Abstract
Cells undergoing developmental processes are characterized by persistent non-genetic alterations in chromatin, termed epigenetic changes, represented by distinct patterns of DNA methylation and histone post-translational modifications. Sirtuins, a group of conserved NAD+-dependent deacetylases or ADP-ribosylases, promote longevity in diverse organisms; however, their molecular mechanisms in aging regulation remain poorly understood. Yeast Sir2, the founding member of the family, establishes and maintains chromatin silencing by removing H4 lysine 16 acetylation and bringing in other silencing proteins. Here we show an age-associated decrease in Sir2 protein abundance accompanied by an increase in H4 lysine 16 acetylation and loss of histones at specific subtelomeric regions in replicatively old yeast cells, which results in compromised transcriptional silencing at these loci. Antagonizing activities of Sir2 and Sas2, a histone acetyltransferase, regulate the replicative lifespan through histone H4 lysine 16 at subtelomeric regions. This pathway, distinct from existing aging models for yeast, may represent an evolutionarily conserved function of Sirtuins in regulation of replicative aging by maintenance of intact telomeric chromatin.
Cells undergo characteristic molecular alterations as organisms age1,2. Studies in model organisms identify conserved genetic pathways that modulate aging, such as insulin signaling, oxidative stress tolerance, and nutrient sensing3,4. Aging and other developmental processes, such as differentiation, apoptosis and gametogenesis, associate with characteristic epigenetic changes at the cellular level, including DNA methylation and histone post-translational modifications5-7. Nevertheless, the functions of these molecular changes during aging remain elusive. Although Sirtuins promote longevity in yeast, worms and flies8, conserved pathways for sirtuins in aging regulation remain controversial. Yeast S. cerevisiae Sir2 (Silencing Information Regulator 2), the founding member of the family, establishes and maintains silencing within yeast heterochromatic-like regions at telomeres, rDNA, and silenced mating type loci (HM) by removing H4 lysine 16 acetylation (H4K16ac) and bringing in other silencing proteins9. Antagonizing activities of Sir2 and a histone acetyltransferase, Sas2, generate a gradient of H4K16ac marking the boundary of silencing chromatin near telomeres10,11.
One cause of yeast aging is nucleolar accumulation of extrachromosomal rDNA circles (ERCs), generated from recombination between rDNA repeats as cells divide12. Deletion of SIR2, results in hyper-recombination within the rDNA and elevated levels of ERCs13; whereas FOB1 deletion reduces rDNA recombination and ERC formation, and extends lifespan14,15. Overexpression of Sir2 also increases lifespan, but does not further increase the lifespan when combined with deletion of FOB1, which has been interpreted to suggest that Sir2-overexpression promotes longevity, in part, by repressing ERCs13. Interestingly, Sir2 overexpression also increases lifespan in worms and flies8; however, ERCs have not been detected in these organisms or in mammalian cells in association with aging, and thus is an unlikely mechanism to underlie aging in diverse organisms16,17.
A central question is whether histone acetylation levels are altered during aging and whether such changes contribute directly to aging. Although histones are nearly the only acetylated proteins reported in yeast18, there are numerous acetylated proteins in metazoans19; in addition, deacetylases, including the Sirtuin class, target both histones and non-histone proteins20. In mammalian cells, Sirt1, 2, 3, and 6 deacetylate histones21-23, but have non-histone substrates, and some of these are promising candidates in aging pathways. For example, p53 promotes apoptosis and Sirt1 deacetylates p53 and extends cell survival23. Hence, despite extensive knowledge about the Sirtuin class of deacetylases, it remains to be established whether deacetylation of H4K16 plays an important role in the Sir2 aging pathway.
In this study we investigated age-associated changes in histone modifications and chromatin. In particular, because ERC accumulation in the nucleolus is not conserved through evolution, we explored chromatin-related changes at Sir2-regulated loci outside of the nucleoli. Our results establish chromatin as a critical target for Sir2 in regulation of lifespan and indicate that Sir2 opposes replicative aging in yeast through histone targets located near telomeres.
Histone modifications change in old cells
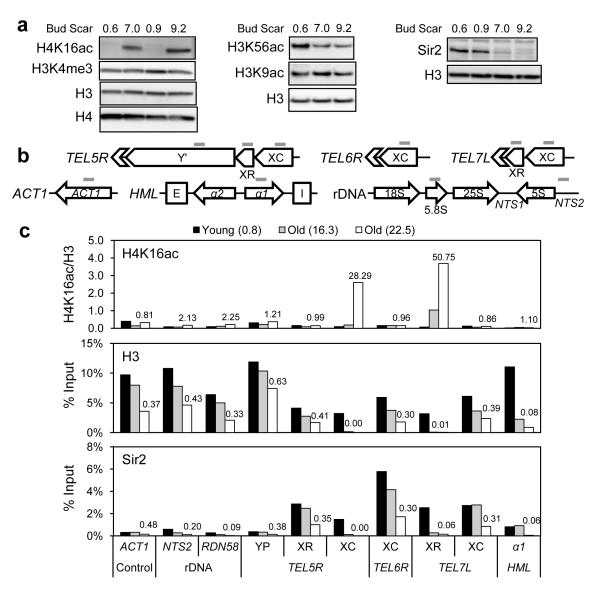
We examined histone post-translational modifications in young and old cells, via isolation of biotin-labeled mother cells24. Progressive aging was evident with increasing bud scars and rDNA copy numbers12 (Supplementary Fig. 1ab). Histone modifications associated with gene transcription, DNA repair, and chromosome condensation were tested by western blotting. Notably, the level of H4K16ac increased with age (Fig 1a left), whereas H3K56ac decreased (Fig. 1a middle). No significant changes were found for other modifications and histone variants tested (Fig. 1a and Supplementary Fig. 2). Sir2 deacetylates H4K16 and H3K569,25, and is linked to aging26; the level of Sir2 protein in age-sorted cells declined opposite the increase in H4K16ac (Fig. 1a right). SIR2 RNA levels remained unchanged in old cells (Supplementary Fig. 1c), as found previously27, indicating that age-dependent changes in Sir2 occur post-transcriptionally.
Figure 1. Chromatin and Sir2 levels change in old cells.
(a) Western analysis for extracts from young and old yeast (strain W1588-4C) with specific antibodies. Consistent results were also observed with strain BY4741 (data not shown). (b) Positions of qPCR primers used in ChIP. (c) ChIP analysis for young and old cells (average bud scar counts in parentheses). Chromatin immunoprecipitated with antibodies against H4K16ac (upper), H3 (middle), or Sir2 (lower) was analyzed by qPCR with primers shown in (b). Fold changes for the oldest sample (white bars) over the young cells (black bars) are indicated above the bars.
Chromatin changes localize to silenced regions
To investigate the genomic location of increasing H4K16ac and decreasing Sir2 in old cells, we performed chromatin immunoprecipitation (ChIP) focusing on Sir2-regulated regions, including rDNA, telomeres, and HM loci (Fig. 1b). These regions, similar to heterochromatin in higher eukaryotes, have low levels of histone acetylation and minimal gene activity28. Compared to young cells, we observed significantly higher levels of H4K16ac in old cells at X core (XC) and X repeat (XR) elements within telomeres TEL5R and TEL7L, but not at other Sir2-regulated sites or control sites (Fig. 1c upper). Interestingly, these hyperacetylated K16 sites co-localized with profoundly reduced histone levels in old cells, while histone levels were decreased to the same, moderate extent at all other sites tested (Fig. 1c middle). Consistent with the western analysis, Sir2 levels decreased at all Sir2-regulated sites in old cells (Fig. 1c lower). We examined six additional telomeres and found that across nine total telomeres, the greatest age-associated increase in H4K16ac was within the X elements, correlating with the strongest decrease in histone levels (Supplementary Table. 1). Since Sir2 was equally lost from all its binding sites, these results suggest that the chromatin landscape was mostly affected at X elements of telomeres as a result of loss of Sir2 in old cells.
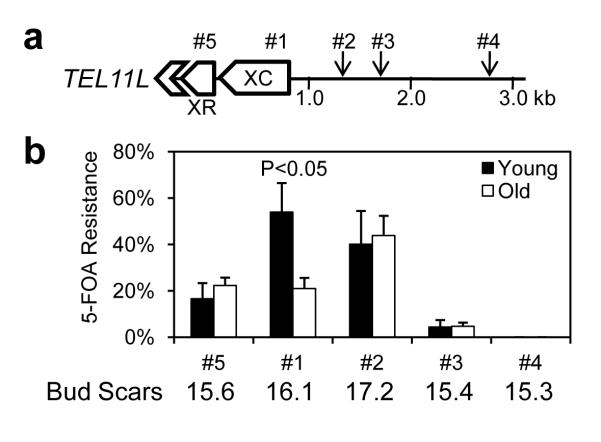
To determine whether the observed age-associated chromatin changes lead to altered gene expression, we assayed the silencing state of a URA3 reporter inserted at various positions near telomere TEL11L29 (Fig. 2a). Silencing of URA3 was measured in young and old cells, by quantifying survival on media containing 5-fluoroorotic acid (5-FOA), which is converted into the toxic 5-fluorouracil by Ura3. Silencing at the XC element, but not elsewhere near the telomere, was significantly reduced in old cells (Fig. 2b), and sensitivity to 5-FOA was not seen in similarly aged ura3Δ cells (Supplementary Fig. 4). These data demonstrate that the increase in H4K16ac, decrease in histone abundance, and reduced Sir2 in aging cells are associated with transcriptional derepression at specific loci near telomeres.
Figure 2. The X core element of telomeres shows silencing defects in old cells.
(a) Schematic showing the positions (#1 to #5) of URA3 gene insertions near telomere TEL11L29. (b) Silencing assay for young and old cells of strains bearing URA3 gene insertions as shown in (a). The extent of silencing is expressed as the fraction of cells resistant to 5-FOA (n=4, error bars showing standard deviations). Average bud scar counts are listed for old cell samples.
Since aging leads to decreased Sir2, we predicted that, in young cells, loss or inhibition of Sir2 might increase H4K16ac and reduce histones at telomeric X elements. As seen previously30, H4K16ac increased at Sir2-regulated sites in sir2Δ cells (Supplementary Fig. 5a); as seen in old wild-type cells, young sir2Δ cells showed decreased histones at X elements in telomeres, but not at rDNA, HML or other control sites (Supplementary Fig. 5b). Similar results were observed for cells treated with nicotinamide (Supplementary Fig. 5cde), a noncompetitive inhibitor of Sir231.
Sas2 has opposing effects on H4K16ac and lifespan compared to Sir2
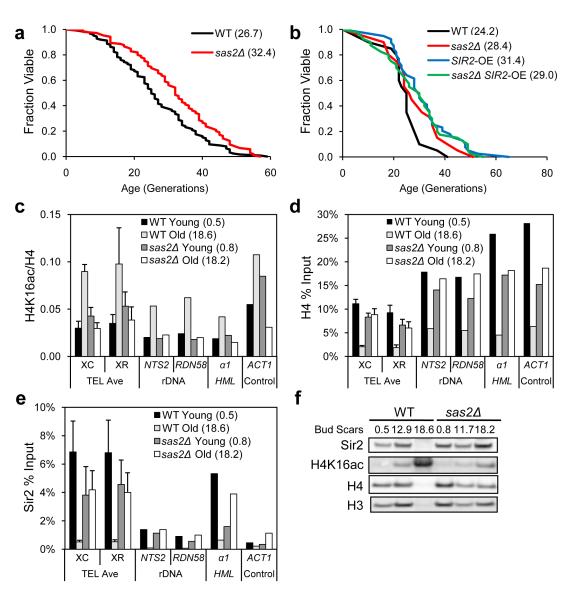
Sas2 is the major H4K16 acetyltransferase to establish boundaries between telomeres and euchromatin10,11; hence, we hypothesized that deletion of SAS2 might extend lifespan. Indeed, the mean lifespan of sas2Δ was greater than wild-type (Fig. 3a). Over-expression of SIR2 (SIR2-OE) by integrating a second copy also extends lifespan13. If this extension is due to enhanced deacetylation of H4K16, then SIR2-OE should not further extend the lifespan of sas2Δ cells. Consistent with this prediction, we found that sas2Δ SIR2-OE cells were not longer-lived than either SIR2-OE or sas2Δ cells (Fig. 3b).
Figure 3. Sas2 antagonizes Sir2 in regulating lifespan and H4K16 acetylation.
(a-b) Replicative lifespan analysis for strains WT, sas2Δ (a) and strains WT, sas2Δ, SIR2-OE, and sas2Δ SIR2-OE (b) with MLS in parenthesis. Lifespan p-values are listed in Supplementary Table 5. (c-e) Old cells were isolated for wild-type and sas2Δ strains (average bud scar counts in parenthesis). Chromatin immunoprecipitated with antibodies specific to H4K16 acetylation (c), H4 (d), or Sir2 (e) was analyzed by qPCR with primers used in Supplementary Table 1. Data for XC positions and XR positions were averaged and standard deviations were shown as error bars. (f) Cellular extracts as in (c-e) were analyzed by western blotting, probed with specific antibodies.
Upon loss of Sir2, Sas2 spreads outward to telomeres to acetylate H4K16, resulting in disruption of telomere silencing10. Since deletion of SAS2 increases lifespan and decreases H4K16ac at subtelomeric regions32, we speculated that Sas2 modulates chromatin changes in old cells. Indeed, relative to wild-type, sas2Δ cells did not show the profound age-associated increase in H4K16ac, decrease in histones, or loss of Sir2 (Fig. 3cde). Western analysis confirmed that deletion of SAS2 reduced overall abundance of H4K16ac and stabilized histones and Sir2 in aged cells (Fig. 3f). Taken together, these data suggest that Sir2 and Sas2 antagonistically modulate lifespan in part through regulating H4K16ac and histone levels at telomeres, and further suggest that Sas2 promotes degradation of Sir2 in old cells through unknown mechanisms.
H4K16 and H3K56 mutations result in shortening of lifespan
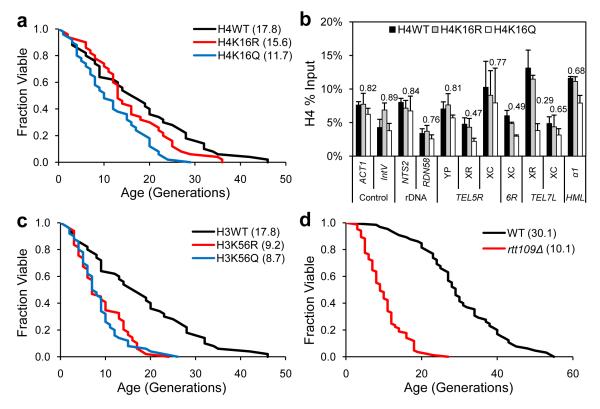
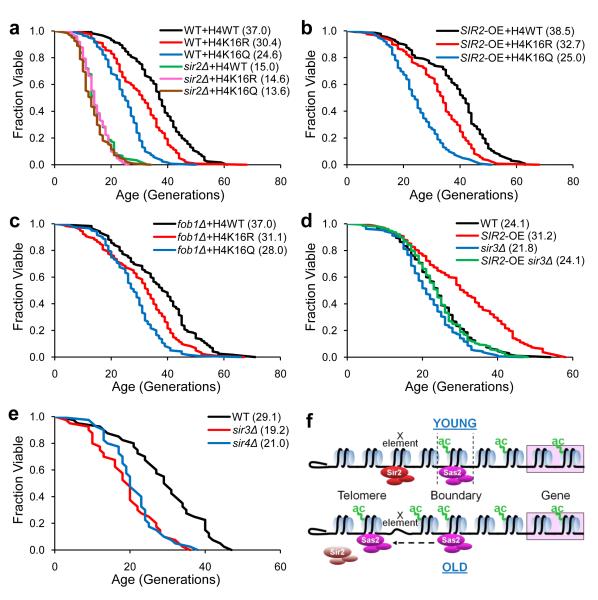
Our observations implicate H4K16ac as a key substrate of Sir2 in opposing replicative aging. To investigate the role of H4K16 in aging more directly, we created strains bearing substitutions of H4K16 to arginine (K16R) or glutamine (K16Q), mimicking the unacetylated or acetylated state. We found that K16Q significantly reduced lifespan, while K16R showed only a marginal effect (Fig. 4a). Neither substitution affected growth (Supplementary Fig. 10), and similar results were observed in a long-lived strain background carrying integrated histone mutations (Figure 5a). It is notable that K16R did not extend lifespan. However, an arginine substitution may not perfectly mimic unacetylated lysine, and global K16R substitution may not have the opposite effect of global K16Q mutation, because H4K16ac could have additional roles in regulating euchromatic genes important for normal lifespan33. Sir2 abundance was unchanged in the mutant strains (Supplementary Fig. 6), indicating that the reduced lifespan is not due to loss of Sir2. Interestingly, we found lowered histone (Fig. 4b) and Sir2 (Supplementary Fig. 7) content at telomeres only in the H4K16Q strain, consistent with H4K16 hyper-acetylation directly causing histone loss at telomeres.
Figure 4. Histone H4K16 and H3K56 mutations affect replicative lifespan through distinct mechanisms.
(a-b) Replicative lifespan analysis (MLS in parenthesis) (a) and ChIP analysis with histone H4 antibody (b) for cells bearing WT histone H4, H4K16R, or H4K16Q plasmids. Fold changes for H4K16Q compared to H4WT in ChIP analysis are indicated above the bars. Error bars show standard deviations (n=3). (c-d) Replicative lifespan analysis for strains carrying WT histone H3, H3K56R, or H3K56Q plasmids (c) and strains WT, rtt109Δ (d) with MLS in parenthesis. Lifespan p-values are listed in Supplementary Table 5.
Figure 5. H4K16 is involved in a Sir2-regulated aging pathway associated with telomere chromatin.
(a-c) Replicative lifespan analysis performed in parallel for strains bearing integrated WT histone H4, H4K16R, or H4K16Q in WT and sir2Δ backgrounds (a), in SIR2-OE background (b), and in fob1Δ background (c) with MLS in parenthesis. (d-e) Replicative lifespan analysis for strains WT, SIR2-OE, sir3Δ, and SIR2-OE sir3Δ (d), and for strains WT, sir3Δ, and sir4Δ (e) with MLS in parenthesis. Lifespan p-values are listed in Supplementary Table 5. (f) Schematic model for telomere chromatin changes in yeast old mother cells.
H3K56ac, regulated by two other Sirtuins, Hst3 and Hst434 and important for genome integrity and replication34-36, was found decreased in old cells (Fig. 1a). In contrast to H4K16, both H3K56R and H3K56Q exhibited similarly reduced lifespans (Fig. 4c). H3K9 substitutions had no effect on lifespan (Supplementary Fig. 8). Deletion of RTT109, the key acetyltransferase for H3K5637, also reduced lifespan (Fig. 4d). Finally, as shown previously38, hst3Δ, hst4Δ, or hst3Δ hst4Δ cells had a significantly decreased lifespan (Supplementary Fig. 9). H3K56R substitution or rtt109Δ causes sensitivity to DNA damage35,36, suggesting that acetylation of K56 is required for genome integrity. Since K56Q, hst3Δ, and hst3Δ hst4Δ, showed similar shortening of lifespan compared to K56R and rtt109Δ, we reasoned that cycling of acetylation/deacetylation of K56 might be important for genome integrity. Indeed, K56Q, but not H4K16 or H3K9, was equally sensitive to MMS or H2O2 as K56R (Supplementary Fig. 10). We conclude that acetylation of H4K16 and H3K56 both influence longevity, but via distinct mechanisms.
Shortening of lifespan by H4K16 mutants is epistatic with Sir2
Deletion of the WRN homolog SGS1 shortens lifespan12,13. However, in contrast to sir2Δ, sgs1Δ did not affect H4K16ac or histone levels (Supplementary Fig. 11). To investigate the genetic pathways of H4K16ac, we compared the lifespan of H4K16 substitutions in sir2Δ and sgs1Δ backgrounds. H4K16 mutations did not further shorten the lifespan of sir2Δ cells (Fig. 5a), indicating that K16 functions in the same pathway as Sir2. In contrast, the K16 substitutions significantly shorted the life span of sgs1Δ cells (Supplementary Fig. 12), suggesting that K16 and Sir2 act in a different pathway than Sgs1. We also examined whether H4K16 is critical for lifespan extension by SIR2 overexpression, and found that K16Q in the SIR2-OE background restricted lifespan similar to K16Q in wild-type (compare Fig. 5b to a). Thus, hypoacetylated H4K16 is required for the lifespan extension by SIR2 overexpression. Overall, these data support the model that Sir2 deacetylates K16 to antagonize aging.
H4K16 functions in a pathway distinct from ERC formation
Our data show age-related Sir2-linked chromatin changes at telomeres, but previous findings have indicated that Sir2 promotes replicative longevity by functioning at the rDNA to inhibit the formation of extrachromosomal rDNA circles (ERCs), which accumulate with age in mother cells and cause senescence13. Fob1 acts antagonistically to Sir2 by promoting rDNA recombination, and deletion of FOB1 reduces ERCs and extends lifespan15. We reasoned that, if K16Q shortens lifespan through effects exclusively at rDNA, then K16Q would not shorten lifespan in a fob1Δ background. However, K16Q reduced the lifespan of fob1Δ cells, and this reduction was greater than the effect of K16R combined with fob1Δ (Fig. 5c). This result is similar to the shorter lifespan of sir2Δ fob1Δ compared to fob1Δ alone13, suggesting that K16Q influences lifespan by a mechanism different from Fob1 and ERCs.
Sir2 forms a complex with Sir3 and Sir4 at telomeres and HM loci, and a distinct complex, RENT, that functions at rDNA39. We found that the lifespan extension by SIR2-OE is suppressed by deletion of SIR3 (Fig. 5d) and that deletion of either SIR3 or SIR4 similarly shortens lifespan (Fig. 5e). Taken together, these data support a model that the Sir2/3/4 complex functions to modulate aging by maintaining telomeric chromatin, and that H4K16ac is a key substrate of Sir2 during replicative aging. Further, our data suggest that the age-associated chromatin alterations at telomeres and other silenced loci results from decreased abundance of Sir2 protein during aging, which can be counteracted by overexpression of Sir2.
Discussion
S. cerevisiae provides a previously unexploited model for analysis of chromatin during aging. Our study identifies a novel role for H4K16 acetylation and telomere chromatin state in determining longevity, mediated by Sas2 acetylation and Sir2 deacetylation. We show an age-associated decrease in Sir2 protein abundance accompanied by an increase in H4K16ac and loss of histones at specific subtelomeric regions in replicatively old cells (Fig. 5f), which results in compromised transcriptional silencing at these loci. Deletion of Sas2, stabilizes Sir2 levels in old cells and extends lifespan. Finally, we demonstrate that mutations to H4K16 negatively affect lifespan, downstream of Sir2, in a pathway at least partially distinct from the accumulation of ERCs in old cells.
Indeed, although extensive evidence supports the idea that replicative lifespan is regulated by Sir2 function at rDNA, a Sir2-related aging mechanism distinct from ERCs has been suggested26,40. Our data indicate that Sir2 also promotes replicative lifespan through at least one additional pathway, apparently localized at telomeres, as depicted in Fig. 4h. In this pathway, Sir2 in young cells maintains low H4K16 acetylation at telomeres and subtelomeric regions and works with Sas2 to establish a silencing boundary10,11. In old cells, due to loss of Sir2 and action of Sas2, H4K16ac rises leading to loss of histones at X elements of telomeres. A role for Sir2 in counteracting H4K16ac specifically in a telomere-mediated aging pathway is potentially evolutionarily conserved, since telomeric changes have a major role in aging and cancer and correlate with chromatin alterations41. However, our data do not rule out non-telomeric location(s) that could be regulated through H4K16ac by Sas2 and deacteylation by the SIR complex.
There are seven mammalian Sirtuins and three (Sirt1, Sirt2 and Sirt3) deacetylate H4K1623. It is possible that reduction of activity or level of these Sirtuins promotes cellular aging through increased H4K16ac. Interestingly, Sirt6 regulates lifespan through deacetylation of H3K9ac22,42, and Sirt6 knock-out mice show genomic instability, including telomeric fusions, and an aging-like phenotype43. Hence, regulation of histone acetylation appears to be a critical function in aging pathways throughout eukaryotes.
Methods Summary
Yeast old mother cells were isolated by 2-4 rounds of sorting; cells were labeled with biotin, cultured overnight, followed by affinity purification24. Mean ages of isolated cells were estimated by counting Calcofluor-stained bud scars. Whole cell extracts of young and old cells were analyzed by western blotting with specific antibodies. Specificity of key antibodies (H4K16ac, H3K56ac, H3K9ac, H3K4me3, and Sir2) used in this study was verified by western blotting with extracts from cells bearing corresponding histone point mutations or gene deletions. Total DNA and mRNA extractions followed by real-time PCR were used to quantify rDNA copy numbers and expression levels of SIR2. Formaldehyde-crosslinked young and old cells were used for chromatin immunoprecipitation (ChIP) with specific antibodies. Immunoprecipitated DNA was quantified by real-time PCR with primer sets targeting specific regions. Telomere silencing assays were performed with strains and methods described elsewhere29. Replicative lifespans of yeast strains were determined as described13. Statistical assessment of lifespan differences was determined by Wilcoxon Rank Sum Test. Yeast strains bearing histone point mutations were generated by deletion of both copies of histone H3 and H4 gene cassettes supplemented by a wild type or mutant copy of H3 and H4 either on a CEN-controlled yeast plasmid, or integrating a copy to its original gene location. Strains, antibodies, and real-time PCR oligos used in this study are listed in Supplementary Tables 2, 3, and 4, respectively.
Methods
Plasmids and Strains
Plasmids pWD23, pWD25, pWD35, pWD36, pWD43, and pWD45 bearing point mutations of H4K16R, H4K16Q, H3K9R, H3K9Q, H3K56R, and H3K56Q, respectively, were generated by QuikChange site-directed mutagenesis (Stratagene) from plasmid pRM204 containing a wild-type copy of histone H3 and H4 genes (HHT2-HHF2) and verified by sequencing. Yeast strains used in this study are listed in Supplementary Table 2. Strains carrying a mutant histone plasmid as the sole source of histone H3 and H4 were made by shuffling the mutant plasmid into strain FY1716, followed by selection on synthetic complete media containing 1 mg/ml 5-FOA to remove the existing wild-type histone plasmid pDM1 from FY1716. Strains bearing an integrated histone mutation were constructed by transforming PCR products containing the mutation into strain FY1716 and selecting on 5-FOA containing media. All histone mutant strains were verified by sequencing.
Isolation of yeast old mother cells
Old mother cells were isolated from exponentially growing cultures in YPD (1% yeast extract, 2% bacto peptone, 2% dextrose) by surface labeling with NHS-LC-Biotin (Thermo Fisher) and affinity purification as previously described44, except that up to 4.8×109 cells were used for labeling for up to four rounds of sorting and Dynabeads Biotin Binder (Invitrogen) was used. Exponential growth before each round of sorting was limited to 6-8 doublings. About 4×108 old mother cells were saved for analysis after each round of sorting. Mean ages of isolated cells (n=50 each) were estimated by counting the number of bud scars, stained with Calcofluor (Sigma) and visualized by fluorescence microscopy.
Preparation of whole cell extracts and western blotting
Whole cell extracts were prepared as described7. Proteins were resolved by SDS-PAGE and transferred to PVDF. Blots were probed with antibodies specific for histones H3, H4, various histone modifications, and Sir2, as listed in Supplementary Table 3.
Quantification of rDNA Copy Number and mRNA Expression Levels
Total DNA was purified typically from 100 μg whole cell extracts of young or old cells with QIAGEN PCR purification kit following sequential 10 μg RNase A treatment and 20 μg Protease K treatment at 37 °C for 1 hour each. Levels of rDNA were quantified by real-time PCR with primers specific to the 5.8S rDNA gene RDN58 and non-transcribed region NTS2, normalized to ACT1 levels. Total mRNA and cDNA were prepared with QIAGEN RNeasy kit and Applied Biosystems reverse transcription kit, and quantified by real-time PCR. Real-time PCR primers are listed in Supplementary Table 4.
Chromatin Immunoprecipitation
Young or old cell samples were crosslinked with 1% formaldehyde at room temperature for 10 minutes immediately after cell sorting or harvesting. Chromatin Immunoprecipitation (ChIP) was performed as described45 with specific antibodies and quantified by real-time PCR. The specificity of all antibodies used in ChIP (listed in Supplementary Table 3) was verified with lysates prepared from strains carrying corresponding histone substitution mutations or gene deletions. Real-time PCR primers are listed in Supplementary Table 4.
Telomere Silencing
Young and old cells were isolated as described above except that the scale was reduced by 20 fold. About 1×107 young and old cells were diluted to 200 μl, followed by 10-fold serial dilutions. Then 10 μl of each dilution was spotted on synthetic complete media (SC) and SC containing 1 mg/ml 5-FOA. Fractions of cells giving rise to 5-FOA resistant colonies are shown.
Yeast Lifespan Determination
Replicative lifespans of yeast strains were determined by micromanipulation as described13. Statistical assessment of lifespan differences was determined by Wilcoxon Rank Sum Test.
Nicotinamide Treatment
Yeast cells were grown at 30 °C overnight in YPD, then transferred to fresh media YPD or YPD containing 5 mM nicotinamide, and allowed to continue growing for 4-5 hours (2 doublings of exponential growth).
Supplementary Material
Acknowledgments
We thank E. Louis for providing strains for the telomere silencing assay, and members of the Kennedy and Kaeberlein labs who participated in lifespan determination studies. This work was funded by NIH grants (S.L.B. and B.K.K.), and an AFAR Julie Martin Mid-Career Award in Aging Research (B.K.K.). M.K. is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Herbig U, et al. Cellular senescence in aging primates. Science (New York, N.Y. 2006;311(5765):1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 3.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annual review of biochemistry. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 4.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408(6809):255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 5.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23(8):413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nature reviews. 2007;8(9):692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamoorthy T, et al. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes & development. 2006;20(18):2580–2592. doi: 10.1101/gad.1457006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126(2):257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Moazed D. Enzymatic activities of Sir2 and chromatin silencing. Current opinion in cell biology. 2001;13(2):232–238. doi: 10.1016/s0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 10.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nature genetics. 2002;32(3):370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 11.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nature genetics. 2002;32(3):378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johzuka K, Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7(2):99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 15.Defossez PA, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Molecular cell. 1999;3(4):447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 16.Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell. 1999;96(2):291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- 17.Laun P, et al. Yeast mother cell-specific ageing, genetic (in)stability, and the somatic mutation theory of ageing. Nucleic acids research. 2007;35(22):7514–7526. doi: 10.1093/nar/gkm919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin YY, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136(6):1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? The EMBO journal. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26(37):5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 21.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes & development. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 22.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26(37):5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 24.Smeal T, et al. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84(4):633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu F, et al. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Molecular cell. 2007;27(6):890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing research reviews. 2007;6(2):128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Lesur I, Campbell JL. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Molecular biology of the cell. 2004;15(3):1297–1312. doi: 10.1091/mbc.E03-10-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annual review of biochemistry. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 29.Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. The EMBO journal. 1999;18(9):2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robyr D, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109(4):437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 31.Grubisha O, Smith BC, Denu JM. Small molecule regulation of Sir2 protein deacetylases. The FEBS journal. 2005;272(18):4607–4616. doi: 10.1111/j.1742-4658.2005.04862.x. [DOI] [PubMed] [Google Scholar]
- 32.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes & development. 2006;20(18):2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(15):5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celic I, et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16(13):1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science (New York, N.Y. 2007;315(5812):649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han J, et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science (New York, N.Y. 2007;315(5812):653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 37.Schneider J, et al. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. The Journal of biological chemistry. 2006;281(49):37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 38.Tsuchiya M, et al. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging cell. 2006;5(6):505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 39.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes & development. 2003;17(17):2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science (New York, N.Y. 2003;299(5613):1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 41.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8(4):299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 42.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 44.Park PU, McVey M, Guarente L. Separation of mother and daughter cells. Methods in enzymology. 2002;351:468–477. doi: 10.1016/s0076-6879(02)51865-6. [DOI] [PubMed] [Google Scholar]
- 45.Wyce A, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Molecular cell. 2007;27(2):275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.