and tanδ are plotted as a function of temperature (γ 2%, ω 1Hz). (B) Dynamic shear storage (G′), loss modulus (G″), and complex viscosity (η*) are plotted as a function of frequency (γ 2%, 37 °C).
Block structure alters the Young's modulus of elastin-mimetic triblock protein polymers
Load-extension curves at 37°C of hydrated B10 films cast either from TFE at 23°C or water at 4°C revealed plastic-like deformation behavior, such that, stress increases linearly with increasing strain until a yield point is reached between 2 − 2.5 MPa, after which elongation occurs with the imposition of a relatively low increment in load. In contrast, hydrated B10 films produced from an aqueous protein solution cast at 23°C displayed rubber-like behavior with homogeneous deformation occurring in response to low stress levels. Corresponding values of Young's modulus were 87 MPa, 60 MPa, and 0.71 MPa for hydrated TFE-23, water-4, and water-23 B10 films, respectively. Of note, these values are two- to 60-fold greater than the Young's modulus measured for B9 films processed under identical casting conditions (Fig. 4, Table 3).
Figure 4.
Uniaxial stress-strain analysis. The Young's modulus was 87 ± 9MPa for TFE-23 and 60 ± 8MPa for water-4 measured from the first linear range, and was 0.71 ± 0.12MPa for water-23 film measured from the first 10% of deformation.
Table 3.
Comparison of Young's Modulus of B9 and B10 Films
| Cast in TFE at 23°C | Cast in water 4°C | Cast in water 23°C | |
|---|---|---|---|
| B10 (MPa) | 87 ± 9 | 60 ± 8 | 0.71 ± 0.12 |
| B9 (MPa) | 35 ± 3 | 1.3 ± 0.3 | 0.01 ∼ 0.03 |
Prior studies of B9 triblock copolymers have demonstrated that solvent type and casting temperature profoundly influences microphase protein block mixing with a commensurate effect on mechanical responses. Specifically, films cast from TFE, which solvates both mid and endblock sequences, promotes significant interphase mixing in cast films. As a result, the hydrophobic, semi-rigid endblocks are organized as a dispersed microphase and thereby contribute to the mechanical response of the material as load bearing elements leading to plastic-like deformation behavior.7 In contrast, water preferentially solvates the hydrophilic midblock. Thus, films cast from water at 23°C display a microphase separated structure with well segregated endblocks that act as relatively discrete virtual crosslinks within an elastomeric protein matrix. Moreover, in casting the aqueous protein solution above the inverse transition temperature of the protein polymer (23°C > 18°C), microphase separation of the endblocks is further promoted due to a coacervation effect. Given the greater degree of microphase separation, the contribution of the elastomeric midblock to the mechanical response of the material is enhanced with a corresponding rubber-like stress-strain profile. The influence of casting temperature is demonstrated by the behavior of films cast from water at 4 °C. In the absence of the coacervation effect present below 18°C, we speculate that films were produced with a lower degree of microphase separated structure and, therefore, displayed a higher Young's modulus. As compared to B9, the presence of substantially larger endblocks and a relatively smaller midblock accentuated the proportion of plastic-like domains in B10 films and, as a consequence, the generation of materials with a higher elastic modulus under all film forming conditions.
Creep responses are modulated by protein block structure
Prior studies characterized creep responses of B9 films cast from water at 4 °C or TFE at 23 °C and revealed substantial deformation responses above 0.2 MPa. As a virtually crosslinked protein network, it is presumed that time-dependent changes in strain in response to stress will be influenced by the density, size, and chemical nature of the physical crosslinks. Thus, we postulated that this could be achieved by increasing both the hydrophobicity of the endblock, as well as the relative size of the endblock by altering the weight ratio of endblock to midblock segments.
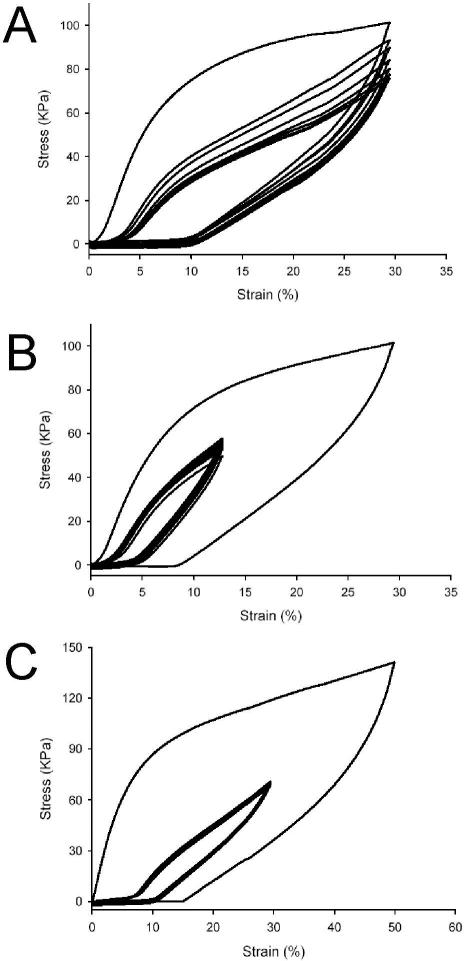
Creep analysis was performed on hydrated B10 films at 37°C that were initially produced under a variety of film casting conditions (Fig. 5). Water cast films produced at 4°C demonstrated limited creep (<10%) over a 20 h observation period at stress levels at or below 0.4 MPa, nearly double the load for B9 films produced under comparable conditions. Films cast from an aqueous solution of B10 at 23°C demonstrated comparable levels of creep, but at stress levels that were one order of magnitude lower. B10 films cast from TFE demonstrated the lowest level of creep with an observed strain of less than 10% when subjected to a stress of 0.8 MPa; an approximately four-fold greater load than that sustained by similarly fabricated B9 films. To our knowledge, creep responses have not been reported for other elastin-like protein polymers in either non-crosslinked or chemically crosslinked forms. Given that the magnitude of the observed deformation response was not directly proportional to the applied stress, these materials behaved as non-linear viscoelastic solids. In summary, these data emphasize that time-dependent mechanical properties of protein-based materials containing large, chemically distinct blocks can be modulated by controlling the tendency for block segregation either by selection of processing conditions or by molecular design.
Figure 5.
Creep analysis of B10 films. (A) Creep of TFE-23 film. From top to bottom, creep was examined as tensile stress was maintained at 1.0MPa, 0.8MPa and 0.6MPa, respectively. (B) Creep of water-4 film. From top to bottom, creep was examined as tensile stress was maintained at 0.8MPa, 0.6MPa and 0.4MPa, respectively. (C) Creep of water-23 films. From top to bottom, creep was examined as tensile stress was maintained at 60KPa, 40KPa and 30KPa, respectively. Under 60KPa stress, creep reached the maximum strain that was allowed on the current testing facility within 12 hours. (D) Comparison of the creep behaviors of water-4 films derived from B10 and B9. The short-term creep behaviors demonstrated that films derived from B10 are more stable under mechanical loading.
Preconditioning by an imposed cyclic stress enhances the resilience of protein polymer films
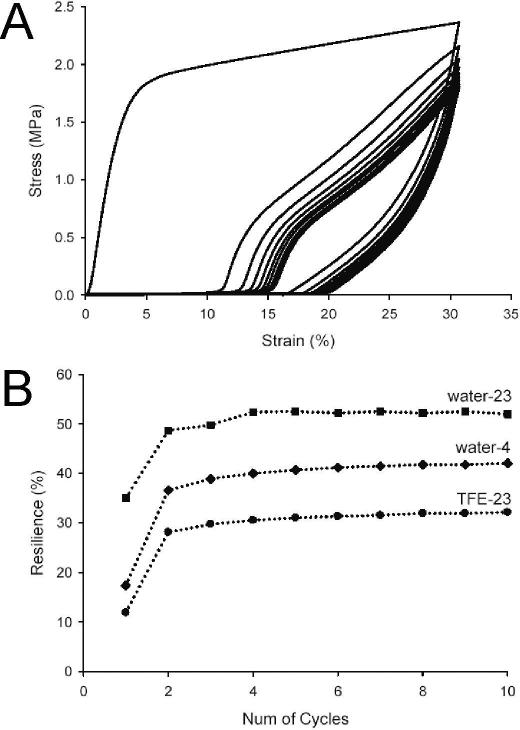
Upon subjecting B10 films to periods of repetitive cyclic deformation to 30% strain, we observed the accumulation of residual deformation and a decline in peak stress that stabilized after several cycles (Fig. 6). In the process, resilience was significantly enhanced over 10 loading cycles with an increase from 11±2% to 30±2% for TFE-23 films, from 18±2% to 39±2% for water-4 samples, and from 35±2% to 51±2% for water-23 films. The greatest increase in resilience largely occurred after the first loading cycle, presumably due to stabilization of load induced changes in microstructure.
Figure 6.
(A) The influence of preconditioning on resilience of water-4 film. A water-4 sample was cyclically stretched to 30% strain, with an off-loading period of 5 minutes between cycles. Plotted are the stress-strain curves from the first ten cycles of stretches, because stress-strain responses were stabilized after the eight cycles of stretch. Similar responses were also observed for TFE-23 and water-23 samples. (B) The dependence of resilience on the number of preconditioning cycles. Samples cast in different conditions are cyclically stretched to 30% strain, with an off-loading period of 5 minutes between cycles. Plotted is resilience after each cycle against the number of the preconditioning cycles.
The effects of varying mechanical preconditioning protocols on resilience were further examined using films initially cast from water at 23 °C (Fig. 7). In all protocols, stabilization of mechanical behavior was largely observed after the initial loading cycle with accumulation of residual strain of 5−10% and a decline in peak stress. As previously stated, the resilience of water-23 films subjected to a repetitive cyclic strain of 30% was 51±2%. When films were subjected to an initial elongation of 30% followed by cyclic stretch at 12% strain, the resilience increased to 58±2%, which was attributed, at least in part, to a reduction in energy dissipation at reduced strain. Nonetheless, the influence of initial deformation history on resilience was evident when films were subjected to an initial strain of 50% and subsequently exposed to 30% cyclic stretch. Although permanent strain was unaffected, as compared to films subjected to 30% cyclic stretch alone, resilience increased to 67±1%. Thus, a significant degree of change in protein microstructure can be induced not only by the conditions of film casting, but also through the effects of mechanical deformation or annealing protocols. As mechanical preconditioning stabilized the microstructures and mechanical properties of protein polymer films, the deformation plasticity tends to decrease. For instance, the yielding behaviors of TFE-23 films diminished and nearly linear behaviors were observed in water-23 films over 10 loading cycles.
Figure 7.
The influence of preconditioning on the resilience of water-23 films. (A) A water-23 sample was cyclically stretched to 30% strain for 21 cycles, with an off-loading period of 5 minutes between cycles. Plotted are the stress-strain curves from the first 10 cycles, because the material response to the external loading is stabilized after 8 cycles of stretch. (B) A water-23 sample was cyclically stretched to 30% strain and then to 12% strain for 20 cycles, with an off-loading period of 5 minutes between cycles. (C) A water-23 sample was cyclically stretched to 50% strain and then to 30% strain for 20 cycles, with an off-loading period of 5 minutes between cycles.
Under what circumstances will physical crosslinks be broken?
Two new tests were designed to examined the capacity of loading conditions to alter mechanical response. Three phases of deformation behavior were observed when preconditioned B10 water-4 films were exposed to cyclic loads of increasing magnitude (Fig. 8). The first phase, which extended up to an imposed load of 1.2 MPa over a 30 h period, was characterized by small elastic deformation responses, as both the total and residual deformations were small. In the second phase, over a load range between 1.2 and 2.7 MPa, both the total and residual deformation increase linearly with increasing magnitude of cyclic stress and appreciable residual deformation was observed. A more rapid increase in the total and residual deformation occurred in the third phase consistent with disruption of physical crosslinks. Films examined under step loading also displayed three similar phases of deformation behavior (Fig. 9). Remarkably, strain levels at each transition point were similar for both protocols, although stress levels were significantly different. Three phases of deformation behavior were also observed for preconditioned films cast from TFE at 23 °C.
Figure 8.
Deformation behaviors of preconditioned water-4 films under cyclic stress of increasing magnitude. (A) A water-4 sample was subjected to cyclic stress of increasing magnitudes (shown in inset), and the deformation history was recorded. Reproducibility was examined on three replicate samples, which were preconditioned at 30% strain for 20 cycles with an off-loading period of 5 minutes between cycles and a two hour recovery time. (B) Deformation at the end of each loading (filled circles) and off-loading (open circles) period were plotted against the magnitude of cyclic stress.
Figure 9.
Deformation behavior of preconditioned water-4 films subjected to a step loading protocol. A water-4 sample was subjected to step stress (shown in inset), and strains at the end of each loading step represented by open circles in water-4 films and by crosses in TFE-23 films were plotted against the magnitude of stress. Reproducibility was examined on three replicate samples, which were preconditioned at 30% strain for 20 cycles with an off-loading period of 5 minutes between cycles.
It bears comment that during off-loading periods water-4 films demonstrated “recovery” of deformation after imposed cyclic loading and, therefore, were able to sustain larger subsequent stresses, when compared to deformation induced by direct step loading. For example, when accumulated strain reached the onset of the second deformation phase, the exerted stress was approximately 1200 kPa and 200 kPa under cyclic and step loading conditions, respectively. The observed recovery effect is likely a consequence of limited polypeptide chain rearrangement, in which the capacity of the material to carry loads was partially recovered and is clearly dependent on the characteristics of both loading and off-loading conditions.
We speculate that in a manner akin to synthetic polymers17, the deformation behavior in the first phase may be attributable to an initial stretching of polypeptide bonds. Inevitably, bond stretch is limited and further deformation must arise from conformational changes in the polymer chain, which likely occurs in the second phase of deformation. Differences in the stress required to induce conformational changes of protein polymer within films processed under different casting conditions are likely related to differences in the mixing of semi-rigid endblocks and flexible midblocks that create energy or stereoelectronic barriers. Under both loading protocols, substantial film deformation was observed after an initial 22−25% strain, which appears to designate the stress level associated with disruption or damage to physical crosslinks. Given that samples were preconditioned at 30% strain for 20 cycles, these data suggest that “new” disruption or damage may occur when deformation approaches or exceeds preconditioning strains. Nonetheless, further experimental analysis will be necessary to identify the dependence of preconditioning state on the onset of this transition, as well as on other time-dependent mechanical responses of elastin-mimetic protein polymers.
CONCLUSIONS
Micro-DSC and rheology studies confirm the presence of an inverse-temperature transition for the elastin-mimetic protein polymer B10 in aqueous solutions with gelation of concentrated solutions at ambient temperatures. Mechanical analysis, particularly studies of creep behavior, demonstrate that physically crosslinked protein networks derived from B10 could withstand significantly greater loads than compared to a triblock copolymer designed with a lower relative content of hydrophobic, plastic-like endblocks. Moreover, we observed that resilience is significantly enhanced by mechanical preconditioning. Newly designed tests consisting of cyclic loading of increasing magnitude and step loading further revealed the presence of three unique phases of deformation behavior, which we speculate correspond to peptide bond stretching, conformational changes of polypeptide chains, and disruption of physical crosslinks. Elastin-mimetic proteins may provide an important vehicle for the design of a variety of medically implanted devices. Thus, it bears emphasis that the retention of material integrity depends upon both preconditioning protocols, as well as the imposed patterns of load.
Supplementary Material
Scheme 1.
Amino Acid Sequence of Protein-Based Block Copolymer B10.
ACKNOWLEDGEMENTS
This work is supported by grants from the NIH and NSF.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figures illustrating stress relaxation of B10 films cast under different conditions can be found in Supporting Information.
REFERENCES
- 1.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389–92. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Cabellio JC, Reguera J, Girotti A, Arias FJ, Alonso M. Adv Polym Sci. 2006;200:119–167. [Google Scholar]
- 3.Haider M, Megeed Z, Ghandehari H. J Control Release. 2004;95:1–26. doi: 10.1016/j.jconrel.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Cappello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, Ferrari F. Biotechnol Prog. 1990;6:198–202. doi: 10.1021/bp00003a006. [DOI] [PubMed] [Google Scholar]
- 5.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. J Biomed Mater Res. 2002;62:195–203. doi: 10.1002/jbm.10272. [DOI] [PubMed] [Google Scholar]
- 6.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Biomacromolecules. 2003;4:602–7. doi: 10.1021/bm0201082. [DOI] [PubMed] [Google Scholar]
- 7.Nagapudi K, Brinkman WT, Leisen J, Thomas BS, Wright ER, Haller C, Wu XY, Apkarian RP, Conticello VP, Chaikof EL. Macromolecules. 2005;38:345–354. [Google Scholar]
- 8.Nagapudi K, Brinkman WT, Thomas BS, Park JO, Srinivasarao M, Wright E, Conticello VP, Chaikof EL. Biomaterials. 2005;26:4695–706. doi: 10.1016/j.biomaterials.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Sallach RE, Wei M, Biswas N, Conticello VP, Lecommandoux S, Dluhy RA, Chaikof EL. J Am Chem Soc. 2006;128:12014–9. doi: 10.1021/ja0638509. [DOI] [PubMed] [Google Scholar]
- 10.Wright ER, Conticello VP. Adv Drug Deliv Rev. 2002;54:1057–73. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Sallach R, Haller CA, Caves JA, Nagapudi K, Conticello VP, Levenston ME, Chaikof EL. Biomacromolecules. 2005;6:3037–44. doi: 10.1021/bm0503468. [DOI] [PubMed] [Google Scholar]
- 12.Daamen WF, Veerkamp JH, van Hest JC, van Kuppevelt TH. Biomaterials. 2007;28:4378–98. doi: 10.1016/j.biomaterials.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Cabellio JC, Reguera J, Girotti A, Arias FJ, Alonso M. Adv Polym Sci. 2006;200:119–167. [Google Scholar]
- 14.Rodriguez-Cabello JC, Rodriguez-Cabello JC, Prieto S, Reguera J, Arias FJ, Ribeiro A. J Biomater Sci Polym Ed. 2007;18:269–86. doi: 10.1163/156856207779996904. [DOI] [PubMed] [Google Scholar]
- 15.Wagenseil JE, Wakatsuki T, Okamoto RJ, Zahalak GI, Elson EL. J Biomech Eng. 2003;125:719–25. doi: 10.1115/1.1614818. [DOI] [PubMed] [Google Scholar]
- 16.Wright ER, McMillan RA, Cooper A, Apkarian RP, Conticello VP. Adv Funct Mater. 2002;12:149–154. [Google Scholar]
- 17.Treloar LRG. The Physics of Rubber Elasticity. 3rd ed. Clarendon Press; Oxford: 1975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.