Abstract
Branching morphogenesis is a developmental process characteristic of many organ systems. Specifically, during renal branching morphogenesis, its been postulated that the final number of nephrons formed is one key clinical factor in the development of hypertension in adulthood. As it has been established that BMPs regulate, in part, renal activity of p38 MAP kinase (p38MAPK) and it has demonstrated that the cytoplasmic protein Neurotrophin Receptor MAGE homologue (NRAGE) augments p38MAPK activation, it was hypothesized that a decrease in the expression of NRAGE during renal branching would result in decreased branching of the UB that correlated with changes in p38MAPK activation. To verify this, the expression of NRAGE was reduced in ex vivo kidney explants cultures using antisense morpholino. Morpholino treated ex vivo kidney explants expression were severely stunted in branching, a trait that was rescued with the addition of exogenous GDNF. Renal explants also demonstrated a precipitous drop in p38MAPK activation that too was reversed in the presence of recombinant GDNF. RNA profiling of NRAGE diminished ex vivo kidney explants resulted in altered expression of GDNF, Ret, BMP7 and BMPRIb mRNAs. Our results suggested that in early kidney development NRAGE might have multiple roles during renal branching morphogenesis through association with both the BMP and GDNF signaling pathways.
Introduction
Essential hypertension, or hypertension with no identifiable cause, is unfortunately a common disease of the Western world (Kearney et al., 2005). In the early 1970's David Barker proposed the “fetal origins of disease hypothesis”, supposing that the prevalence of many adult diseases, including hypertension, is a result of abnormal fetal development (Barker et al., 1970). Brenner later refined this hypothesis by proposing that lower nephron numbers predisposed individuals to essential hypertension (Brenner et al., 1988). Since reports based on the Brenner-Barker hypothesis suggest a link between kidney development and hypertension (Langley and Jackson, 1994; Levitt et al., 1996; Woodall et al., 1996), elucidating the molecular mechanisms that govern kidney development could elucidate the key factors affecting the development of hypertension later in life.
The development of the kidney begins with renal branching morphogenesis (RBM). During RBM reciprocal inductive interactions, between the ureteric bud (UB) and the surrounding metanephric mesenchyme (MM) result in the development of the collecting ducts and the nephrons. The precise molecular signals that control RBM are currently unknown and still actively pursued. Bone morphogenic proteins (BMPs) are members of the transforming growth factor beta (TGFβ) superfamily of signaling molecules and have been implicated in a diverse array of biological processes, including cell growth, differentiation and apoptosis (Hogan, 1996). BMPs play crucial roles during RBM, transducing their signal either through the canonical SMAD-mediated pathway, and/or through the non-canonical BMP signaling cascade of MAP kinases, TAK1, TAB1, and p38MAPK (Nohe et al., 2004; Oxburgh et al., 2004; Winnier et al., 1995).
We recently demonstrated that NRAGE is a potential member of the non-canonical BMP pathway utilizing the multipotential neural progenitor cells resulting in BMP instructive apoptosis (Kendall et al., 2005). It has been suggested that the same non-canonical BMP signaling pathway also mediates branching of the UB (Hu et al., 2004) suggesting a potential role for NRAGE during embryonic renal branching morphogenesis. It was hypothesized that a decrease in the expression of NRAGE during RBM would result in altered branching of the UB and potentially in cell viability.
Utilizing NRAGE morpholinos (Kendall et al., 2005), we attenuated NRAGE protein expression in ex vivo kidney culture explants to determine if decreased NRAGE expression affects p38MAPK activation and consequently branching of the UB. We also investigated the global ramifications of lowering NRAGE expression in the developing explants in hopes of elucidating other pathways and mechanisms that NRAGE may regulate during renal development. As predicted, lowering NRAGE expression severely retarded the growth and branching of the UB. What was surprising and unexpected was that gene profiling revealed that lowering NRAGE levels lead to a reduction in the expression of BMPR1b, Ret, GDNF, and BMP7 in the developing kidney. Rescue experiments demonstrated that exogenously applied recombinant GDNF corrected the deficiency in branching in ex vivo explants cultures, with GDNF being more robust to promote growth and branching than BMP7. These results demonstrate the importance of NRAGE in affecting the maximal response during branching morphogenesis.
Methods
Cell Culture
mIMCD-3 (ATCC, Virginia, USA) cells were cultured in DMEM/F12 (Invitrogen, California, USA) supplemented with 10% fetal bovine serum (Hyclone, Utah, USA) in a 37°C and 5%CO2 humidified incubator. In branching experiments, 1×106 cells were plated in a collagen matrix as described by Piscione without modification (Piscione et al., 2001). The matrix was assembled on ice and plated with various doses of GDNF (0-10ng/ml) (R&D Systems, Minnesota, USA), BMP7 (0-10ng/ml) (R&D Systems, Minnesota, USA), no supplementation, or TGFβ (0-25ng/ml) (R&D Systems, Minnesota, USA) for 3-14 days with media refreshed every morning.
Co-immunoprecipitation and immunoblotting
Cell lysates were generated from mIMCD-3 cells that were treated with and without 10 ng/ml BMP7 (R&D Systems, Minnesota, USA) for 1 hour in DMEM-F12 (Invitrogen, California, USA). Cells were lysed in 350 μl of NPB lysis buffer consisting of: 20 mM Tris, pH 7.5, containing 300 mM sucrose, 60 mM KCl, 15 mM NaCl, 5% (v/v) glycerol, 2 mM EDTA, 1% (v/v) Triton X-100, with protease inhibitor cocktail I (Sigma-Aldrich, Missouri, USA) for 20 minutes on ice. Lysates were immunoprecipitated using 50 μl of G-sepharose beads (Amersham Biosciences-GE Healthcare, USA) and 2 μg of NRAGE (1:1000) or Ret antibody (1:2000) (Upstate-Millipore, Massacheusetts, USA; R&D Systems, Minnesota, USA) overnight at 4°C. The beads were collected by centrifugation at 12,000 RPMs for 5 minutes and washed three times with fresh ice-cold lysis buffer. The samples were subjected to 12% SDS-PAGE under reducing conditions. After transferring the resolved proteins to Hybond C membrane (Amersham Biosciences-GE Healthcare, USA), blots were probed for NRAGE (1:1000) (Upstate-Millipore, Massachusetts, USA), TAK1 (1:1000) (Upstate-Millipore, Massachusetts, USA), Tab1 (1:1000) (Pro-Sci, California, USA), XIAP (1:1000) (Cell Signaling Technology, Massachusetts, USA) or β-actin (1:2500) (Sigma-Aldrich, Missouri, USA) antibodies. Blots were developed using an appropriate horseradish peroxidase conjugated goat anti-mouse or rabbit IgG (Bio-Rad) and the ECL detection system (Amersham Biosciences-GE Healthcare, USA).
Kidney Organ Culture
Kidney organ culture was performed as previously described by Nikopoulos et al, 2008 (Nikopoulos et al., 2008) using kidneys from E11.5 Hoxb7-GFP mice (Srinivas et al., 1999) or E11.5 ICR mouse embryos (Taconic, New York, USA). Morpholino sequences used in this study are as follows, NRAGE morpholino: GGTTTCTGAGCCATAGCTCTCGTC and for the negative control morpholino: CCTCTTACCTCAGTTACAATTTATA (Gene-Tools, Oregon, USA). BMP7 or GDNF (R&D Systems, Minnesota, USA) was added at concentrations and time described for each experiment. Kidneys were analyzed under a Leica stereomicroscope (Leica, USA) or subjected to immunofluorescent staining using: TOPRO-3 (1:10000; Invitrogen, California, USA) to identify nuclei, or Ki67 (1:1000; AbCam, Massachusetts, USA) to identify proliferating cells, Dolichous Bifluorous Agglutinin to identify cells of the ureteric bud (DBA; 1:1000; Vector Labs, California, USA) or anti-laminin to also identify cells of the ureteric bud (Sigma-Aldrich, Missouri, USA). Kidney explants stained with Ki67 were visualized using a Leica TCS-SP confocal microscope (Leica, USA). The number of Ki67 positive cells was determined by counting the Ki67 positive nuclei in a given field for each kidney analyzed and calculating the number of cells per μm3 using the Leica TCS software.
TUNEL analysis
E11.5 Hoxb7-GFP kidney explants were cultured with either NRAGE morpholino or negative control for 72 hours, in DMEM/F12 media + 10% FBS prior to being in 4% PFA overnight at 4°C. After consecutive washes in PBS, kidneys were incubated with 20μg/ml of proteinase-K (37°C) and were subjected to Terminal deoxynucleotidyl Transferase Biotin-dUTP Nick End Labeling (TUNEL) analysis to detect apoptotic cells using the TetraMethylRhodamine red (TMR) in situ cell death detection kit (Roche) and counterstained with 1:10,000 dilution of TOPRO-3 (Invitrogen, California, USA) overnight. Kidneys were mounted on glass slides with Prolong Gold (Invitrogen, Calfornia, USA) and analyzed using a Leica SP-TCS confocal microscope (Leica, USA). TUNEL positive nuclei in the metanephric mesenchyme, as delineated by the ureteric bud specific GFP expression, were counted and the calculated as the number of TUNEL positive cells per unit volume (μm3) for comparison between NRAGE and Negative Control treated kidneys.
RNA Isolation and Quantitative PCR of Kidney Organ Cultures and mIMCD-3 cells
E11.5 mouse (Taconic, New York, USA) kidney explants were cultured with NRAGE morpholino, negative control morpholino, or Endo-Porter only. Total RNA was extracted from six kidney organ cultures for each treatment for each day of culture and pooled into one RNA sample using the RNAqueous-micro kit (Ambion, California, USA). RNA samples were treated with 1 unit of DNAse I (37°C, 15 minutes) then inactivated using Ambion DNAase inactivation slurry. mIMCD-3 cells were incubated with NRAGE morpholino, negative control morpholino, and p38MAPK phosphorylation inhibitor SB203580, DMSO, or Endo-Porter only, for 48 hours. Total RNA was extracted from mIMCD-3 cells using the RNAqueous®-4PCR (Ambion, California, USA) and also treated with 1 unit of DNAse I (37°C, 15 minutes). DNAase was inactivated as above. cDNA was synthesized from all RNA samples with the first-strand synthesis reaction kit (SuperArray-SABiosceinces, Maryland, USA). Verified quantitative real-time PCR (qPCR) primers for all genes were obtained from SuperArray. qRT-PCR was performed in triplicate for each gene for each day, and three independent repeats of the experiment was performed using SYBR-Green I as per manufacturers instructions using an iCycler IQ (Bio-Rad, California, USA). Relative fold change in gene expression was calculated using the means of all experiments for each gene on each day with each treatment using the 2(-ΔΔCT) method as previously described (Livak and Schmittgen, 2001). One sample t-test for each gene was performed using Prism 4.0 Software (GraphPad, California, USA).
Hoxb7-NRAGECherry Transgenic Mice
Utilizing the same promoter sequence used by Costantini and colleagues to generate mice that specifically expressed GFP in the ureteric bud of the kidney; we constructed a transgene where full length NRAGE was fused in frame with the fluorescent protein mCherry (Shaner et al., 2004). mCherry was chosen as the fluorescent marker due to its low level of cell toxicity and high photostability. Furthermore, it has been previously utilized in transgenic animals to track development, where it was shown to have no effect on normal development (Renn and Winkler, 2009; Shcherbo et al., 2009; Winnier et al., 1995). After the digestion of the plasmid, we performed pro-nuclear injections on 60 fertilized eggs. We fixed and cryo-preserved the isolated kidneys from at E17 for fluorescent microscopic analysis. To identify cells of the ureteric bud of the kidney we utilized Dolichos biflorous agglutinin (1:1000, Vector Labs), a lectin that specifically stains cells of the ureteric bud of the kidney, or subjected them to p38MAPK staining. p38MAPK staining was performed by using a polyclonal antibody to p38MAPK (1:1000 dilution; Cell Signaling Technology, Massachusetts, USA) or a polyclonal phospho-p38MAPK antibody (1:1000; Cell Signaling Technology, Massachusetts, USA), and each received a rabbit Alexa-546 (1:5000; Invitrogen, California, USA) secondary antibody. Nuclei were identified by either TOPRO-3 (Invitrogen, California, USA) or DAPI (Vector Labs,
mIMCD-3 Cells in Collagen Matrix
Utilizing rat tail collagen (Invitrogen, California, USA), 10,000 mIMCD-3 cells were embedded into three-dimensional culture system as described previously (Piscione et al., 1997). The mIMCD-3 cells were treated with 10ng/ml BMP7, 25ngml GDNF or morpholino. When treated with morpholino, the mIMCD3 cells were treated for 48 hours in an adherent cell culture dish to suppress NRAGE expression before the beginning of the tubulogenesis assay. We defined a branch point as a pixel that has three or more neighboring pixels on our phase contrast microscope. When there are no longer two neighboring pixels, these branches are labeled 2° branches.
Results
NRAGE co-immunoprecipitates with members of the non-canonical BMP signaling pathway
Previously, we have shown that NRAGE interacts with members within the non-canonical BMP signaling pathway, specifically TAK, TAB and XIAP (Kendall et al., 2005). This association correlates with increased p38MAPK activation and caspase dependent apoptosis in neural progenitors (Kendall et al., 2005). In the kidney, it has been shown that NRAGE mRNA is expressed in cells of UB epithelium and the MM (Bertrand et al., 2004). Since non-canonical BMP signaling is also associated with increased p38MAPK activation, culminating in robust branching of the developing UB, it was hypothesized that NRAGE may be involved in the non-canonical BMP signaling pathway mediating p38MAPK activation during RBM. To establish NRAGE's association with the members of the non-canonical BMP pathway we utilized mIMCD-3 cells, differentiated epithelial cells from the collecting duct of the adult murine kidney. While mIMCD-3s cells are derived from adult tissues, they have been used to approximate the cellular environment of UB cells. We performed immunoprecipitation experiments using lysates generated from mIMCD-3 cells and our antibodies to NRAGE (Salehi et al., 2000). Western blot analysis demonstrated that TAK1, TAB1, and XIAP co-immunoprecipitates with NRAGE (Figure 1A) just as it does within populations of multipotential neural progenitors. MALDI-TOFF mass spectrometry verified the identity of the captured proteins in these co-immunoprecipitation experiments as TAK1, TAB1, and XIAP.
Figure 1. NRAGE morpholino dampens NRAGE expression and p38MAPK activation.
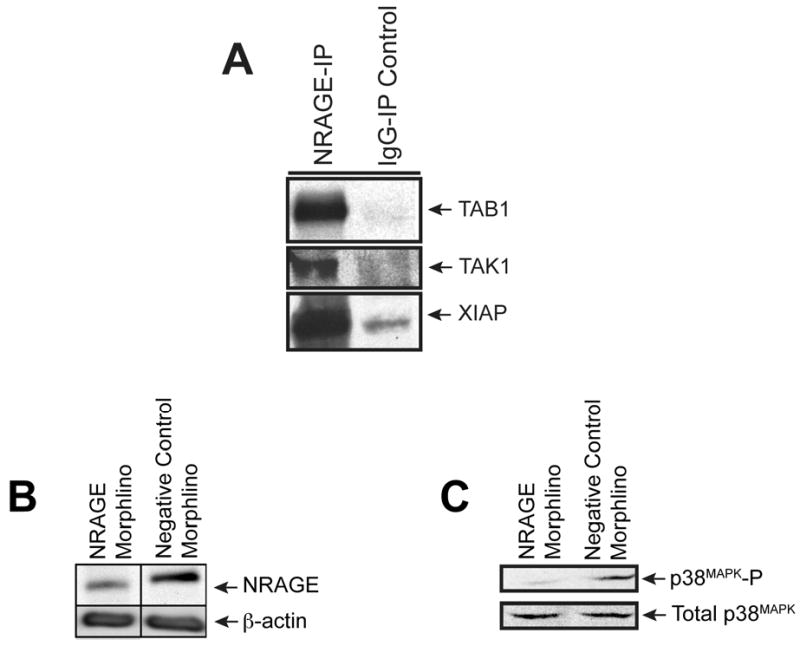
(A) NRAGE co-immunoprecipitates with members of the non-canonical BMP pathway. Lysates from 10 ng/ml BMP7 treated mIMCD-3 cells were co-immunoprecipitated with NRAGE antibody and subjected to western blot analysis. The resulting blots were probed for TAB1, TAK1, and XIAP. (B) NRAGE expression is dampened upon treatment with morpholinos against NRAGE. Immunoblot analysis of lysates generated from mIMCD-3 cells treated for 1 hour with 10 ng/ml recombinant BMP7 after being treated with either NRAGE morpholino or with negative control morpholino for 48 hours. (C) p38MAPK activation (p38MAPK-P) is decreased with NRAGE morpholino treatment. Immunoblot analysis of lysates generated from mIMCD-3 cells treated for 1 hour with 10 ng/ml recombinant BMP7 after being treated with either NRAGE morpholino or with negative control morpholino for 48 hours.
Treatment of mIMCD-3 cells with low doses (10 ng/ml) of BMP7 stimulates tubulogenesis of these cells via the non-canonical BMP signaling pathway (Piscione et al., 2001). Using NRAGE morpholino, we examined whether NRAGE was required for p38MAPK phosphorylation, a key step in non-canonical BMP signaling, in the presence of a low dose of BMP7 in mIMCD-3 cells. Cells were treated with NRAGE morpholino or negative control morpholino for 48 hours prior to the addition of 10 ng/ml BMP7 for one hour. Western blot analysis demonstrated that in NRAGE morpholino treated mIMCD-3 cells there was a significant decrease in NRAGE protein expression (Figure 1B). Western blot analysis also illustrated a decrease in p38MAPK phosphorylation (Figure 1C). The treatment of mIMCD-3 cells with NRAGE morpholino does not result in complete suppression of NRAGE protein expression. However, the level of inhibition of NRAGE expression is sufficient to inhibit p38MAPK, consistent with our previous studies in multipotential neural progenitor cells (Kendall et al., 2005). This result suggests that NRAGE expression and a complete non-canonical BMP signaling pathway might be required for the maximal phosphorylation of p38MAPK in mIMCD-3 cells.
Renal branching is inhibited upon decreased NRAGE expression and decreased p38MAPK phosphorylation
Since p38MAPK phosphorylation is important in normal branching of the UB of the kidney (Hida et al., 2002), and based on our last experiment (see Figure 1C) we investigated whether a decrease in NRAGE protein expression would have an effect upon the branching of the UB ex vivo.
It has been shown that gene expression can be altered using an ex vivo kidney organ culture model and antisense morpholino (Gross et al., 2003; Quaggin et al., 1998). Employing our published method of ex vivo kidney culture (Nikopoulos et al., 2008) to suppress renal protein expression, we cultured E11.5 kidney explants with NRAGE morpholino or with a negative control morpholino for 72 hours prior to initiating our biochemical analyses. Three kidneys explants were pooled per time point, and this was repeated three times to collect three pools of kidney lysates per time point as representative protein lysates. As shown in Figure 2, NRAGE protein expression was maximally inhibited at the 72-hour time point. The level of NRAGE expression began to decline significantly after the 24th hour but it did not reach maximal suppression until the 72nd hour. Rosenblum has already demonstrated that the level of p38MAPK phosphorylation can be directly correlated with the level of tubulogenesis with mIMCD-3 cells embedded in a collagen matrix (Hu et al., 2004). However, mIMCD-3 cells in a three-dimensional collagen matrix system, is intended to approximate RBM but not to replicate it. We attempted to circumvent this nuance by utilizing an ex vivo kidney culture system (Nikopoulos et al., 2008) to analyze branching morphogenesis in kidneys with diminished NRAGE expression and utilizing an antisense NRAGE specific morpholino. To this end, we used explants isolated from E11.5 Hoxb7-GFP mice (Srinivas et al., 1999), which express green fluorescent protein (GFP) in the UB. Treating the explants with either NRAGE morpholino or negative control morpholino, we recorded the progress of UB branching over 72 hours. Analysis of the kidney branching pattern by fluorescent microscopy demonstrated a significant decrease (two tailed t-test p<0.0001) in the number of endpoint branches in kidneys treated with NRAGE morpholino (7.13 branches ± 0.38) as compared to kidneys treated with negative control morpholino (10.2 branches ± 0.58) (Figure 3). This supported the hypothesis that decreased NRAGE expression would result in decreased branching of the UB.
Figure 2. NRAGE morpholino is most effective after culturing kidney explants with morpholino for 72 hours.
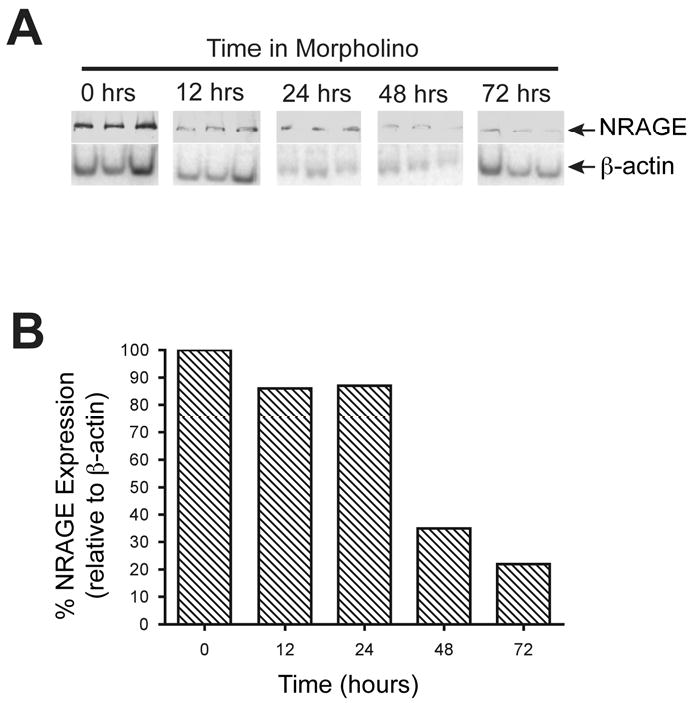
(A) Immunoblot analysis of three pooled lysates per time point, each pool derived from three kidney explants, cultured with NRAGE morpholino for up to 72 hours. (B) Densitometry analysis of panel A. Note the gradual decrease in NRAGE expression, most pronounced at 72 hours after addition of morpholino.
Figure 3. Attenuated NRAGE expression results in stunted growth of the ureteric bud.
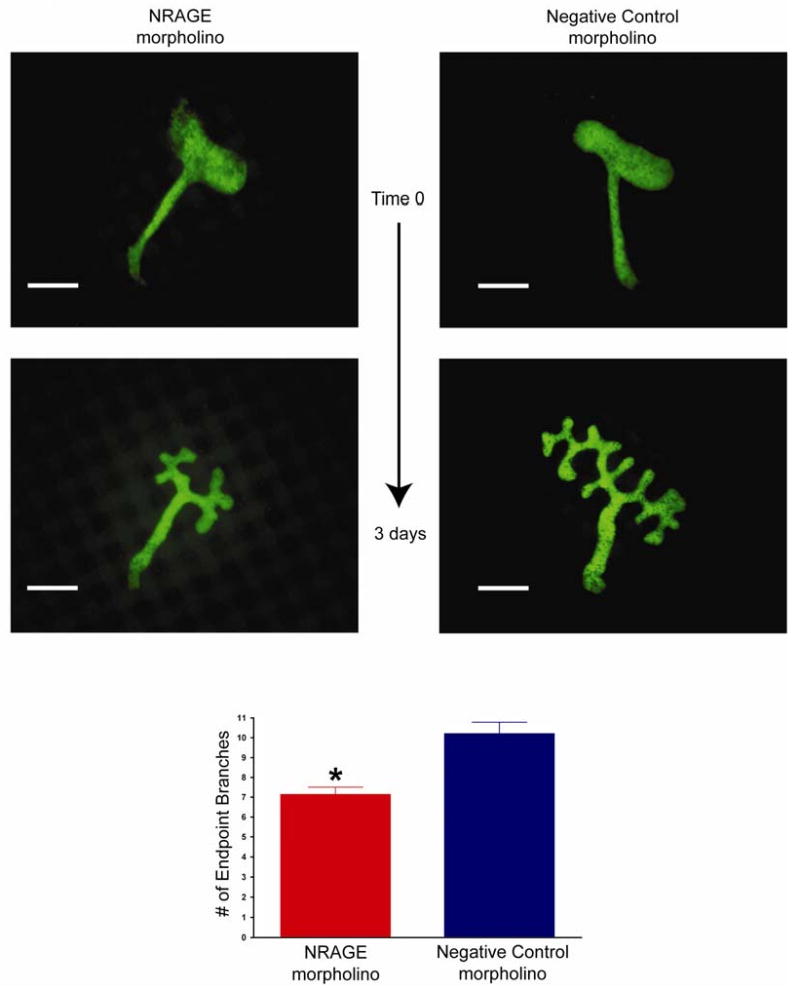
E11.5 kidneys from Hoxb7-EGFP mice were cultured with NRAGE morpholino or a negative control morpholino for three days. Extent of the effect of NRAGE morpholino on ureteric bud branching was determined by counting endpoint branches per explant and the means of 30 experiments are presented. Note: Number of endpoint branches when kidneys are treated with NRAGE morpholino: 7.13 branches ± 0.38. Number of endpoint branches when kidneys are treated with NRAGE morpholino: 10.20 branches ± 0.58. *NRAGE morpholino versus negative control morpholino unpaired two-tailed t-test p<0.0001. Scale bar = 200 microns.
Phosphorylated p38MAPK is observed in cells of the ureteric bud when NRAGE is overexpressed using Hoxb7-NRAGECherry transgenic mice
Confident that NRAGE was a member of the non-canonical BMP pathway in the kidney, we attempted to take our observations one step further and produced a Hoxb7-NRAGECherry transgenic mouse to examine NRAGE's role during renal development in vivo, by over expressing NRAGE in the UB, under the control of the Hobx7 promoter element. Three independent founders were produced and used to create viable independent lines. None of the three lines demonstrated remarkable changes to overall kidney development in terms of morphology and weight. At E17, two kidneys per embryo were collected and analyzed. Immunofluorescent analysis demonstrated that Hobx7NRAGECherry mice expressed mCherry in the UB domain, as expected (Figure 4A-B). Quantitative real-time PCR (qPCR) analyses revealed that there was not a significant difference in the level of p38MAPK mRNA between the E17 kidneys of Hoxb7-NRAGECherry (2(-ΔΔCT)=0.02) and wild-type embryos (2(-ΔΔCT) =0.018). This was true whether the analyses were performed on individual kidneys (N=9) or a combination of three aged matched kidneys (pools of 3 kidneys). Using antibodies against total p38MAPK and phosphorylated p38MAPK, we were able to determine that there was activated p38MAPK in the domain of the UB (Figure 4K; note the co-expression of phosphorylated p38MAPK and the mCherry fluorescent protein as a yellow overlay). While we were not able to quantify a difference between the level of phosphorylated p38MAPK in Hoxb7-NRAGECherry transgenic mice and their wild-type siblings, we were confident that when taken together with our initial in vitro data, there was a relationship between p38MAPK activation and NRAGE expression levels in the kidney.
Figure 4. Phosphorylated p38MAPK is observed in cells of the ureteric bud when NRAGE is overexpressed using Hoxb7-NRAGECherry transgenic mice.
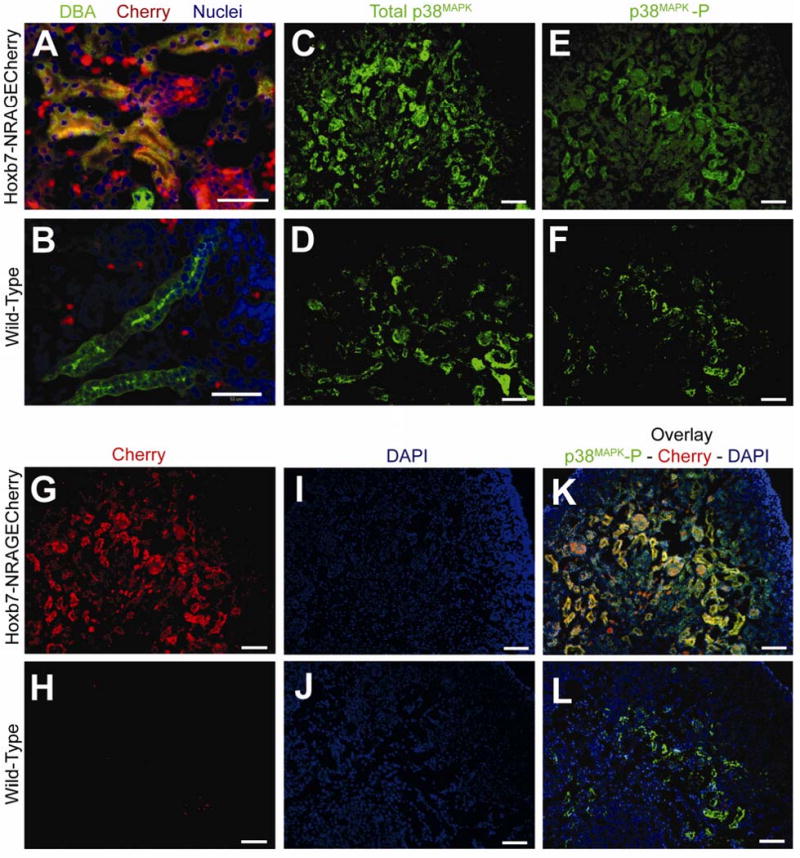
Transgenic mice were generated to overexpress NRAGE in cells of the ureteric bud using the Hoxb7 promoter fragment and tagged with the mCherry fluorescent protein (Hobx7-NRAGECherry). Presented are representative images from the immunofluorescent staining of kidneys from E17 transgenic mice. (A-B) Immunofluorescent staining of E17 Kidneys with Dolichos Bifluorous (Green) and TOPRO-3 (Blue). (A) Hoxb7-NRAGECherry E17 kidneys stained with Dolichos Bifluorous Agglutinin (Green; ureteric bud cells), and TOPRO-3 (Blue; nuclei). Note the overlay of mCherry fluorescent protein (Red) with Dolichos Bifluorous Agglutinin (Green) as Yellow, demonstrating that mCherry is indeed expressed in cells of the ureteric bud. (B) Wild-type E17 kidneys stained with Dolichos Bifluorous Agglutinin (Green; ureteric bud cells) and TOPRO-3 (Blue; nuclei). Note that no mCherry protein (Red) is expressed in cells of the ureteric bud (Green). (D,F,H,J,L) Wild-Type E17 kidneys: (D) stained with antibody to p38MAPK (p38MAPK; Green), (F) stained with antibody to phosphorylated p38MAPK (p38MAPK-P; Green), (H) expression of mCherry fluorescent protein (Red), (J) stained with DAPI (nuclei; Blue), (L) overlay of panels F (phosphorylated p38MAPK), H (mCherry), and J (DAPI). Note panels F, H, J, and L are all images taken from the same section of a wild-type E17 kidney. (C,E,G,I,K) Hoxb7-NRAGECherry E17 kidneys: (C) stained with antibody to p38MAPK (E) stained with antibody to phosphorylated p38MAPK (p38MAPK-P), (G) expression of mCherry fluorescent protein, (I) stained with DAPI, (K) overlay of panels E (phosphorylated p38MAPK), G (mCherry), and I (DAPI). Note panels E, G, I, and K are all images taken from the same section of a transgenic Hoxb7-NRAGECherry E17 kidney. Note: (Panels A and B): ureteric bud cells (Green; DBA), mCherry fluorescent protein (Red), Nuclei (Blue; TOPRO-3). (Panels C through L): p38MAPK (Green; Alexa-546), phsophorylated p38MAPK (Green; Alexa-546), mCherry fluorescent protein (Red), Nuclei (Blue; DAPI), Overlay of phosphorylated p38MAPK and mCherry (Red) as Yellow. Scale bar = 50 microns.
Decreased NRAGE expression results in decreased apoptosis and proliferation
NRAGE mediates apoptosis in a p38MAPK dependent manner in primary cortical stem cells (Kendall et al., 2005). We investigated whether the inhibition of NRAGE protein expression leads to alterations in apoptosis and or proliferation in cells of the UB and the MM surrounding the UB. We subjected E11.5 kidney explants from Hoxb7-GFP embryos to three days in culture with NRAGE morpholino or negative control morpholino. TUNEL analysis demonstrated that kidneys treated with NRAGE morpholino displayed a significant decrease (unpaired two tailed t-test, p=0.0045) in the number of TUNEL positive MM cells, averaging 0.014 ± 0.006 TUNEL positive cells per μm3 (N=3), in comparison with kidney explants treated with negative control morpholino, which averaged 0.171 ± 0.027 TUNEL positive cells μm3 (N=3). There was no significant difference in TUNEL staining between the treatment groups in cells of the UB (Figures 5A-B).
Figure 5. NRAGE expression mediates both apoptosis and proliferation in the developing Kidney.
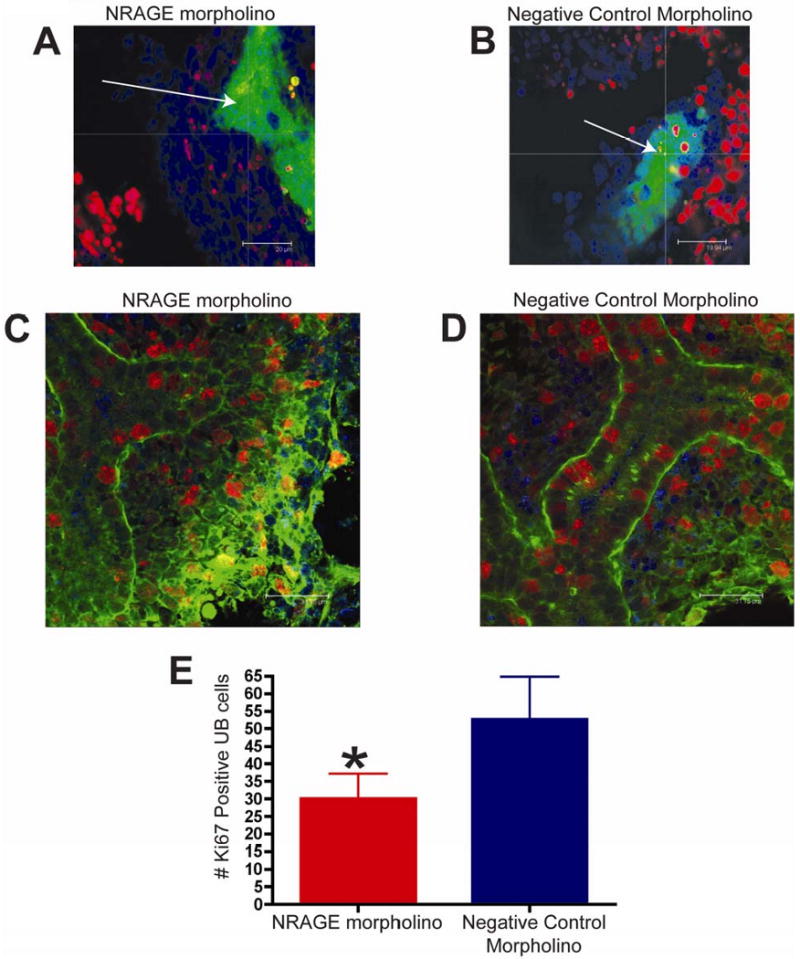
(A-B) TUNEL analysis of E11.5 kidneys from the same Hoxb7-GFP embryo, one cultured with NRAGE morpholino (A) and the other kidney with negative control morpholino (B) for 72 hours (average projection of z-stack of all optical sections collected). Note (A-B): Green: ureteric bud (GFP), Red: TUNEL positive nuclei (TMR), Blue: all nuclei (TOPRO-3). (C-E) Proliferating cells were identified by whole mount Ki67 immunofluorescent staining of E11.5 kidneys, from the same ICR embryo, one cultured with antisense morpholino to NRAGE (C) and the other kidney with negative control morpholino (D) (optical section 2 μm). (E) There were significantly fewer Ki67 positive cells in the ureteric bud (*unpaired two-tailed t-test p=0.0024) in NRAGE morpholino treated explants in comparison to negative control treated explants. Note (C-D): Green: ureteric bud (Dolichos Bifluorous Aggultinin-Alexa488), Red: Ki67 positive nuclei (Ki67-Alexa546), Blue: all nuclei (TOPRO-3). Arrow: ureteric bud branches.
To complement the TUNEL analysis, we utilized an antibody to Ki67 to quantify the number of proliferating UB and MM cells in both NRAGE and negative control morpholino treated kidneys. We counted the number of Ki67 positive cells in the UB of kidneys explants treated with either NRAGE or negative control morpholino. Figure 5C-E illustrates a significant decrease (unpaired two-tailed t-test, p=0.0024) in the number of Ki67 positive UB cells in NRAGE morpholino kidney explants (28 ± 3.6 Ki67 positive UB cells; N=6) versus the negative control treated explants (55 ± 5.6 Ki67 positive UB cells; N=6). Moreover, to ensure that we were counting the cells in equal areas, we determined the number of Ki67 positive cells was, on average, 0.025 ± 0.001 cells per μm3 (N=6) in NRAGE morpholino treated kidneys and 0.050 ± 0.002 Ki67 positive cells per μm3 (N=6) in negative control treated kidneys. Comparing the number of Ki67 positive cells per μm3 in each treatment group, we confirmed that there was a significant decrease in Ki67 positive cells in NRAGE morpholino treated kidneys (unpaired two-tailed t-test, p<0.0001). There was no significant difference in the number of Ki67 positive cells in the MM.
In summary, decreased NRAGE expression lead to decreased apoptosis in cells of the MM but not in cells of the UB. In contrast, diminished NRAGE expression lead to decreased proliferation in cells of the UB but had no impact upon proliferation in cells of the MM. These results suggest that NRAGE may mediate apoptosis and proliferation in both cells of the UB and MM during RBM. However, since there are many pathways by which cells undergo proliferation or apoptosis, we followed up on these results by establishing gene expression profiles for NRAGE morpholino and negative control morpholino, respectively.
Decreased NRAGE expression in the kidney results in altered expression of renal genes during kidney development
A targeted gene expression profile of NRAGE depleted E11.5 ex vivo kidney explant cultures was undertaken to determine the extent of NRAGE's potential involvement in other signaling pathways or developmental processes. Murine E11.5 ICR kidney explants were treated with NRAGE morpholino or with a negative control morpholino for three days. Three days of culture was selected as our end point because, we had previously demonstrated that the maximum inhibition of NRAGE protein expression in kidney explants treated with NRAGE morpholino is at the 72-hour time point (see Figure 2). We isolated and pooled the RNA of six treated kidneys, per treatment, at the 72 hours and performed qPCR to assess the changes in mRNA expression of genes involved in renal development and BMP signaling.
The data collected from kidneys treated with NRAGE morpholino identified a significant change in the expression of only four of the 28 genes examined that are implicated in renal development (see Table1 for a complete list of genes examined). The expression of two sets of ligand and corresponding receptor proteins displayed the highest change in expression: GDNF, Ret, and BMP7 and BMPRIb (Table 2). There was no significant change in gene expression in kidneys compared to those treated with negative control morpholino or treated with Endo-Porter only (data not shown). Western blot analysis confirmed the change in protein levels (Figure 7A), this was important because of GDNF-Ret signaling is critically important to overall kidney development. To determine if the change in gene expression in kidney organ cultures was specific to events surrounding the development of the UB, we repeated the analysis using NRAGE depsleted mIMCD-3 cells cultured with NRAGE morpholino for 72 hours. The qPCR data from mIMCD-3 cells cultured with NRAGE morpholino identified six genes whose expressions were significantly changed: Slit2, ATF2, SMAD5, SMAD7, BMPRIb, BMPR2 (Table 3). Oddly, GDNF and BMP7 did not reach significance in these analyses. This could be explained as a different cellular context in the differentiated mIMCD-3 cells versus whole kidney explants. It should be noted that it has been shown that both GDNF and BMP7 influence the growth and organization of mIMCD-3 cells. To this point, branching experiments were performed using mIMCD-3 cells embedded in a collagen matrix. Both 10ng/ml BMP7 (Figure 6C-D) and 25ng/ml GDNF (Figure 6E-F) had a robust effect on branching in vitro. Consequently, the most parsimonious explanation for RET and GDNF appearing in the kidney explant screen and not the mIMCD-3 screen is that NRAGE interacts with multiple partners in multiple pathways. These interactions are context dependent and depend upon factors such as the developmental time point analyzed, for example early development, as seen in the E11 kidney explants, versus mature differentiated cells, as seen in the mIMCD-3 cells. It also depends on the specific cell type considered, for example UB cells versus MM cells.
Table 1. List of genes analysed in qRT-PCR expression profile of embryonic kidney explants and mIMCD-3 cells in culture.
| TCF7- Transcription factor 7 T-cell specific | Robo2 - Roundabout homolog 2 | Grem1 - Gremlin |
| Lef1 - Lymphoid enhancer binding factor 1 | ATF - Activating transcription factor 2 | FGF7,8 - FGF - Fibroblast growth factor |
| Ctnnb1 – Beta-Catenin | BMP4 - Bone morphogenetic protein 4 | Spry1 - Sprouty homolog 1 (Drosophila) |
| Myc - Myelocytomatosis oncogene | BMP7 - Bone morphogenetic protein 7 | Ngfr - Nerve growth factor receptor (TNFR superfamily) |
| Wnt 4 - Wingless-related MMTV integration site 4 | SMAD1,2,3,5,7 - MAD homolog | |
| Wnt11 Wnt 11 - Wingless-related MMTV integration site 11 | Nfkb1 - Nuclear factor of kappa light chain gene enhancer in B-cells 1 | |
| Pax2 - Paired box gene 2 | Lhx1 - LIM homeobox protein 1 | |
| WT1 - Wilms tumor homolog | BMPR1a - Bone morphogenetic protein receptor, type 1A | |
| GDNF - Glial cell line derived neurotrophic factor | BMPR1b - Bone morphogenetic protein receptor, type IB | |
| Ret - Ret proto-oncogene | BMPR2 – Bone morphogenic protein receptor, type II | |
| Slit2 - Slit homolog 2 | Nog - Noggin |
Table 2. Genes identified as having a significant change in expression after three days of kidney organ culture with NRAGE morpholino.
Genes identified as having significant change in gene expression after three days of kidney organ culture with NRAGE morpholino, normalized to negative control morpholino treated kidneys. The normalized value is 1 and shown is the difference of [(Fold Change Gene X) − 1].
| Genes With Altered Expression | Fold Change In Gene Expression Normalized to Negative Control ([Fold Change GeneX] - 1) - decrease, + increase |
P value, Number in Group |
|---|---|---|
| GDNF | - 0.149 | p<0.001, N=9 |
| Ret | + 0.662 | p=0.0022, N=9 |
| BMP7 | + 0.198 | p=0.0035, N=9 |
| BMPRIb | - 0.159 | p=0.0454 N=9 |
Figure 7. NRAGE modulated both the Ret and BMP signaling in the developing kidney.
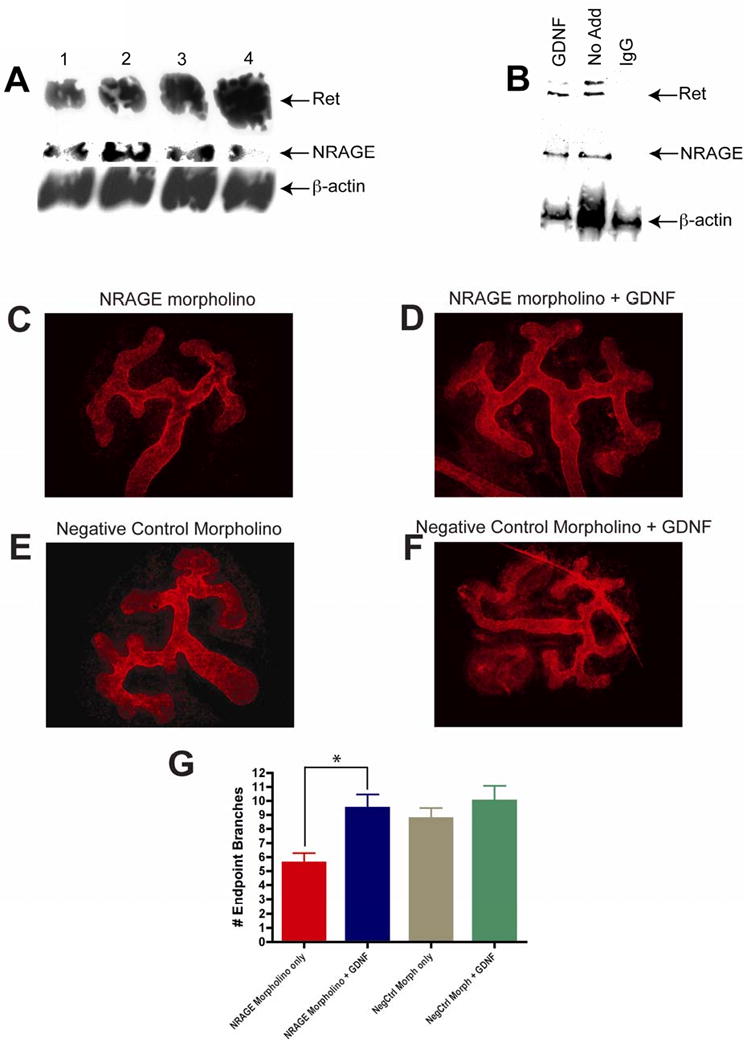
(A) Ret protein expression increases as NRAGE expression decreases. Lysates were generated from three E11.5 kidney explants per well cultured with 10 μM of NRAGE morpholino for three days and analyzed via western blot analysis. Antibodies to Ret, NRAGE, and β-actin were utilized. (B) NRAGE interacts with Ret receptor in mIMCD-3 receptors. Lysates were generated from mIMCD-3 cells treated with and without 25 ng/ml of GDNF for 1 hour. Immunoprecipitation with NRAGE and subsequent western blot analysis demonstrates that NRAGE interacts with Ret with and without GNDF addition. (C-G) Addition of GDNF to NRAGE morpholino treated kidneys stimulates branching of the UB. GDNF was added to NRAGE morpholino (D) or negative control morpholino (F) treated E11.5 ICR kidney explants for three days. Extent of the effect of GDNF addition on morpholino treated kidneys was determined by anti-laminin staining and counting branch end points of each explants. (G) There was a significant increase in the number of branch endpoints in kidney explants treated with NRAGE morpholino and GDNF (*unpaired two-tailed t-test p=0.011). Presented are the means of six explants per treatment.
Table 3. Genes identified as having a significant change in mIMCD-3 cells after 48 hours with NRAGE morpholino treatment.
Genes identified as having significant change in gene expression mIMCD-3 cells cultured with NRAGE morpholino for 48 hours, normalized to negative control morpholino treated kidneys. The normalized value is 1 and shown is the difference of [(Fold Change Gene X) − 1].
| Genes With Altered Expression | Fold Change In Gene Expression Normalized to Negative Control ([Fold Change GeneX] - 1) - decrease, + increase) |
P value, Number in Group |
|---|---|---|
| Slit2 | - 0.034 | p=0.0093, N=9 |
| ATF2 | - 0.039 | p=0.0025, N=9 |
| SMAD5 | - 0.081 | p=0.0024, N=9 |
| SMAD7 | - 0.212 | p=0.0121, N=9 |
| BMPRIb | - 0.109 | p=0.0005, N=9 |
| BMPR2 | - 0.095 | p=0.0028, N=9 |
Figure 6. BMP and GDNF treatment of mIMCD-3 cells embedded in a collagen matrix results in an increase in the number of branches.
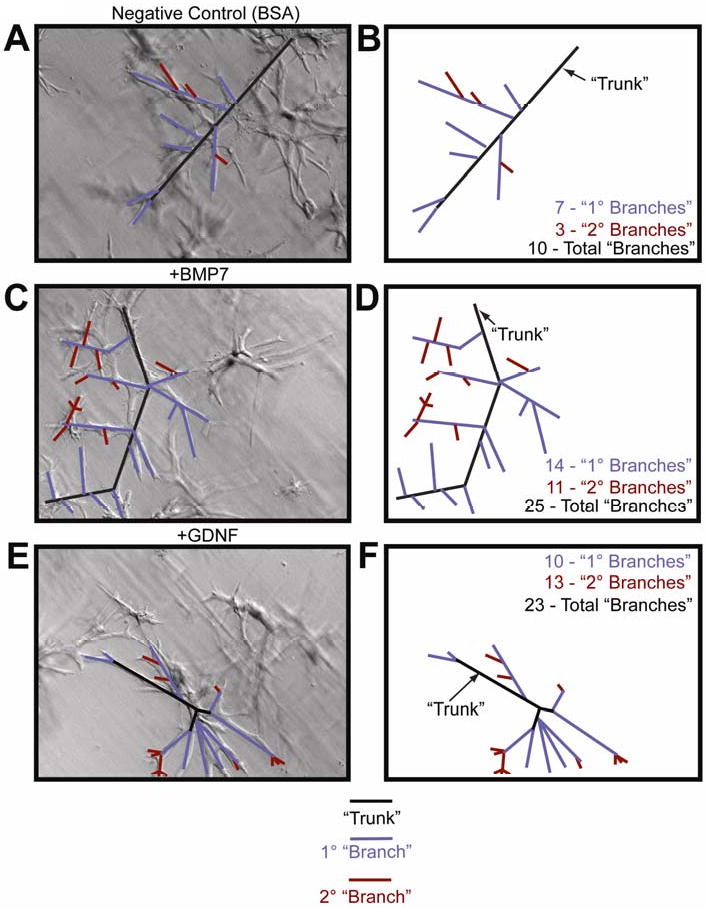
mIMCD-3 cells embedded in a three-dimensional collagen matrix were subjected to treatments with 10ng/ml BMP7 and 25ng/ml GDNF, resulting branches were counted. (A-B) Negative Control (BSA) treated mIMCD-3 cells, (C-D) mIMCD-3 cells treated with 10ng/ml BMP, (E-F) mIMCD-3 cells treated with 25ng/ml GDNF.
Addition of GDNF to NRAGE morpholino treated kidneys stimulates branching of the ureteric bud
GDNF is required for normal development of the kidney (Sanchez et al., 1996). Therefore, a decrease in GDNF transcript levels, as seen in our qPCR gene expression, should result in significantly inhibited branching of the UB. We attempted to stimulate UB branching in kidney explants treated with NRAGE morpholino by the addition of recombinant GDNF. E11.5 ICR kidney explants were cultured with NRAGE morpholino for 72 hours, with 25ng/ml of recombinant GDNF added for the last 48 hours. Branching of the UB was stimulated in kidney explants with decreased NRAGE expression when 25ng/ml GDNF was added to the culture (Figure 7C-G). These results suggest that NRAGE may not only be involved in solely mediating the map kinase cascade downstream of the non-canonical BMP pathway, but could also be involved in the GDNF-Ret signaling pathway. It is unknown if NRAGE impacts solely upon the ligand, GDNF, or the receptor Ret, which are expressed in the cells of the MM and UB, respectively. To address this point, we used lysates from mIMCD-3 cells treated with 25ng/ml of GDNF for 1 hour to co-immunoprecipitate the Ret receptor with NRAGE. As demonstrated in Figure 7B, NRAGE and RET were co-immunoprecipitated. It is not hard to envision a system where NRAGE is employed as a rheostat to bolster Ret signaling to enhance renal growth and depending on the cellular environment to bolster non-canonical BMP signaling to enhance branching.
Discussion
Our focus has been on NRAGE and its role in RBM because the risk of developing hypertension in adult life increases with abnormal RBM. There have been several genes that have been shown to have abnormal RBM phenotypes in knock-out mice for their respective genes, including WT1 (Kreidberg et al., 1993), Pax2 (Torres et al., 1995), Eya-1 (Ding et al., 1999), Six-1 (Xu et al., 1999), Lim-1 (Shawlot and Behringer, 1995), GDNF (Shakya et al., 2005), c-Ret (Schuchardt et al., 1996), Gremlin (Michos et al., 2007) and we anxiously await and believe NRAGE will join this list. Reports have shown that BMP signaling during RBM is dependent upon the non-canonical BMP signaling pathway. Rosenblum's group has demonstrated that p38MAPK phosphorylation is a key event in tubulogenesis of mIMCD-3 cells (Hu et al., 2004), an observation we have replicated and confirmed using kidney explants. Other groups have demonstrated the canonical BMP pathway is not necessary for the normal branching of the UB (Chu et al., 2004) using mice that were lacking SMAD4 expression in their UB. These observations, in conjunction with our findings that NRAGE mediates p38MAPK phosphorylation in neural progenitors (Kendall et al., 2005), suggested a role for NRAGE in RBM as part of the non-canonical BMP pathway.
The data presented herein suggests that NRAGE has a role in RBM, but it is more complex than we originally envisioned, with different roles in the cells of the UB versus cells of the MM. In mIMCD-3 cells, NRAGE associates with members of the non-canonical BMP pathway and the c-Ret receptor. In our kidney explant cultures, NRAGE expression impacts directly upon the expression of GDNF and Ret. Since the BMP and GDNF-Ret signaling pathways are two key signaling pathways in RBM, it is not hard to envision a model in which NRAGE act as a rheostat to influence both pathways. In one situation, NRAGE would be a part of the signaling pathway that provides maximum phosphorylation of p38MAPK to help facilitate UB branching. In another context, NRAGE would to contribute to the GDNF-Ret signaling pathway to assist in growth and maturation of the kidney. In both situations, NRAGE' effects would impact upon UB cells and MM cells (see Figure 8 for proposed model). Further analysis using transgenic and knock-out mice with cell specific control of expression of NRAGE in vivo will provide further clarification as to the role of NRAGE in RBM and its impact upon hypertension in adult life.
Figure 8. A model for NRAGE in kidney development.
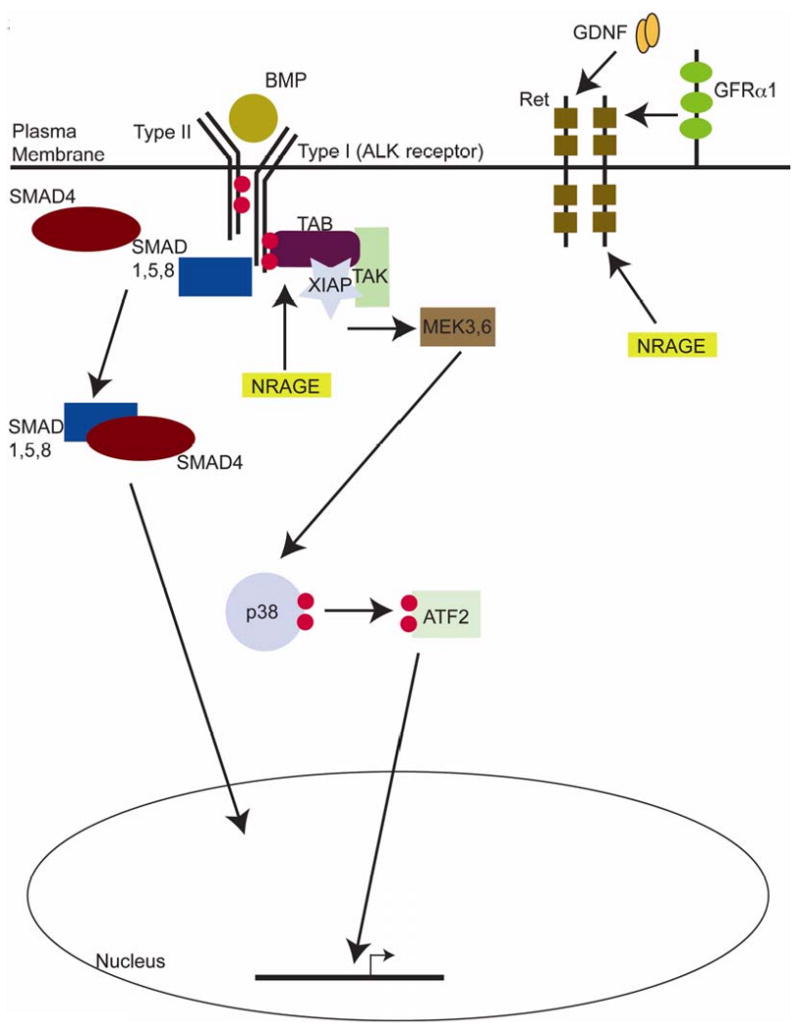
NRAGE acts as a member of the non-canonical BMP pathway, mediating the activation state of p38MAPK. NRAGE also acts through the Ret-GDNF pathway via an association with the Ret receptor in cells of the UB and possibly through regulating GDNF expression in cells of the MM.
Acknowledgments
GNN is supported by a pre-doctoral fellowship from the American Heart Association. JMV is supported by NIH R01 NS055304 and in part by NIH COBRE in Stem and Progenitor Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker DM, Sutherland LE, Jaffe D, Dahl LD. Effects of chronic excess salt ingestion. Juxtaglomerular granulation in kidneys of rats with differing genetic susceptibilities to hypertension. Arch Pathol. 1970;89:247–58. [PubMed] [Google Scholar]
- Bertrand M, Huijbers I, Chomez P, De Backer O. Comparative expression analysis of the MAGED genes during embryogenesis and brain development. Dev Dyn. 2004;230:325–34. doi: 10.1002/dvdy.20026. [DOI] [PubMed] [Google Scholar]
- Brenner BM, Garcia DL, Anderson S. Glomeruli and blood pressure. Less of one, more the other? Am J Hypertens. 1988;1:335–47. doi: 10.1093/ajh/1.4.335. [DOI] [PubMed] [Google Scholar]
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–12. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- Ding B, Huang SL, Zhang SQ, Li YX. Inhibitory effect of MAP kinase antisense oligonucleotide on angiotensin II-induced c-myc gene expression and proliferation of rat cardiac fibroblast. Zhongguo Yao Li Xue Bao. 1999;20:934–40. [PubMed] [Google Scholar]
- Gross I, Morrison DJ, Hyink DP, Georgas K, English MA, Mericskay M, Hosono S, Sassoon D, Wilson PD, Little M, Licht JD. The receptor tyrosine kinase regulator Sprouty1 is a target of the tumor suppressor WT1 and important for kidney development. J Biol Chem. 2003;278:41420–30. doi: 10.1074/jbc.M306425200. [DOI] [PubMed] [Google Scholar]
- Hida M, Omori S, Awazu M. ERK and p38 MAP kinase are required for rat renal development. Kidney Int. 2002;61:1252–62. doi: 10.1046/j.1523-1755.2002.00273.x. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6:432–8. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- Hu MC, Wasserman D, Hartwig S, Rosenblum ND. p38MAPK acts in the BMP7-dependent stimulatory pathway during epithelial cell morphogenesis and is regulated by Smad1. J Biol Chem. 2004;279:12051–9. doi: 10.1074/jbc.M310526200. [DOI] [PubMed] [Google Scholar]
- Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–23. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- Kendall SE, Battelli C, Irwin S, Mitchell JG, Glackin CA, Verdi JM. NRAGE mediates p38 activation and neural progenitor apoptosis via the bone morphogenetic protein signaling cascade. Mol Cell Biol. 2005;25:7711–24. doi: 10.1128/MCB.25.17.7711-7724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–91. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Langley SC, Jackson AA. Increased systolic blood pressure in adult rats induced by fetal exposure to maternal low protein diets. Clin Sci (Lond) 1994;86:217–22. doi: 10.1042/cs0860217. discussion 121. [DOI] [PubMed] [Google Scholar]
- Levitt NS, Lindsay RS, Holmes MC, Seckl JR. Dexamethasone in the last week of pregnancy attenuates hippocampal glucocorticoid receptor gene expression and elevates blood pressure in the adult offspring in the rat. Neuroendocrinology. 1996;64:412–8. doi: 10.1159/000127146. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- Nikopoulos GN, Adams TL, Adams D, Oxburgh L, Prudovsky I, Verdi JM. The use of Endo-Porter to deliver morpholinos in kidney organ culture. Biotechniques. 2008;44:547–549. doi: 10.2144/000112725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 2004;16:291–9. doi: 10.1016/j.cellsig.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Oxburgh L, Chu GC, Michael SK, Robertson EJ. TGFbeta superfamily signals are required for morphogenesis of the kidney mesenchyme progenitor population. Development. 2004;131:4593–605. doi: 10.1242/dev.01324. [DOI] [PubMed] [Google Scholar]
- Piscione TD, Phan T, Rosenblum ND. BMP7 controls collecting tubule cell proliferation and apoptosis via Smad1-dependent and -independent pathways. Am J Physiol Renal Physiol. 2001;280:F19–33. doi: 10.1152/ajprenal.2001.280.1.F19. [DOI] [PubMed] [Google Scholar]
- Piscione TD, Yager TD, Gupta IR, Grinfeld B, Pei Y, Attisano L, Wrana JL, Rosenblum ND. BMP-2 and OP-1 exert direct and opposite effects on renal branching morphogenesis. Am J Physiol. 1997;273:F961–75. doi: 10.1152/ajprenal.1997.273.6.F961. [DOI] [PubMed] [Google Scholar]
- Quaggin SE, Vanden Heuvel GB, Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech Dev. 1998;71:37–48. doi: 10.1016/s0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- Renn J, Winkler C. Osterix-mCherry transgenic medaka for in vivo imaging of bone formation. Dev Dyn. 2009;238:241–8. doi: 10.1002/dvdy.21836. [DOI] [PubMed] [Google Scholar]
- Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL, Verdi JM, Barker PA. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 2000;27:279–88. doi: 10.1016/s0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–3. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- Schuchardt A, D'Agati V, Pachnis V, Costantini F. Renal agenesis and hypodysplasia in ret-k- mutant mice result from defects in ureteric bud development. Development. 1996;122:1919–29. doi: 10.1242/dev.122.6.1919. [DOI] [PubMed] [Google Scholar]
- Shakya R, Watanabe T, Costantini F. The Role of GDNF/Ret Signaling in Ureteric Bud Cell Fate and Branching Morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–30. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009 doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Goldberg MR, Watanabe T, D'Agati V, al-Awqati Q, Costantini F. Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev Genet. 1999;24:241–51. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<241::AID-DVG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–65. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Woodall SM, Johnston BM, Breier BH, Gluckman PD. Chronic maternal undernutrition in the rat leads to delayed postnatal growth and elevated blood pressure of offspring. Pediatr Res. 1996;40:438–43. doi: 10.1203/00006450-199609000-00012. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–7. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]


