Abstract
Catalysis by the zinc metalloenzyme human carbonic anhydrase II (HCA II) is limited in maximal velocity by proton transfer between His64 and the zinc-bound solvent molecule. Asn62 extends into the active site cavity of HCA II adjacent to His64 and has been shown to be one of several hydrophilic residues participating in a hydrogen-bonded solvent network within the active site. We compared several site-specific mutants of HCA II with replacements at position 62 (Ala, Val, Leu, Thr, and Asp). The efficiency of catalysis in the hydration of CO2 for the resulting mutants has been characterized by 18O exchange, and the structures of the mutants determined by X-ray crystallography to 1.5 – 1.7 Å resolution. Each of these mutants maintained the ordered water structure observed by x-ray crystallography in the active-site cavity of wild-type HCA II; hence, this water structure was not a variable in comparing with wild type the activities of mutants at residue 62. Crystal structures of wild-type and N62T HCA II showed both an inward and outward orientation of the side chain of His64; however, other mutants in this study showed predominantly inward (N62A, V, L) or predominantly outward (N62D) orientations of His64. A significant role of Asn62 in HCA II is to permit two conformations of the side chain of His64, the inward and outward, that contributes to maximal efficiency of proton transfer between the active site and solution. The site-specific mutant N62D had a mainly outward orientation of His64, yet the difference in pKa between the proton donor His64 and zinc-bound hydroxide was near zero, as in wild-type HCA II. The rate of proton transfer in catalysis by N62D HCA II was 5% that of wild type showing that His64 mainly in the outward orientation is associated with inefficient proton transfer compared with His64 in wild type which shows both inward and outward orientations. These results emphasize the roles of the residues of the hydrophilic side of the active-site cavity in maintaining efficient catalysis by carbonic anhydrase.
The carbonic anhydrases comprise several well studied classes among which is the α class including the human and animal enzymes (1, 2). The zinc metalloenzyme human carbonic anhydrase II (HCA II) is the most extensively studied of these enzymes and catalyzes the hydration of CO2 by a two stage mechanism (3-5). The first is the reaction of CO2 with zinc-bound hydroxide to form and dissociate bicarbonate (eq 1). The second is the regeneration of the zinc-bound hydroxide by proton transfer to solution (eq 2).
| (1) |
| (2) |
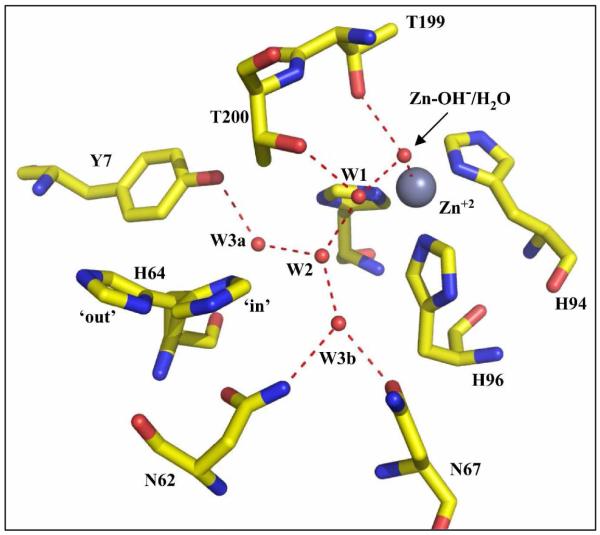
Here B is an exogenous proton acceptor and His64 is the internal proton shuttle residue (6). The rate limiting step in maximal velocity of this catalysis is the transfer of a proton between His64 and the zinc-bound solvent (6, 7). His64 is located on the rim of the active-site cavity with its side chain extending into the cavity (Figure 1)(5, 8, 9). In crystal structures two orientations are observed for this side chain (9, 10), one is an inward conformation with the side chain oriented towards the zinc, and a second is an outward conformation with His64 oriented towards the mouth of the active site cavity and external solution.
Figure 1.
The active site of HCA II from the data of Fisher et al. (38). The side chain of His64 is shown in both the inward and outward conformations. The red spheres represent oxygen including the oxygens of ordered water molecules numbered W1, W2, W3a, and W3b. Dashed red lines indicate presumed hydrogen bonds. This figure was generated and rendered with PyMOL (www.pymol.org).
The Nδ1 of the imidazole side chain of His64 in HCA II in the inward conformation is too far from the zinc (about 7.5 Å; (10, 11)) for direct proton transfer, and solvent hydrogen isotope effects are consistent with proton transfer through intervening hydrogen-bonded solvent bridges (12). Crystal structures of the isozymes of CA in the α class exhibit networks of ordered, apparently hydrogen-bonded water molecules between His64 and the zinc-bound water in the active site cavity (Figure 1). In the Grotthuss mechanism, high proton mobility is achieved by proton shuttling along hydrogen bonds without requiring much motion from their oxygen atoms. However, the apparently hydrogen bonded water structure shows at best weak hydrogen bonding between the Nδ1 atom of the side chain of His64 (inward orientation) and the oxygen atom of the nearest water molecule W2, a distance which is 3.2  (Figure 1) (11). The proton migration in CA is of great current interest as a model for proton transfer in more complex systems, and is also under study in several labs using computational methods (13-17). These continuing studies of CA are significant in understanding the role of water in facilitating proton transfers in proteins and in catalysis.
(Figure 1) (11). The proton migration in CA is of great current interest as a model for proton transfer in more complex systems, and is also under study in several labs using computational methods (13-17). These continuing studies of CA are significant in understanding the role of water in facilitating proton transfers in proteins and in catalysis.
Recent attention has focused on the significance of the ordered solvent observed in the crystal structures of CA. Several residues have been shown to stabilize this ordered solvent structure, among them Tyr7, Asn62, and Asn67 (Figure 1)(18, 19). Fisher et al. (18) replaced these hydrophilic residues with hydrophobic residues (Y7F, N62L, and N67L) and observed several changes in structure and catalysis. These mutations had different effects on the orientation of the side chain of His64 (Y7F and N62L predominantly inward and N67L predominantly outward), altered the pKa of His64, and affected the ordered water structure. The observed rate constants for proton transfer between His64 and the zinc-bound hydroxide were changed for these mutants, but there was little effect on the rate constant for conversion of CO2 into bicarbonate (18). Among these the mutant N62L was interesting since the X-ray crystal structure showed His64 in the inward orientation and the ordered water structure intact as in the wild type (18).
The side chain of Asn62 in HCA II is extended into the active-site cavity about 9 Å from the zinc and as close at 3.3 Å from the side chain of His64 (Figure 1)(8-10). This residue appears conserved in many species of CA II, for example in mouse and chicken as well as in many other isozymes of carbonic anhydrase (20). Computations of conformational states of active-site residues in HCA II point out steric interactions between the side chains of His64 and Asn62 that contribute to the orientation of His64 (17, 19). In previous experimental studies, the pH profile of catalysis by N62L HCA II appeared irregular and difficult to interpret (18). However, additional amino acid substitutions at position 62 in this study have provided a more straightforward interpretation. The replacement of Asn62 with other amino acids was shown to cause changes in the orientation of the proton shuttle His64 and the pKa of its imidazole side chain, but had little or no effect on the structure of ordered solvent in the active-site cavity. The results point out the capacity of His64 to participate in proton transfer more efficiently in the inward than in the outward conformations, and the role of residue 62 in fine tuning the values of pKa of His64 and the zinc-bound solvent molecule.
MATERIALS AND METHODS
Expression and Purification of HCA II mutants
The mutants N62D, N62A, N62V and N62T of HCA II were made by site-directed mutagenesis using an expression vector containing wild type HCA II coding region (6). The point mutations were made using the QuikChange II Kit (Stratagene, LaJolla) for site-directed mutagenesis. The single mutants were confirmed by DNA sequencing over the entire coding region of HCA II. Expression of each mutant was done by transforming mutated vectors into Escherichia coli BL21(DE3)pLysS cells, which do not express any indigenous CA under the following conditions. The transformed cells were expressed at 37 °C in LB medium containing 100 μg/ml ampicillin. HCA II production was induced by the addition of isopropyl thiogalactoside to a final concentration of 1mM when the bacterial culture reached an OD600 of 0.6. The cells were harvested 4 hrs after induction. The cell pellets were lysed and HCA II was purified through affinity chromatography using p-(aminomethyl)benzenesulfonamide coupled to agarose beads (21). Electrophoresis on a 10% polyacrylamide gel stained with Coomassie Blue was used to confirm the purity of enzyme samples, which were found to be greater than 96% pure. HCA II and the mutants studied here bound sulfonamides tightly; therefore, enzyme concentrations were determined by titration of active sites with ethoxzolamide while measuring the catalyzed depletion of 18O from CO2 and analyzing data with the Henderson approach (22).
Crystallography
Crystals of the HCAII single-site mutants of N62A, N62V, N62T and N62D were obtained using the hanging-drop vapor diffusion method (23). The crystallization drops were prepared by mixing 5 μL of protein (10mg/mL) in 50 mM Tris-HCl (pH 7.0) with 5 μL of the precipitant solution 50 mM Tris-HCl (pH 9.0) and 1.3 M sodium citrate at 20 °C against 1 ml of the precipitant solution. The pH of the crystallization solutions was about 7.9. Useful crystals were observed 4 days after the crystallization setup. Previous studies had shown the binding of sulfate at the zinc in N62L HCA II (18); hence sulfate was not used as a precipitant in crystallizations reported here. The X-ray diffraction datasets for all of the mutant HCAII crystals were obtained at room temperature, using an R-AXIS IV++ image plate system with Osmic mirrors and a Rigaku RU-H3R Cu rotating anode operating at 50 kV and 100 mA. The detector-crystal distance was set to 80 mm. Each dataset was collected at room temperature with the crystals mounted in quartz capillaries. The oscillation steps were 1° with a 7 min exposure per image. X-ray data processing was performed using DENZO and scaled and reduced with SCALEPACK (24). All manual model building was performed using Coot (25) and refinement was carried out with the crystallography and nuclear magnetic resonance system (CNS) suite of programs, version 1.1 (26).
The wild-type HCA II crystal space group [Protein Data Bank (PDB) accession code 2CBA (11)] was isomorphous with all of the datasets collected, and was used to phase the datasets. To avoid phase bias of the model, the zinc ion, mutated residues, and water molecules were removed. After one cycle of rigid-body refinement, annealing by heating to 3000 K with gradual cooling, geometry-restrained position refinement, and temperature-factor refinement, 2Fo - Fc and Fo - Fc Fourier electron-density maps were generated. These electron-density maps clearly showed the position of the zinc and mutated residues, which were subsequently built into their respective models. After several cycles of refinement, solvent molecules were incorporated into the models using the automatic waterpicking program in CNS until no more water molecules were found at a 2.0σ level. Refinement of the models continued until convergence of Rcryst and Rfree was reached (see Table 1).
Table 1.
Crystal structure data and refinement statistics of four variants of HCA II.
| N62A | N62V | N62T | N62D | |
|---|---|---|---|---|
| unit cell dimensions, a, b, c (Å), β ( ° ) |
42.8 41.7 72.9 104.6 |
42.8 41.7 72.9 104.6 |
42.7 41.6 72.8 104.6 |
42.7 41.7 72.8 104.5 |
| resolution (Å) | 50 – 1.7 (1.76-1.7) |
50 - 1.5 (1.55-1.5) |
50 - 1.6 (1.66-1.6) |
50 – 1.6 (1.66-1.6) |
| No. of unique reflections |
25972(2500)a | 37008(3472) | 31194(2971) | 31546(2992) |
| completeness (%) | 93.8(91.5) | 92.2(87.0) | 94.3(90.6) | 95.2(91.1) |
| redundancy | 2.4(2.3) | 2.6(2.5) | 2.9(2.9) | 2.9(2.7) |
| Rsymmb(%) | 0.08(0.45) | 0.074(0.42) | 0.065(0.349) | 0.054(0.33) |
| Rcrystc /Rfreed(%) | 17.9 / 19.5 | 18.4 / 19.7 | 17.05 / 19.5 | 17.9 / 18.3 |
| No of protein/solvent atoms | 2055/110 | 2057/201 | 2057/179 | 2058/289 |
| average B factors(Å2) main/side chain Zn/solvent |
18.2/21.3 11.9/36.3 |
15.8/19.0 9.5/35.7 |
13.1/16.2 6.8/29.3 |
15.3/18.3 9.3/42.3 |
| Ramachandran Plot (%). most favored/ additionally allowed, |
96.9/3.1 | 96.9/3.1 | 96.1/3.9 | 96.5/3.5 |
| eRMSD (Å) | 0.089 | 0.092 | 0.08 | 0.087 |
| I / σ(I) | 25.3 (3.1) | 31.3 (3.2) | 12.9 (3.1) | 28.8 (4.6) |
| R.m.s.d. for bond lengths (Å) / angles ( ° ) |
0.007 / 1.3 | 0.004 / 1.3 | 0.004 / 1.3 | 0.005 / 1.3 |
| PDB accession number | 3DV7 | 3DVB | 3DVC | 3DVD |
Values in parenthesis are for the highest resolution shell.
Rsymm = Σ |I - <I>| / Σ <I> ×100
Rcryst = Σ |Fo| — |Fc| / Σ |Fobs| × 100
Rfree is calculated the same as Rcryst, except it uses 5% of reflection data omitted from refinement.
The root mean square deviation of Cα positions as compared to the wild-type crystal structure of HCAII (PDB ID: 2CBA)(11).
Oxygen-18 Exchange
This method is based on the measurement by membrane inlet mass spectrometry of the depletion of 18O from species of CO2 (27, 28). A continuous measure of isotopic content of CO2 is provided by CO2 passing across the membrane where it enters a mass spectrometer (Extrel EXM-200). In the first of two independent stages of catalysis, the dehydration of labeled bicarbonate has a probability of transiently labeling the active site with 18O (eq 3). In a second stage, the protonation of the zinc-bound 18O-labeled hydroxide results in the release of H218O to the solvent (eq 4)
| (3) |
| (4) |
Two rates for the 18O exchange catalyzed by carbonic anhydrase are obtained by this method (27). The first is R1, the rate of exchange of CO2 and HCO3- at chemical equilibrium, as shown in eq 5. Here kcatex is a rate constant for maximal interconversion of substrate and product, KeffS is an apparent binding constant for substrate to enzyme, and [S] is the concentration of substrate, either CO2 or bicarbonate (29). The ratio kcatex/ KeffCO2 is, in theory and in practice, equal to kcat/Km for hydration obtained by steady-state methods.
| (5) |
RH2O, the rate of release from the enzyme of water bearing substrate oxygen (eq 4), is the second rate determined by the 18O-exchange method. This is the component of the 18O exchange that is dependent upon the donation of protons to the 18O-labeled zinc-bound hydroxide (6, 27). In such a step, His64 as a predominant proton donor in the catalysis provides a proton (eq 4). In eq 6, kB is the rate constant for proton transfer to the zinc-bound hydroxide and (Ka)donor and (Ka)ZnH2O are the ionization constants of the proton donor and zinc-bound water molecule. The determination of the kinetic constant kB and ionization constants of eq 6 was carried out by nonlinear least-squares methods (Enzifitter, Elsevier-Biosoft, Cambrige, U.K.)
| (6) |
The catalyzed and uncatalyzed exchange of 18O between CO2 and water at chemical equilibrium were measured in the absence of buffer at a total substrate concentration of 25 mM using membrane-inlet mass spectrometry (27). The temperature was 25 °C and the total ionic strength of solution was kept at a minimum of 0.2 M by the addition of Na2SO4.
Esterase activity
The hydrolysis of 4-nitrophenylacetate uncatalyzed and catalyzed by variants of carbonic anhydrase was measured by the method of Verpoorte et al. (30). The increase in absorbance at 348 nm was measured; this is the isosbestic point of the product nitrophenol and the nitrophenolate anion with an absorbtivity of 5.0 × 103 M-1cm-1. Initial velocities were measured using a Beckman Coulter DU 800 spectrophotometer.
RESULTS
We examined the crystal structures and catalytic properties of four mutants of HCA II containing substitutions at position 62, namely, N62A, D, V, and T. In addition, these properties for N62L were reported previously (18).
Crystallography
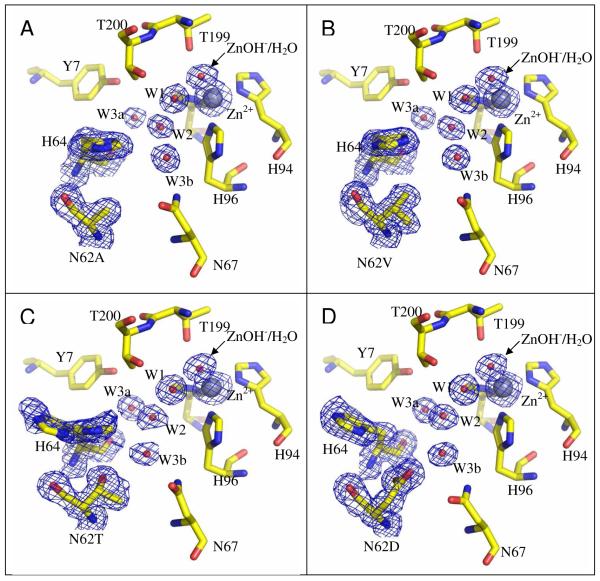
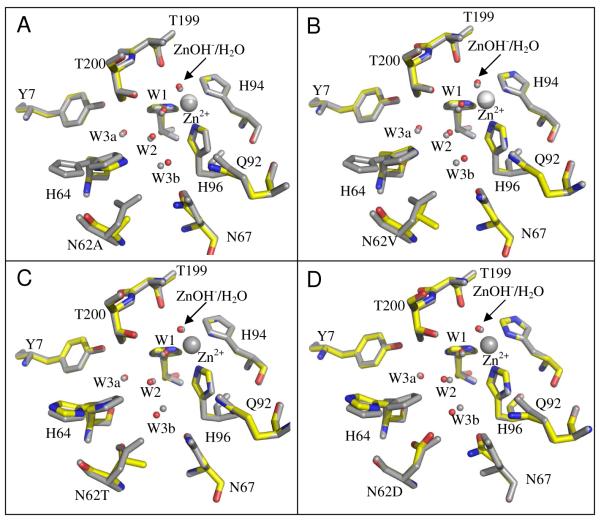
The crystals of the four mutant forms of HCA II were isomorphous with the wild type enzyme with mean unit cell dimensions of a = 42.7 ± 0.1 Å, b = 41.7 ± 0.1 Å, c = 72.8 ± 0.1 Å and β = 104.6 ± 0.1° (Table 1, Figures 2,3). All data sets were greater than 92% complete and were processed to 1.7 - 1.5 Å resolution (Table 1). A least squares superposition of these mutants with the crystal structure of wild-type HCA II (PDB ID: 2CBA; (11)) gave an average RMSD of Cα atoms 0.09 ±0.01 Å. The polypeptide backbone at position 62 was shifted slightly into the active-site cavity for the mutants N62A, N62V and N62T with a Cα movement of 0.6, 0.5, and 0.5 Å respectively.
Figure 2.
The active site of site-specific mutants (A) N62A, (B) N62V, (C) N62T, and (D) N62D HCA II. Residues are labeled and shown as stick models; the zinc atom is depicted as a white sphere and the oxygens of solvent water molecules as red spheres. Note the side chain of His64 is orientated inward for N62A and N62V, both inward and outward for N62T, and outward for N62D HCA II. The Fo-Fc electron density map was generated by omitting the residues at position 62, 64, and five water molecules in the active site. The map is depicted as a blue mesh and contoured at 3 σ for each site-specific mutant. This figure was generated and rendered with PyMOL (www.pymol.org).
Figure 3.
Structural superposition of the active site of the site-specific mutants (A) N62A, (B) N62V, (C) N62T, and (D) N62D with wild-type HCA II. Each panel has the wild-type HCA II (grey) structurally aligned with the respective mutant. Residues are labeled and shown as stick models, the zinc atom is depicted as a white sphere and the oxygens of solvent water molecules as red spheres. The structure of wild-type HCA II used was from PDB accession number 2CBA (11). This figure was generated and rendered with PyMOL (www.pymol.org).
The side chain of the proton shuttle His64 is of particular interest and its dual orientation in HCA II is well documented (9, 10). The inward and outward conformers of His64 in the wild-type structure of Hakansson et al (10) were refined to an occupancy of approximately 70% and 20% respectively, whereas these conformers of His64 in N62T were refined with equal occupancy of 50% each. Full occupancy was assigned to His64 in the case of the mutants that report only a single conformer of His64 (N62A,V,L), although small populations (≤ 10 %) of His64 may be undetected and may assume an alternative conformation. Appropriate B factors are given in Table 2.
Table 2.
Mean B-factors (Å2) for the ordered side chains of His64 and amino acids in its immediate vicinity in the active-site cavity of HCA II and variants.
| Wild typea | N62A | N62V | N62T | N62D | |
|---|---|---|---|---|---|
| His64 | 13.2 b/ 8.5 c | 24.9 b | 19.6 b | 12.5 b / 14.7 c | 18.2 c |
| Trp5 | 11.3 | 20.4 | 16.8 | 14.3 | 17.2 |
| Try7 | 8.9 | 14.9 | 13.0 | 9.5 | 12.1 |
Data from the Protein Data Bank, accession code 2CBA solved to a comparable resolution of 1.5 Å.
‘In’ conformer.
‘Out’ conformer.
In wild type HCA II the inward orientation has dihedral angles χ1 and χ2 near 44° and 95°, respectively, while the outward orientation has angles of -39° and 98°. The dihedral angle χ1 describes the inward and outward orientations of the side chain of His64 representing rotation about the Cα-Cβ bond. For N62A and N62V HCA II, His64 was observed only in the inward conformation, and for N62D only in the outward conformation (Supporting Information, Table SI1). In contrast, the mutant N62T HCA II resembled wild type enzyme by displaying two superimposed conformations for the His64 side chain, the inward and outward. These two side-chain conformations were equally present in the crystal structure of N62T HCA II.
None of the mutations significantly disrupted the active site solvent network compared with wild-type HCA II, although a slight shift of 0.7 Å in the position of the water molecule W3b was observed in all the mutant forms (Figures 2,3). In the case of the hydrophobic substitutions of Ala and Val at position 62, water molecule W3b moved away from the side chain of residue 62 (Figures 3A and B). Hydrophilic substitutions of Asp and Thr caused W3b to move towards the polar side chain of residue 62 (Figures 3C and D). Besides these small positional perturbations, the network of ordered solvent remained conserved (Figure 3). Of all the mutant structures, N62A has slightly higher B factors for the ordered water structure, but this is most likely due to its slightly lower resolution than the others (Supporting Information, Table SI2).
The replacement of Asn62 with Asp and Ala induced shifts in the side chains of nearby residues Asn67 and Gln92 (Supporting Information, Table SI1). The side chain atoms, Oδ1 and Nδ2 of Asn67 in N62D were shifted by 1.4 Å and 0.3 Å respectively towards His64 (Δχ1 ≈ 25°; Δχ2 ≈ 37°). In the case of N62A, the same atoms were shifted by 0.8 Å and 0.2 Å respectively towards His64 (Δχ1 ≈ 25°; Δχ2 ≈ 31°). The side chain atoms Nη2 of Gln92 in N62D and N62A were shifted by 1.3 Å (Δχ3 ≈ 33°) and 0.8 Å (Δχ3 ≈ 20°) respectively.
Catalysis
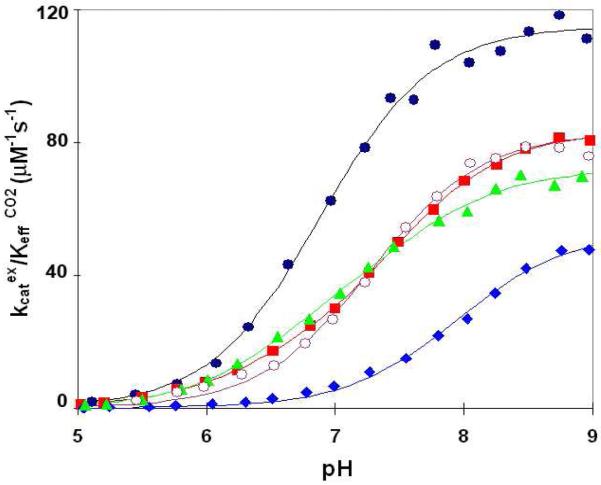
The catalysis by carbonic anhydrase of 18O exchange between CO2 and water yields two rates, as described in Methods. The first is R1 of eq 5 from which we determined kcatex/KeffCO2 for the hydration direction. The pH profile of this rate constant for wild type and mutants at residue 62 were adequately fit to the titration of a single residue (Figure 4) with resulting values of the kinetic pKa, an estimate of the pKa of the zinc-bound water (3, 29), given in Table 3. These data show a maximum at high pH corresponding to the reactivity of the zinc-bound hydroxide form of the enzyme in the hydration direction, with the maximal values of kcatex/KeffCO2 given in Table 4. The pKa values were all close to pKa 7.0 as found in wild type, with the exception of N62D HCA II for which the pKa was elevated to 7.9 (Table 3, column 5). However, the maximal, pH independent values of kcatex/KeffCO2 for the four mutants showed considerable variation with each mutant less than kcatex/KeffCO2 for wild type; the value for N62D was the smallest (Table 4). A somewhat more precise fit of the data was achieved by introduction of a second ionization; however, this had minor effects on the resulting parameters of Tables 3 and 4. The values of KeffCO2 are large in these experiments, exceeding the solubility of CO2 (about 34 mM) under our conditions. Hence, kcatex was not able to be determined.
Figure 4.
The pH profiles for kcatex/KeffCO2 for the hydration of CO2 catalyzed by the following variants of HCA II: wild type (●); N62V (○); N62A (■); N62T (▲); and N62D (◆). Data were obtained by 18O exchange between CO2 and water measured at 25 °C in solutions containing 25 mM of all species of CO2 and at sufficient Na2SO4 to maintain 0.2 M ionic strength. No buffers were added.
Table 3.
Apparent values of pKa obtained by various kinetic measurements of catalysis by HCA II and mutants.
| Enzyme | Orientation of His64 |
pKa His64a (eq 6) |
pKa ZnH2Oa (eq 6) |
pKa ZnH2Ob (eq 5) |
pKa ZnH2O (esterase) |
|---|---|---|---|---|---|
| wild-type | in/out | 7.2 | 6.8 | 6.9 | 6.9 |
| N62T | in/out | 7.0 | 7.0 | 7.0 | 7.0 |
| N62D | out | 8.4 | 7.6 | 7.9 | 7.7 |
| N62A | in | 6.2 | 6.5 | 7.2 | 7.1 |
| N62V | in | 5.9 | 5.9 | 7.3 | 7.1 |
| N62Lc | in | 6.0 d | 6.0 d | 7.3 | 7.1 |
Measured from the fits of eq 6 to data of Figure 5. The values of pKa have standard errors generally near ± 0.1 and no greater than ± 0.2.
Measured from the data of Figure 4 using eq 5. The standard errors in pKa are mostly ± 0.1 and no greater than ± 0.2.
These data from Fisher et al. (18)
These values estimated from poorly resolved pH dependence shown in Fisher et al. (18)
Table 4.
Maximal values of rate constants for hydration of CO2, for hydrolysis of p-nitrophenylacetate, and for proton transfer in the dehydration direction catalyzed by HCA II and variants.a
The values of pKa determined from kcatex/KeffCO2 in the 18O exchange experiment are very similar to the values of pKa determined from the pH dependence of kcat/Km for the catalyzed hydrolysis of 4-nitrophenylacetate (Table 3, column 6). The variation in the maximal values of kcat /Km for ester hydrolysis was less that for kcatex/KeffCO2; however, kcat /Km for N62D HCA II was the smallest in both sets of data (Table 4). Again, Km for this ester hydrolysis is too large to determine kcat.
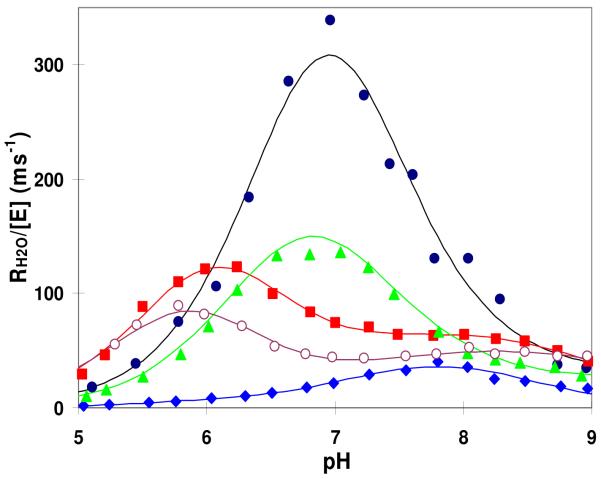
The values of RH2O/[E] provide an independent set of parameters that represent a rate constant for the proton transfer dependent release of H218O from the active site (eq 4). RH2O/[E], the rate of release of H218O from the active site, is determined by intramolecular proton transfer as verified by pH profiles, kinetic isotope effects, and chemical rescue experiments (28, 31). The pH profiles of RH2O/[E] appear bell shaped for much of the pH range of these studies (Figure 5), a feature that has been interpreted in terms of eq 6 and the transfer of a proton from one prominent donor His64 to the zinc-bound hydroxide (28, 31). The solid lines for the bell-shaped regions of Figure 5 represent fits of eq 6 to the data with the resulting parameters appearing in Tables 3 and 4. In order to apply eq 6 correctly, the assignment of the values of pKa for the donor and acceptor needs to be established. This was achieved using the values of pKa for the zinc-bound water pKa ZnH2O as determined by kcatex/KeffCO2 from the 18O-exchange experiment and by kcat/Km for the ester hydrolysis, which are in agreement (Table 3, columns 5,6). The 18O-exchange experiments were carried out at chemical equilibrium in the absence of added buffer. Buffers can participate in the proton transfer and complicate interpretation.
Figure 5.
The pH profiles of RH2O/[E], the rate constant for the release of H218O from the enzyme, catalyzed by these variants of HCA II: wild type (●); N62V (○); N62A (■); N62T (▲); and N62D (◆). Conditions were as described for Figure 4.
The values of pKa ZnH2O and pKa His64 for wild-type and N62T HCA II were nearly identical (Table 3); however, the value of the rate constant for proton transfer kB for N62T was half that of wild type (Table 4). For N62D HCAII the value of pKa ZnH2O is elevated to 7.6 -7.9 (Table 3) with the value of pKa His64 at 8.4 also elevated compared with wild type. This corresponds to His64 predominantly in the outward orientation with kB very low at 0.04 μs-1, about 5% of the value of wild type (Table 4).
The remaining variants of Tables 3 and 4, specifically N62A,V, and L, are not as straightforward because the values of pKa ZnH2O near 7.2 determined from kcatex/KeffCO2 of the 18O exchange experiment and kcat/Km from the ester hydrolysis (Table 3, columns 5,6) do not agree with the values of pKaZnH2O determined from RH2O/[E] which are near 6.0 (Table 3, column 4). The bell-shaped curves for these variants are clearly shifted to lower pH (Figure 5) consistent with values of pKa ZnH2O and pKa His64 both near 6.0 (Table 3). The resulting values of kB are in the range of 0.2 to 0.4 μs-1, not very different than the corresponding values of the other enzymes of Table 4 (except N62D). The data for N62L HCA II are from a previous report (18) and were not interpreted there because of their complicated pH profile for RH2O/[E]. However, the data of Figure 5 show pH profiles for N62A and N62V that are less complicated but show features shared by N62L. With this background eq 6 was applied to all three of these variants N62A,V, and L. We note that each of the mutants N62A and N62V (Figure 5), as well as N62L (18), have a secondary, very small maximum for RH2O/[E] at pH near 8 that resembles the data for N62D. This presumably indicates a second, less efficient 18O exchange pathway.
DISCUSSION
The goal of this work was to elucidate the role in catalysis of residue 62 on the hydrophilic side of the active-site cavity in HCA II, in particular its influence on the structure of ordered solvent, on the properties of the proton shuttle His64, and on the kinetics of catalysis. An advantage with the amino-acid substitutions we made at position 62 was that each of the resulting mutants had an ordered solvent structure in the active-site cavity very similar to that of wild-type HCA II. This appears to remove solvent structure as a variable in comparing catalysis by these mutants. As a result, these mutants allow this study to focus more completely on the properties of the side chain of His64.
The crystal structures of the variants at residue 62 are closely superimposable with wild-type HCA II (Figure 3). A major difference is in the orientation of the side chain of the proton shuttle residue His64 which appears for certain mutants in an entirely inward or entirely outward conformation, within experimental error (Figure 2, Supporting Information Table SI1). In wild-type HCA II these conformers interchange with a rotation about χ1 of about 100°. In any case, the kinetic barrier between inward and outward orientations is near 6 kcal/mol in wild-type HCA II according to computations, and is not expected to contribute to rate of catalysis (with an energy barrier near 10 kcal/mol)(32).
For the mutation N62D HCA II, His64 was observed entirely in the outward conformation, within experimental uncertainty (Figures 2, 3). In this case the pKa for the imidazole of the side chain of His64, determined from pH profiles of catalysis, was elevated to 8.4 compared with wild type which is near 7.2 (Table 3)(33). The elevated pKa of His64 in the N62D mutants is likely due to the nearby Asp62, possibly in an undetected inward orientation. This is also consistent with the pH dependence of rotamer populations in wild-type HCA II showing the outward orientation favored at lower pH (9, 10). In N62D HCA II, the pKa of the zinc-bound water is also elevated to 7.6 - 7.9 (Table 3), which is also anticipated based on the shift to the outward orientation of the partially positively charged side chain of His64.
The ordered water structure appears unchanged in N62D compared with wild type although His64 is in the outward orientation in this mutant (Figure 2). It is significant that ΔpKa between proton donor and acceptor (pKa ZnH2O — pKa His64) is small, near 0.4 – 0.8, for both wild type and N62D HCA II, yet the rate constant for proton transfer kB for N62D is about 20-fold less than wild type (Table 4). This shows that the lower rate constant kB for N62D is not due primarily to changes in pKa, but rather to the much lower capacity for proton transfer of His64, presumably because of its greater distance from the zinc (about 12 Å). This is consistent with accumulated evidence from other studies of the distance dependence of the proton transfer in carbonic anhydrase (34). The longer distance between His64 and the zinc-bound hydroxide in N62D (∼ 12 Å) is suggested to have a slower proton transfer at least in part because of the energy inherent in constructing longer water wires for proton transfer (32). In addition, computations show that the outward orientation of His64 is associated with shorter lifetimes of the ordered water molecules in the active-site cavity (19). Hence, the lower rate constant for proton transfer kB for N62D can be attributed to His64 in the outward orientation. However, data on this topic are complex. Some computations indicate that proton transfer in carbonic anhydrase is dominated by electrostatics and is not expected to be dependent on distance (15, 35). Introducing a charged group such as Asp62 near the proton transfer pathway would be expected to alter the electrostatic properties of the donor and acceptor, as noted above, but also the apparent pKa values of water molecules in the proton transfer pathway. This could contribute to the low efficiency of proton transfer in catalysis by N62D. The orientation of His64 in T200S HCA II is outward yet the steady-state constants for catalysis are very similar to those of wild-type HCA II (36).
For each of the three mutants in which His64 is in the inward orientation (N62A, N62V, N62L) the pKa for His64 determined from RH2O is decreased to near 6.0 (Table 3), due in part to the position of the imidazole ring in a more hydrophobic environment and more sequestered from bulk solvent (Figure 2). The pKa for the zinc-bound water determined in these mutants from RH2O/[E] is considerably smaller than in wild type (Table 3). This decrease is expected in these three mutants because of greater stabilization of the zinc-bound hydroxide by the inward oriented imidazolium form of His64. However, in contrast to the smaller value of pKa near 6 determined from RH2O/[E], the pKa values for zinc-bound water observed from kcatex/KeffCO2 and of kcat/Km of the esterase catalysis are near 7 (Table 3). These data imply that in the transition state for proton transfer (in N62A, N62V, N62L), the pKa values of the proton acceptor is low compared with the pKa determined from the first stage of catalysis (eq 3). In each case however, the rate constant for proton transfer appears close to the same (0.20 to 0.39 μs-1, Table 4). This probably indicates that it is the inward orientation that is responsible for proton transfer between His64 and the zinc-bound solvent molecule in catalysis.
Finally, the values of pKa for donor and acceptor for N62T HCA II appear very close to those for wild type (Table 3). Both of these enzymes show appreciable occupancy of His64 in the inward and outward orientations. Perhaps it is significant that these pKa values are midway between those found for His64 in the mutants for which this residue lies entirely in the inward and entirely in the outward orientations. However, the rate constant for proton transfer kB is less by about two-fold for N62T compared with wild type (Table 4).
An interesting feature of the data of Figure 4 compared with an earlier study (18), is that in this study replacement of Asn62 with hydrophobic residues caused substantial changes in kcatex/KeffCO2 (Table 4). In the case of N62D, a decrease as much as two-fold in the maximal values of kcatex/KeffCO2 for hydration was observed (Table 4). This is not explained for N62A,V, and L by values of the pKa of the zinc-bound water which are arguably the same as in wild type. Moreover, the decrease in kcatex/KeffCO2 for N62D is associated with a more basic pKa for the zinc-bound water than in wild type (Table 3). Since these mutations alter the charge density in the active-site cavity, electrostatic effects on the transition state for the nucleophilic attack on CO2 or on the dissociation of bicarbonate could be responsible.
Conclusions
Replacment of Asn62 with a number of amino-acids maintained the ordered water structure observed by x-ray crystallography in the active-site cavity of HCA II. This removed water structure as a variable in comparing with wild type the activities of mutants at residue 62. A significant role of Asn62 in HCA II is to permit two conformations of the side chain of His64, the inward and outward, that are necessary for maximal efficiency of this enzyme in transferring protons between the active site and solution (34, 37). The site-specific mutant N62D was observed to have an outward orientation of His64, yet the difference in pKa between the proton donor His64 and zinc-bound hydroxide was near zero, as in wild-type HCA II. The rate of proton transfer in catalysis by N62D HCA II in the dehydration direction was 5% that of wild type, showing that His64 in this mutant is inefficient in proton transfer compared with wild type because of its predominantly outward orientation compared with wild type which shows both inward and outward orientations. Additional contributions to inefficient proton transfer could arise from nonspecific electrostatic effects due to introduction of a charged group Asp62 near the proton transfer pathway. These results emphasize the role of Asn62 among the residues on the hydrophilic side of the active-site cavity in maintaining efficient catalysis by carbonic anhydrase.
Supplementary Material
Abbreviations
- HCA II
isozyme II of human carbonic anhydrase
- N62A
a site-specific mutant in which Asn62 is replaced by Ala
- RMSD
root mean square deviation
Footnotes
The coordinates of the following mutants of human carbonic anhydrase II have been deposited in the Protein Data Bank with these accession numbers: N62A (3DV7); N62V (3DVB); N62T (3DVC); and N62D (3DVD).
This work was funded by grants from the National Institutes of Health (Gm25154) and the Thomas H. Maren Foundation.
SUPPORTING INFORMATION AVAILABLE
A table containing the dihedral angles of three residues (His64, Asn67, Gln92) in the active-site cavity of wild-type HCA II and the variants N62A,V,D,T. A second table containing the B factors for the ordered water molecules in the active-site cavity of the same variants. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chegwidden WR, Carter ND, Edwards YH. The Carbonic Anhydrases New Horizons. Birkhauser Verlag; Basel: 2000. [Google Scholar]
- 2.Supuran CT, Scozzafava A, Conway J. Carbonic Anhydrase - Its Inhibitors and Activators. CRC Press; Boca Raton: 2004. [Google Scholar]
- 3.Lindskog S. Structure and mechanism of carbonic anhydrase. Pharmacol Ther. 1997;74:1–20. doi: 10.1016/s0163-7258(96)00198-2. [DOI] [PubMed] [Google Scholar]
- 4.Silverman DN, Lindskog S. The Catalytic Mechanism of Carbonic-Anhydrase - Implications of a Rate-Limiting Protolysis of Water. Accounts of Chemical Research. 1988;21:30–36. [Google Scholar]
- 5.Christianson DW, Fierke CA. Carbonic anhydrase: Evolution of the zinc binding site by nature and by design. Accounts of Chemical Research. 1996;29:331–339. [Google Scholar]
- 6.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 7.Steiner H, Jonsson BH, Lindskog S. Catalytic Mechanism of Carbonic-Anhydrase - Hydrogen-Isotope Effects on Kinetic-Parameters of Human C Isoenzyme. European Journal of Biochemistry. 1975;59:253–259. doi: 10.1111/j.1432-1033.1975.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 8.Liljas A, Lovgren S, Bergsten PC, Carlbom U, Petef M, Waara I, Strandbe B, Fridborg K, Jarup L, Kannan KK. Crystal-Structure of Human Carbonic Anhydrase-C. Nature-New Biology. 1972;235:131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- 9.Fisher SZ, Maupin CM, Budayova-Spano M, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: Insights into the proton transfer mechanism. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 10.Nair SK, Christianson DW. Unexpected pH-Dependent Conformation of His-64, the Proton Shuttle of Carbonic Anhydrase-II. Journal of the American Chemical Society. 1991;113:9455–9458. [Google Scholar]
- 11.Hakansson K, Carlsson M, Svensson LA, Liljas A. Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J Mol Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- 12.Venkatasubban KS, Silverman DN. Carbon-Dioxide Hydration Activity of Carbonic-Anhydrase in Mixtures of Water and Deuterium-Oxide. Biochemistry. 1980;19:4984–4989. doi: 10.1021/bi00563a008. [DOI] [PubMed] [Google Scholar]
- 13.Fisher SZ, Maupin CM, Budayova-Spano M, Govindasamy L, Tu CK, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: Insights into the proton transfer mechanism. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 14.Cui Q, Karplus M. Is a “proton wire” concerted or stepwise? A model study of proton transfer in carbonic anhydrase. Journal of Physical Chemistry B. 2003;107:1071–1078. [Google Scholar]
- 15.Braun-Sand S, Strajbl M, Warshel A. Studies of proton translocations in biological systems: simulating proton transport in carbonic anhydrase by EVB-based models. Biophys J. 2004;87:2221–2239. doi: 10.1529/biophysj.104.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riccardi D, Konig P, Prat-Resina X, Yu H, Elstner M, Frauenheim T, Cui Q. “Proton holes” in long-range proton transfer reactions in solution and enzymes: A theoretical analysis. J Am Chem Soc. 2006;128:16302–16311. doi: 10.1021/ja065451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy A, Taraphder S. Identification of proton-transfer pathways in human carbonic anhydrase II. Journal of Physical Chemistry B. 2007;111:10563–10576. doi: 10.1021/jp073499t. [DOI] [PubMed] [Google Scholar]
- 18.Fisher SZ, Tu CK, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Speeding up proton transfer in a fast enzyme: kinetic and crystallographic studies on the effect of hydrophobic amino acid substitution in the active site of human carbonic anhydrase II. Biochemistry. 2007;42:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 19.Maupin CM, Saunders MG, Thorpe IF, McKenna R, Silverman DN, Voth GA. Origins of enhanced proton transport in the Y7F mutant of human carbonic anhydrase II. J. Am. Chem. Soc. 2008;130:11399–11408. doi: 10.1021/ja802264j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewett-Emmett D, Tashian RE. Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Molecular Phylogenetics and Evolution. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 21.Khalifah RG, Strader DJ, Bryant SH, Gibson SM. C-13 Nuclear Magnetic-Resonance Probe of Active-Site Ionizations in Human Carbonic-Anhydrase B. Biochemistry. 1977;16:2241–2247. doi: 10.1021/bi00629a031. [DOI] [PubMed] [Google Scholar]
- 22.Segel IH. Enzyme Kinetics. Wiley-Interscience; New York: 1975. [Google Scholar]
- 23.McPherson A. Preparation and Analysis of Protein Crystals. Wiley; New York: 1982. [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Silverman DN. Carbonic anhydrase: oxygen-18 exchange catalyzed by an enzyme with rate-contributing proton-transfer steps. Methods Enzymol. 1982;87:732–752. doi: 10.1016/s0076-6879(82)87037-7. [DOI] [PubMed] [Google Scholar]
- 28.Silverman DN, Tu C, Chen X, Tanhauser SM, Kresge AJ, Laipis PJ. Rate-equilibria relationships in intramolecular proton transfer in human carbonic anhydrase III. Biochemistry. 1993;32:10757–10762. doi: 10.1021/bi00091a029. [DOI] [PubMed] [Google Scholar]
- 29.Simonsson I, Jonsson BH, Lindskog S. C-13 NMR study of carbon dioxide-bicarbonate exchange catalyzed by human carbonic anhydrase-C at chemical-equilibrium. European Journal of Biochemistry. 1979;93:409–417. doi: 10.1111/j.1432-1033.1979.tb12837.x. [DOI] [PubMed] [Google Scholar]
- 30.Verpoorte JA, Mehta S, Edsall JT. Esterase Activities of Human Carbonic Anhydrases B and C. Journal of Biological Chemistry. 1967;242:4221–4229. [PubMed] [Google Scholar]
- 31.An H, Tu C, Duda D, Montanez-Clemente I, Math K, Laipis PJ, McKenna R, Silverman DN. Chemical rescue in catalysis by human carbonic anhydrases II and III. Biochemistry. 2002;41:3235–3242. doi: 10.1021/bi0120695. [DOI] [PubMed] [Google Scholar]
- 32.Maupin CM, Voth GA. Preferred orientations of His64 in human carbonic anhydrase II. Biochemistry. 2007;46:2938–2947. doi: 10.1021/bi062170f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell ID, Lindskog S, White AI. A study of the histidine residues of human carbonic anhydrase C using 270 MHz proton magnetic resonance. J Mol Biol. 1975;98:597–614. doi: 10.1016/s0022-2836(75)80089-1. [DOI] [PubMed] [Google Scholar]
- 34.Silverman DN, McKenna R. Solvent-Mediated Proton Transfer in Catalysis by Carbonic Anhydrase. Acc Chem Res. 2007;40:669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 35.Riccardi D, Konig P, Guo H, Cui Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochemistry. 2008;47:2369–2378. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 36.Krebs JF, Fierke CA, Alexander RS, Christianson DW. Conformational Mobility of His-64 in the Thr200Ser Mutant of Human Carbonic Anhydrase-II. Biochemistry. 1991;30:9153–9160. doi: 10.1021/bi00102a005. [DOI] [PubMed] [Google Scholar]
- 37.Jude KM, Wright SK, Tu C, Silverman DN, Viola RE, Christianson DW. Crystal structure of F65A/Y131C-methylimidazole carbonic anhydrase V reveals architectural features of an engineered proton shuttle. Biochemistry. 2002;41:2485–2491. doi: 10.1021/bi015808q. [DOI] [PubMed] [Google Scholar]
- 38.Fisher Z, Prada J. A. Hernandez, Tu C, Duda D, Yoshioka C, An H, Govindasamy L, Silverman DN, McKenna R. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry. 2005;44:1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.