Abstract
We used the whole genome approach to identify major functional categories of genes whose expression depends on gestational age. Using microarray analysis, we compared gene expression profiles in the villous tissues of 1st (45-59 days) and 2nd trimester (109-115 days) placentae versus C-section term placentae. We found that in 1st trimester placentae, genes related to cell cycle, DNA, amino acids, and carbohydrate metabolism were significantly overrepresented, while genes related to signal transduction were underrepresented. Among genes involved in organism defense, we identified genes involved in chemical response, metabolism, and transport. Analysis of signal transduction pathways suggested, and subsequently confirmed independently, that the Wnt pathway was changed with gestational age leading to inhibition of β-catenin protein expression. Our study will serve as a reference database to gain insight into the regulation of gene expression in the developing placentae and to compare with gene expression in placentae from complicated pregnancies.
Keywords: Placenta, gestational age, microarray, Wnt pathway, xenobiotic transporters, metabolizing enzymes
Introduction
The placenta plays a critical role in the development of the fetus 1, 2. Among its many functions, the placenta delivers nutrients and oxygen to the fetus, as well as removes waste products from the fetal compartment. The placenta is also a source of hormones and growth factors necessary for a successful pregnancy. The placenta protects the fetus by acting as a physical, biochemical, and immunological barrier to xenobiotics and microorganisms. For example, it is capable of reducing the amount of xenobiotics reaching the fetal circulation through metabolism and fetal-maternal transport of the xenobiotics 3. Nevertheless, there are numerous examples of xenobiotics that can cross the placenta and are teratogenic, especially when administered early in gestation during organogenesis when the fetus is most vulnerable to xenobiotic toxicity 4. In addition, defects in placenta development can lead to preeclampsia, intrauterine growth restriction, and other pathological conditions that may cause fetal morbidity or mortality 1. However, the molecular mechanisms responsible for these pathological conditions and xenobiotic teratogenesis are poorly understood. Therefore, the goal of our study was to identify changes in gene expression patterns during normal development of the human placenta (first trimester, second trimester and term) using a microarray approach. We reasoned that identification of pathways regulated by factors changing with gestational age, could be an important step towards understanding placental function, as well as for future comparison with gene expression in placentae from complicated pregnancies such as those in women experiencing gestational diabetes, preeclampsia and teratogenic sequelae.
Methods
Specimen Collection
Collection of human placentae was approved by the Institutional Review Board of the University of Washington. Subjects were excluded if they had any chronic disease, were on chronic medication, were smokers, abused alcohol or drugs. Term human placenta samples were obtained from scheduled uncomplicated C-sections performed at the University of Washington Medical Center, Seattle, WA. The 1st (45-59 days) and 2nd (109–115 days) trimester placentae, from uncomplicated elective termination, were provided by Birth Defects Laboratory of the University of Washington. Any differences in drug exposure for the termination or cesarean sections were not taken in consideration. After collection, the tissues were kept on ice for ∼45 min until the placental villus could be dissected and snap-frozen in liquid nitrogen and stored at −80° C. RNA was extracted from the placental tissue using the RNAesy kit (Quigen, Valencia, CA) according to the manufacturer's protocol.
Microarray processing methods for the Affymetrix microarray platform
Placental gene expression was determined by the CEEH Functional Genomics Laboratory of the University of Washington using the GeneChip platform by Affymetrix (Santa Clara, CA) and the manufacturer's protocol. Briefly, the first- and second-strand cDNAs were synthesized, the double-stranded cDNA purified and the cRNA synthesized by in vitro transcription (IVT), the biotin-labeled cRNA recovered and quantified, the cRNA fragmented and hybridized to the microarray slide (Human Genome U133 Plus 2.0 Array), and the hybridized cRNAs detected using streptavidin-coupled fluorescent dye.
Scanning and analysis of Affymetrix arrays
The hybridized Affymetrix arrays were scanned with an Affymetrix GeneChip® 3000 scanner. Image generation and feature extraction were performed using Affymetrix GCOS Software. Cel files were further processed in GeneTraffic, a microarray database and management system (Stratagene, La Jolla, CA). The data were normalized using the RMA (Robust Multi Array) normalization method 5. The two tail unpaired t-test was used to calculate p values. Changes in gene expression between gestational ages of more than 2-fold (p<0.05) were considered as differentially expressed genes and were analyzed further. Differentially expressed genes were analyzed using GoMiner (http://discover.nci.nih.gov/gominer/htgm.jsp) 6 to determine Gene Ontology categories 7 which were differentially regulated and significantly overrepresented. The database for Annotation, Visualization and Integrated Discovery (http://apps1.niaid.nih.gov/david) was used to identify known pathways. Microarray data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE9984.
Western blot analysis
Proteins were extracted from frozen material using lysis buffer (Cell Signaling) supplemented with 1mM PMSF. Western blot was performed using standard procedures described elsewhere 8. Proteins (30-60 μg) were separated in 4% to 15% Tris–HCl gradient polyacrylamide gels (Bio-Rad, Hercules, CA). The separated proteins were transferred by electroblotting to an Immun-Blot PVDF membrane (Bio-Rad) using Tris–glycine buffer containing 20% methanol. Membranes were blocked with 5% nonfat dry milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.02% Tween-20) for 30 minutes and incubated overnight with a primary antibody to β–catenin (BD Biosciences, San Jose, CA; 1:5000 dilution) at 4°C. Blotted antigens were detected with an anti-mouse secondary antibody (1:5000 dilution, Pierce, Rockford, IL) using a chemiluminescence detection kit (Pierce). A β–actin antibody (1:5000 dilution, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used as an internal control.
Immunohistochemistry
Cryostat sections were fixed in ice-cold acetone-methanol (1:1) for 10 min and allowed to air dry. Sections were incubated in 0.1% hydrogenium peroxide for 30 min, washed in PBS and blocked in normal serum for 1 hour before overnight incubation with a β-catenin antibody (BD Biosciences, San Jose, CA; 1:200 dilution). Tissue sections were washed in PBS and antibody-antigen complexes were detected using a biotinylated secondary antibody, VECTASTAIN® Elite ABC kit (Vector Laboratories, Burlingame, CA) and diaminobenzidine. A total of 4 samples of each gestational age were used in the study. Sections processed without primary antibody were used as a negative control.
To verify β-catenin subcellular localization, β-catenin was visualized with a secondary goat anti-mouse antibody conjugated with AlexaFluor 568 (Molecular Probes; 1:2000 dilution). Confocal images were taken on a Zeiss LSM510META confocal microscope and Photoshop software (Adobe Systems, San Jose, CA) was used to process the figure montage from the imported images.
Results
Identification of differentially expressed genes
RNA samples from 12 independent placentae of 3 different gestational ages (four biological replicates per group) were extracted, passed quality control, and used for hybridization. All hybridization data points were collected and processed for normalization and filtering. The hybridization signals, collected from the individual arrays, demonstrated a normal distribution and were considered suitable for statistical analysis and comparison of expression profiles (data not shown). The quality of the arrays was evaluated by determining the distribution of the gene expression signals and by comparing the magnitude of the average signal and background on each array. To identify the genes that may be linked to placental development, we estimated the number of genes that were up- or down- regulated in placentae of different gestational ages. A gene-by-gene comparison of the mean relative expression level of these genes in placentae of 1st or 2nd trimester versus term placentae was performed.
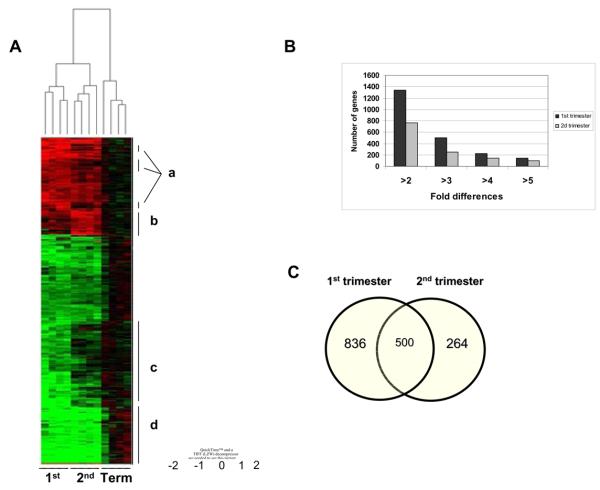
Hierarchical two dimensional clustering of tissue samples was performed using the expression profiles of all differentially expressed genes. Tissue samples or genes with similar expression patterns were clustered adjacent to one another. Figure 1A shows that samples of each gestational age were clustered together, indicating a high level of correlation in gene expression pattern amongst placentae of similar gestational age. Clustering demonstrates that the overall number of genes downregulated in early pregnancy is larger compared to the number of genes upregulated relative to term placentae. Among upregulated genes, smaller subclusters of genes were predominantly expressed in the 1st or 2nd trimester (clusters a and b). Among downregulated genes, we found cluster of genes (cluster c) predominantly downregulated in the 1st trimester, and to a lesser extent, in the 2nd trimester. Subcluster d consists of genes that are coordinately and most prominently downregulated in both 1st and 2nd trimester vs term placentae.
Figure 1.
A. Unsupervised hierarchical clustering of genes in placentae of different gestational age using average correlation linkage analysis. Cluster has been built using genes that demonstrated at least 2 fold differential expression in 1st and 2nd trimester compared to term placentae and p<0.05. Each placental sample is represented by an individual branch on the top of the panel. The branches cluster together by gestational age. The average expression value for term placentae samples for a given gene was used as the reference point and is shown in black. Genes overexpressed relative to average expression value of term placentae are shown in red. Genes expressed lower to relative average expression value of term placentae are shown in green. Subclusters of genes that demonstrated predominant differences in the levels of expression in the 1st and/or 2nd trimester placentae vs term placentae are shown with vertical lines and the corresponding letter (a-d).
B. Number of genes whose expression between 1st trimester vs. term or 2nd trimester vs. term or both vs. term is different by at least two-fold (p<0.05).
In first and second trimester placentae, expression of 1336 and 764 genes respectively differed by at least 2-fold (p<0.05) from that in the term placentae. Among these genes, 500 were common between 1st and 2nd trimester while 836 were 1st trimester-specific and 264 were 2nd trimester-specific (Figure 1B). These results indicate that there are significant quantitative differences between 1st and 2nd trimester placentae in their transcriptome profile. Moreover, between 1st and 2nd trimester there was ∼3-fold reduction in genes whose expression is dependent on gestational age. The genes that were differentially expressed genes changed at least 2 fold (p<0.05) in the 1st and 2nd trimester placentae vs term placentae are provided as Supplemental Tables S1 and S2.
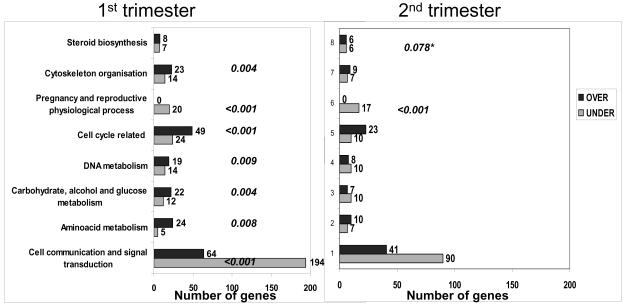
To determine functional groups of differentially expressed genes as defined by Gene Ontology (GO) annotations we used the GoMiner software. For GoMiner analysis we used the list of those genes whose expression was different by at least two-fold (p<0.05) in 1st or 2nd trimester placentae vs term placentae. Then, the one-sided Fisher's exact test to determine p values was corrected for multiple comparisons to identify significant enrichment within gene classes that are up-regulated or down-regulated. This p value or false discovery rate (FDR) identified a fraction of categories that are changed by random chance. In the present study, the FDR was set at 0.1. GO categories with FDR <0.1 were considered significantly enriched. Using this approach, the top 8 GO categories identified in 1st or 2nd trimester that were significantly enriched are shown in Figure 2. Among these identified categories for each relevant functional theme shown, FDR met cut-off level (<0.1) for at least one gestational age vs. term. In 1st and 2nd trimester we observed changes in similar subsets of differentially expressed genes in any given category. However, in 2nd trimester placentae, the number of differentially expressed genes in each category was reduced and therefore did not meet FDR cut-off level. GO analysis and category clusters for the 1st and 2nd trimester placentae are shown below. Because of the large size of the clusters, these results are provided in Supplemental figures.
Figure 2.
Top 7 GeneOntology categories identified in 1st and 2nd trimester using GoMiner software to determine enriched groups of differentially expressed genes. Categories united by common functional theme are shown on the left. Number of genes overexpressed (over) or underexpressed (under) in 1st trimester (left panel) or 2nd trimester (right panel) placentae vs term placentae are shown. Computed False Discovery Rate (FDR) below cut-off level (0.1) for each over or underexpressed group of genes is shown in italics next to the each bar. FDR values that did not meet cut-off levels are not shown. *-FDR is for group of genes related to steroid biosynthesis that demonstrated balanced change (number of upregulated genes is equal to number of downregulated genes) in 2nd trimester vs term placentae.
Genes that are overexpressed in the 1st trimester vs term placentae
Supplemental figure 1A and 1B shows clusters of enriched gene categories (66) and corresponding genes (147) that are overexpressed in 1st trimester vs. term placentae. We identified 3 major enriched clusters of genes. Two clusters (1a and 1b) are associated with terms “cell division”, “mitosis” and “regulation of cell cycle”. Cluster 1a consists of 28 genes. Representative examples are cyclins A2, B1 and B2. Expression of these genes is increased in S phase of cell cycle. Another gene NEK2 (see Supplemental Tables S1 and S2 for definition) accumulates throughout S phase and shows maximal levels in late G2. This expression pattern is highly reminiscent of that of A and B cyclins. Similarly, TPX2 is exclusively expressed in proliferating cells from the transition G1/S until the end of cytokinesis. Other genes that control and are involved in reorganization to the kinetochore/centromere and the spindle formation during normal mitosis (KNTC1, MAD2L1, PTTG1, CENPF) were also upregulated in 1st trimester vs term placentae.
Cluster 1b consists of 17 genes, such as CDKN3, TCF7L2, VEGF, E2F3. Consistent with mitotic activity in placenta was the overexpression of the transcription factors E2F3 and TCF7L2. TCF7L2 complexes with β-catenin and participates in activation of downstream genes associated with cell cycle progression. While there is clear evidence of mitotic activity in placenta, CDKN3 may play a role in negative regulation of cell cycle. VEGF plays a role in proliferation of endothelial cells which is consistent with placental vessel development. However, overlapping gene categories related to angiogenesis such as “blood vessel morphogenesis”, “blood vessel development”, “vasculature development”, consisting of 12 genes, were downregulated in 1st trimester compared with term placentae.
The next largest cluster (cluster 2) is associated with GO terms “amine metabolism”, “nitrogen compound metabolism”, “amino acid metabolism”, “DNA metabolism”. This cluster consist of 22 genes that play a role in nitric oxide generation (DDAH1), polyamine (ODC), tyrosine (FAH), aminocystein (SCLY), arginine (Arg2) metabolisms, valine, leucine and isoleucine degradation (MCCC2, BCAT1), biosynthesis of purines, thymidylate, methionine, histidine, pantothenate, formyl tRNA-Met (MTHFD1) and oxidative degradation of sulfur-containing amino acids (SUOX). Consistent with amino acid metabolism we observed change in cationic amino acids (arginine, lysine and ornithine) transport (SLC7A1) and ornithine mitochondrial transport (SLC35A15).
Clusters 3a and 3b consist of 22 genes and are associated with the terms “carbohydrate metabolism”, “hexose” and “glucose” metabolism, “alcohol metabolism”, “glycolysis”. These clusters consist of genes that are involved in different steps related to glucose metabolism (GSY1, GPI, FUT4). Some genes within this cluster are known to be regulated by genes that are related to cell cycle progression or provide a link to other pathways. For example, enolase 1 (ENO1), in addition to its role in glycolysis, plays a part in various processes such as growth control, hypoxia tolerance, and allergic responses. Another gene, APOA2, involved in glucose metabolism, may stabilize high density lipoprotein (HDL) structure by its association with lipids, and affects the HDL metabolism during placentae development.
The above data demonstrate significant proliferative activity in 1st trimester placenta associated with increase in DNA, amino acid and glucose metabolism. It is necessary to note that similar gene categories were found differentially expressed in 2nd trimester placentae versus term. However, the number of differentially expressed genes was reduced in 2nd trimester and did not pass FDR cut-off level.
Genes that are underexpressed in the 1st trimester vs term placentae
Supplemental figure 2 shows clusters of enriched categories and corresponding genes that are down regulated in the 1st trimester placentae vs term placenta. Fourteen identified categories associated with 257 genes are shown in supplemental figure 2A as cluster 1. Two largest overlapping GO categories are “signal transduction” (221 genes), “cell communication” (240 genes) and daughter categories. Overlapping genes within these categories that met FDR cut off levels were also associated with “protein kinase cascade” (45 genes), “intracellular signaling cascade” (103 genes),“G-protein coupled receptor signaling” (44 genes), “cell surface receptor linked protein signaling pathways” (105 genes). Detection of these categories is consistent with regulation of signal transduction during pregnancy through differential expression and post-translation modification of proteins. For example, RapGEF and AHRGEF are two guanine nucleotide exchange proteins that activate Rap1/Rap2 and Rho proteins respectively. Rap proteins counteract the mitogenic function of Ras. Reduced levels of Rap activity therefore will be consistent with high proliferative activity observed in 1st trimester placentae.
Clearly, the above data indicate that multiple signal transduction cascades are subjected to gestational-age dependent regulation. We next asked which signaling pathways were affected. A pathway which is significantly down-regulated is the NF-kappa-B cascade (12 genes). Amongst these genes we found TLR 1, 3, 4, 8, TNFSF10 and CASP3. Toll-like receptors (TLR) participate in the innate immune response leading to NF-kappa-B activation, cytokine secretion and the inflammatory response. TNFSF10 is a cytokine that binds to TRAIL Receptor and may induce apoptosis through caspase activation. Of note, we found 21 downregulated genes relative to term placentae in category “positive regulators of apoptosis”, including proapoptotic genes like CASP1 and 4. On the other hand, inhibitors of apoptosis like BCL2L1 were also reduced, probably to offset reduced levels of proapoptotic factors. More information on genes involved in organism defense is provided below. We also identified 10 transcripts which belong to MAPKKK cascade although this pathway did not meet FDR cut off levels. Amongst genes within this pathway are dual specificity phosphatases DUSP 4, 16 and 22 that are involved in regulation of ERK, p38 and JNK pathways activity. GADD45 B and G respond to stress, and upon activation by DNA damage lead to cell cycle arrest.
Genes located in Cluster 2 are associated with overlapping categories “pregnancy” (17 genes), “interaction between organisms” (26 genes) and “reproductive organismal physiological process” (22 genes). Amongst these genes are pregnancy specific beta-1-glycoproteins (PSG1, 3, 4, 6, 9, 11) known to be expressed in differentiating cytotrophoblasts, although their function remains unknown. PAPPA, pregnancy associated plasma protein, known to be expressed in the placenta, STS is known to participate in estrogen synthesis, STAT5b and PRLR are involved in signal transduction initiated by prolactin 9, or by IL2 10 . Interestingly, JAK2, non-receptor tyrosine kinase that activates STAT5 11 transcription factors phosphorylation was also found downregulated in the “signal transduction” category described above. Essentially, all genes associated with cluster 2 were found significantly underexpressed in 2nd trimester placentae vs term placentae (see below and Figure 2).
Genes that are changed in 2nd trimester placentae vs term placentae
Supplemental figure 3 shows cluster of 2nd trimester genes associated with GO categories that met FDR cut off level. All identified 5 categories were similar to cluster 2 (Pregnancy) in Supplemental figure 2A. These GO categories are: “pregnancy” (13 genes), “reproductive (organismal) physiological process” (17 genes), “physiological interaction between organisms” (15 genes), and “interaction between organisms” (16 genes).
Analysis of other gene categories in 2nd trimester that did not pass cut-off FDR indicated that they are virtually the same as in 1st trimester but with a reduced number of genes. Our data indicate that gestational age dependent regulation of genes is most pronounced in the 1st trimester.
Change in genes related to defense
Placenta is a barrier which protects the fetus from toxins that may enter the mother's circulation. Therefore, we investigated differential expression of genes related to organism defense. Table 1 shows a number of differentially regulated genes related to the response to abiotic and biotic stimulus. The majority of genes in this category were related to response to chemicals. In the latter category, chemotaxis represented the largest gene subcategory. Genes involved in the response to drugs, xenobiotics and toxins included downregulation AHR, BCAR3 and DEFA1. For example, AHR mediates biochemical and toxic effects of halogenated aromatic hydrocarbons. It also probably plays a role in the development and maturation of many tissues. We found only 5 transporters which were differentially expressed in 1st and/or 2nd trimesters as compared to term (Tables 2 and 3). Among them only one, OATP2A1, was downregulated in both 1st and 2nd trimester placentae versus term placentae (p<0.05). Other transporters (OATP2B2, ABCG2 and CNT2) were differentially regulated in 1st or 2nd trimester versus term. Cytochrome 450 enzymes are involved in metabolic activation or inactivation of exogenous and endogenous chemicals. Among all the CYPs, we found 3 that are differentially regulated in 1st and 2nd trimester placentae versus term (Tables 4 and 5). In the 2nd trimester placentae CYP24A1 was found to be up-regulated as compared to term placentae (Table 5). CYP24A1 is a peripheral inner mitochondrial membrane protein that regulates the metabolism and biological function of vitamin D and many of its related analogs.
Table 1.
Number and direction of differentially expressed gene related to response to abiotic and biotic (bottom) stimulus
| 1st trimester | 2nd trimester | |||
|---|---|---|---|---|
| Category | over | under | over | under |
| Response to abiotic stimulus | 13 | 24 | 14 | 12 |
| Response to chemicals | 13 | 20 | 14 | 9 |
| chemotaxis | 6 | 6 | 9 | 2 |
| response to drugs | 2 | 2 | 1 | 1 |
| response to xenobiotics | 0 | 3 | 1 | 2 |
| response to toxins | 0 | 0 | 1 | 0 |
| drug transport | 1 | 0 | 1 | 0 |
| Response to biotic stimulus | 23 | 61 | 20 | 28 |
| Defense response | 21 | 60 | 19 | 28 |
| Immune response | 21 | 53 | 19 | 22 |
| inflamatory response | 8 | 18 | 8 | 3 |
| humoral immune response | 9 | 15 | 3 | 5 |
| immune cell activation | 2 | 9 | 2 | 6 |
| innate immune response | 4 | 11 | 2 | 2 |
| cytokine metabolism | 1 | 7 | 0 | 4 |
| cytokine production | 1 | 7 | 0 | 4 |
Linked GO categories shown in bold. Subcategories for categories “Response to chemical” and “Immune response” shown in italics.
Table 2.
Drug transporters differentially expressed in the 1st trimester versus term placentae by at least 2-fold (p<0.05).
| ID | Symbol | Gene name | Fold change |
SD* |
|---|---|---|---|---|
| 203473_at | OATP2B1 | solute carrier organic anion transporter family, member 2B1 |
−2.5 | 0.72 |
| 211113_s_at | ABCG2 | ATP-binding cassette, sub-family G (WHITE), member 2 |
−2.4 | 0.68 |
| 204368_at | OATP2A1 | solute carrier organic anion transporter family, member 2A1 |
−2.86 | 0.08 |
SD – Standard deviation.
Table 3.
Drug transporters differentially expressed in the 2nd trimester versus term placentae by at least 2-fold (p<0.05).
| ID | Symbol | Gene name | Fold change |
SD |
|---|---|---|---|---|
| 204368_at | OATP2A1 | solute carrier organic anion transporter family, member 2A1 |
−2.89 | 0.5 |
| 207249_s_at | CNT2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 |
2.8 | 1.4 |
Table 4.
Cytochrome P450 enzymes differentially expressed in the 1st trimester versus term placentae by at least 2-fold (p<0.05).
| ID | Symbol | Gene name | Fold change |
SD |
|---|---|---|---|---|
| 232292_at | CYP19A1 | Cytochrome P450, family 19, subfamily A, polypeptide 1 |
−8.5 | 5.75 |
| 204309_at | CYP11A1 | cytochrome P450, family 11, subfamily A, polypeptide 1 |
−3.2 | 1 |
| 202314_at | CYP51A1 | cytochrome P450, family 51, subfamily A, polypeptide 1 |
2.3 | 0.54 |
Table 5.
Cytochrome P450 enzymes differentially expressed in the 2nd trimester versus term placentae by at least 2-fold (p<0.05).
| ID | Symbol | Gene name | Fold change |
SD |
|---|---|---|---|---|
| 232292_at | CYP19A1 | Cytochrome P450, family 19, subfamily A, polypeptide 1 |
−9.3 | 5.6 |
| 204309_at | CYP11A1 | cytochrome P450, family 11, subfamily A, polypeptide 1 |
−4.5 | 1.4 |
| 206504_at | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 |
2.1 | 0.73 |
| 202314_at | CYP51A1 | cytochrome P450, family 51, subfamily A, polypeptide 1 |
2.3 | 0.38 |
Defense response to biotic stimulus was mostly associated with genes related to immune response (Table 1). Associated categories were represented by inflammatory response, humoral response and innate immune response. It is worth noting that genes related to cytokine metabolism and production were downregulated in 1st and 2nd trimester placentae compared to term. Functional activity of cytokines depends on the status of signal transduction proteins, which we showed before were significantly downregulated. Therefore, it is reasonable to suggest that cytokine activity correlates with gestational age.
Identification and analysis of gestational regulation of the Wnt pathway
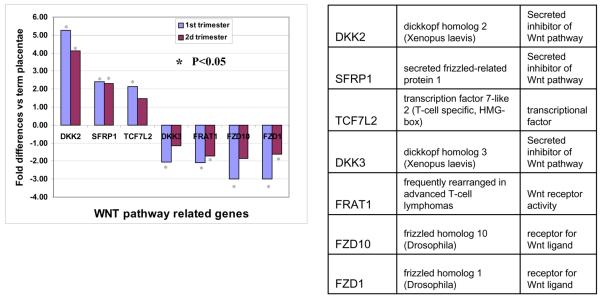
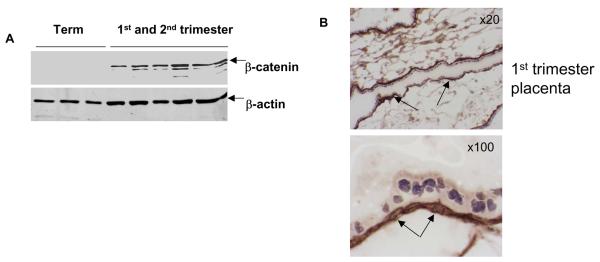
Analysis of differentially expressed genes related to signal transduction in connection with cell proliferation identified gestational age dependent regulation of components of Wnt pathway. Figure 3 demonstrates that the same set of genes is differentially regulated in both 1st and 2nd trimesters versus term placentae. Although some transcripts lose statistical significance in the 2nd trimester or fold-change is less then 2, the direction of change remains the same. Among these genes, we did not find differential expression of Wnt ligands. However, we found inhibition of two Frizzled receptor (Fzd 1 and 10) expressions and activation of negative regulators of Wnt pathway (DKK-2 and SFRP). Previously, it was shown that activation of negative regulator of Wnt pathway expression, DKK-1, is increased following activation of canonical Wnt pathway because its promoter element contains binding sites for β-catenin dependent transcription 12. We verified that DKK-1, while it did not reach statistical significance due to variability in expression, demonstrated 6.7 fold increased expression in 1st trimester versus term. Activation of canonical Wnt pathway leads to increased cell proliferation which is consistent with increased mitotic activity in early stage placentae. The hallmark of Wnt pathway activation is stabilization of β-catenin and its translocation into the nucleus 13. Therefore, to verify the status of gestational age dependent regulation of canonical Wnt pathway we used Western blot analysis to determine steady state levels of β–catenin in the placentae of different gestational ages. As expected, there was a marked decrease in the levels of β-catenin protein expression in term placentae (Fig. 4A) compared to early gestational age placentae. We further confirmed strong β-catenin expression in the cytoplasm and nuclei of villous cytotrophoblasts in the 1st trimester placentae (Fig.4B). Confocal microscopy analysis of β-catenin localization in the 1st trimester placentae (Fig.4C) also showed presence of a certain level of β-catenin in the nucleus of cytotrophoblasts. While syncytiotrophoblasts showed some expression of β-catenin, their expression levels were much less than seen in cytotrophoblasts and was predominantly found in the cytoplasm. Higher magnification images (Fig.4B, panels d and f) as well as confocal images (Fig.4C) detected nuclear localization of β-catenin in the syncytiotrophoblasts of early placentae, while in the term placentae, nuclear β-catenin was below the detection limit in the epithelial compartment. However, β-catenin was detectable in the mesenchymal compartment of the term placentae. Thus, the nuclear localization and the overall higher levels of β-catenin in the 1st and 2nd trimester placentae are consistent with the activation of canonical Wnt pathway in villous cytotrophoblasts.
Figure 3.
Differential regulation of genes related to canonical Wnt pathway in 1st and 2nd trimester placentae versus term placentae. A. The fold-difference and direction of change in Wnt pathway gene expression vs. term placenta. *-significant (p<0.05) compared to term placentae. Gene names and function are shown in the table.
Figure 4.
A. Reduced levels of β-catenin expression in term placentae compared to 1st or 2nd trimester placentae. The β-catenin expression is detectable in term placentae after longer exposure (not shown). Lower panel shows levels of β-actin expression used for normalization of the Western blot. B. Immunohistochemical analysis shows that β-catenin in 1st (panel a) and 2nd trimester placentae (panel b) is highly expressed in the nuclei and cytoplasm of cytotrophoblasts (shown with long arrows) and to a lesser extent in the cytoplasm of syncytiotrophoblasts (higher magnification, panel d, shown with short arrows). Very weak signal for β-catenin is also detected in the nuclei of at least some syncytiotrophoblasts (panel d, dashed arrows). In the term placentae β-catenin is detected only in the mesenchymal compartment (panels e and f, shown with arrowheads) while in epithelial component β-catenin is undetectable. Panel c shows 1st trimester placentae processed for immunohistochemical staining with primary antibody omitted, C. Confocal images of the 1st trimester placentae stained with β-catenin antibody and visualized with secondary antibody conjugated with AlexaFluor 568. Nuclei were stained with DAPI (1 ug/ml) and presented as green pseudo-color for better contrast (left panel). In the cytotrophoblasts (solid arrows) β-catenin (red) is clearly localized in the nuclei as demonstrated in the z-stack images (right panel). In the syncytiotrophoblasts β-catenin is localized in the cytoplasm. Weak β-catenin signal is also detected in the nuclei of some syncytiotrophoblasts (dashed arrow). Scale bar = 22 um.
Discussion
The placenta is a complex organ which plays multiple functions in fetal development. Placenta insufficiency may lead to fetal morbidity or mortality as well as leading to or resulting from pathological conditions such as preeclampsia and intrauterine growth restriction.
There is limited information about global expression profiling of genes that influence placenta development. Therefore, we undertook a genomic approach in profiling the genes expressed in placentae of different gestational ages. The ultimate goal of this study was to determine major changes in placental gene expression observed at various gestational stages through identification of functional categories that are over or under represented using analysis and ranking of biologically coherent categories instead of gene-by-gene approach. We found that in early gestation placentae, genes related to cell cycle, DNA, aminoacids and carbohydrate metabolism were significantly overrepresented compared to term placentae. Genes related to different signal transductions were underrepresented in early placentae compared to term placentae. Most pronounced changes in gene expression were observed during the first trimester versus term placentae. Our results show that while 1st and 2nd trimester have similar categories of genes differentially expressed vs term placentae, in the 2nd trimester placentae, the number of genes with > 2-fold difference in expression in most categories were reduced. As a result, 2nd trimester placentae lost gene categories which met cut-off levels for FDR. Such interpretation is supported by the fact that genes involved in the category “pregnancy” and “interaction between organisms” retain significant overrepresentation in both 1st and 2nd trimester compared to term placentae while other gene categories are over or underrepresented only in the 1st trimester placentae. Most likely pregnancy related genes are independent of those categories (cell proliferation, signal transduction) found in the 1st trimester placentae.
Analysis of individual genes indicated that, among identified genes, there are many transcripts important for placenta development. It is known that knock out of some of these genes induces lethal phenotype because of defects in placenta 14-17. For example, knockout of p38alpha gene results in embryonic lethality due to insufficient oxygen and nutrients transfer across the placenta 14.
Our results show that early gestational-age placentae is characterized by increased cell cycle proliferation, which is consistent with increased DNA, carbohydrate, and amino acid metabolism observed in fast growing tissues. Gestational dependent reduction of placenta proliferative rate has previously been determined by immunostaining with K67 18. This study revealed that within the placenta, cytotrophoblasts have the highest proliferative rate. Significant changes in cell proliferation were also found in early mouse placenta (E10.5 and E12.5) 19. However, in contrast to our findings, in the mouse placentae, metabolic pathways were not significantly enriched in early gestation. Changes in placentae cell proliferation is reminiscent of those that occur in cancer. However in the placentae, amongst genes that promote cell growth we observed increased expression of cyclin dependent kinase (CDK) inhibitor CIP2. Higher level of CDK inhibitors expression in the placenta is likely an important mechanism of control of proliferative activity. This mechanism is frequently lost in cancer. High proliferative activity is associated with increased rate of apoptosis. Previously, it was reported 18 that the rate of apoptosis in the placenta is reduced with gestational age, an observation consistent with higher cell turnover rate in the early gestation placenta. In our study, we found reduced expression of both positive regulators and inhibitors of apoptosis. While these results are consistent with changes in apoptotic process, understanding the precise biological outcomes from such changes requires additional studies.
Angiogenesis is a necessary component of placenta development. VEGF is involved in regulation of cytotrophoblast survival and its deregulation is observed in preeclampsia 20. We found that with the exception of VEGF 21, all other genes related to vasculature development and morphogenesis were down regulated in the 1st trimester placenta. These data indicate that as the placenta increases in size, vessel growth in early gestational-age placentae may depend primarily on VEGF. At later gestational age, expression of VEGF is replaced by other genes important for vasculature development. These results are consistent with changes in VEGF expression in human placenta reported by others 20,22.
In addition to placental cell proliferation, we found broad and significant reduction of expression of genes involved in various signal transduction pathways. Analysis of these pathways indicated that positive and negative regulators of such pathways are affected in the same direction. As a result, a conclusive answer whether a given pathway is activated or inhibited requires independent approaches. Using canonical Wnt pathway as an example, we determined that there is significant gestational-age dependent downregulation of this pathway. Wnt pathway is crucial in organism development 23 and activation of this pathway was shown to be important in colon cancer cell transformation 24. Published data indicate that the Wnt pathway plays a role in the placenta development. Mice defective in Wnt-2 ligand showed a defect in placenta development 25. Activation of Wnt pathway leads to inhibition of GSK-3β activity associated with serine 9 phosphorylation. Active GSK-3β is a component of destructive complex that is required for β-catenin binding and phosphorylation. Phosphorylated β-catenin is subjected to subsequent proteasomal degradation. Loss of GSK-3β activity leads to reduced β-catenin phosphorylation and stabilization. Stabilized β-catenin translocates to the nuclei. In the nuclei, β-catenin associates with TCF/Lef transcription factors resulting in activation of downstream targets. Expression of TCF7L2 transcription factor, which belongs to TCF/Lef family of transcription factors, is also increased in 1st trimester placentae versus term placentae suggesting activation of Wnt pathway. We further confirmed increased levels of the β-catenin in the proliferating cytotrophoblasts. We also found β-catenin expression and nuclear localization in syncytiotrophoblasts, although nuclear expression was very weak. Clear differences in the nuclear β-catenin expression in cytotrophoblasts compared to syncytiotrophoblasts is consistent with the differential role of Wnt signaling pathways in these cell types. Therefore, levels of β-catenin and its binding partner TCF7L2 correlate during gestation, a finding consistent with activation of the Wnt pathway. In addition, increased Wnt pathway activity in 1st and 2nd trimester is consistent with proliferative activity and invasiveness. While our results are consistent with Wnt pathway activation, we were not able to detect differential expression of the Wnt ligands that are known to activate the pathway. The most likely explanation for this observation is that regulation of Wnt pathway is dependent on the receptor availability (FZD 1 and 10), negative regulators of the Wnt ligands binding to receptors (DKK family of proteins, SFRP1). The role of the Wnt pathway in trophoblasts invasion was recently demonstrated in vitro 26. Therefore, inhibition of Wnt pathways with gestational age would predict reduction of trophoblasts invasiveness. On the other hand, β-catenin is mainly a binding partner for E-cadherin that is required for the formation of adherent junctions 27. It is interesting to note that E-cadherin expression is also reduced with increase in gestational age 28. In addition, higher levels of the nuclear β-catenin were found in placentae of complete hydatidiform mole pregnancies as compared to normal placentas 26. Thus, regulation of Wnt pathways appears necessary for normal placental development.
The placenta is an important organ, which protects the fetus from a variety of endogenous and exogenous compounds in the mother's circulation. The most pronounced changes were found in genes related to the immune response. We found relatively few changes in genes related to response to chemicals. Amongst the CYP enzymes, we found an increase in CYP11A1 and 19A1 expression with gestational age. Because these enzymes are involved in steroid metabolism, correlation of expression with gestational age is consistent with increased requirements in steroid hormones. CYP24A1 catalyzes the NADPH-dependent 24-hydroxylation of 25-hydroxyvitamin D resulting in abrogation of vitamin D mediated growth control and therefore consistent with increased cell cycling in early gestation placentae. Amongst transporters, OATP2A1 is downregulated in 1st and 2nd trimester placentae. OATP2A1 mediates the release of newly synthesized prostaglandins from cells, the transepithelial transport of prostaglandins, and the clearance of prostaglandins from the circulation.
Microarray analysis of mouse placentae identified cell cycle genes, DNA metabolism genes and signal transduction genes overrepresented in early placentae vs late stage of placentae development 19, 29. These results are consistent with main groups of genes that are regulated by gestational age in human placentae.
While this manuscript was in preparation, gene expression profiling of the human maternal-fetal interface during gestation was published 30. This study used basal plate as a source of RNA. Despite the fact that our study used villous tissue, both studies identified the number of genes belonging to overlapping functional categories. These categories include cell cycle, carbohydrate and protein metabolism, angiogenesis/vasculogenesis, apoptosis and Wnt pathway. The analysis of genes identified in Winn's study, and in our study, clearly shows that within any given functional category there is significant but not absolute overlapping in the expression of individual genes. For example, among 7 genes that are assigned to Wnt pathway, only SFRP1, FRAT1, TCF7L2 overlapped with the Winn study. While we identified differential expression two FZD receptors, DKK-2 and 3 in villous tissue with gestation, Winn's study identified Kremen 1 receptor and DKK-1 in basal membrane. Thus, the differences in individual genes are likely to reflect the type of cells within placentae taken for analysis. However, the biological significance of differential expression of the individual genes within any given category remains unknown.
Our study addresses an important process of normal placenta growth and identified major changes in the pattern of placental gene expression with progression of pregnancy. Further investigation is needed to explain the significance of gestational age dependent regulation of different gene categories. Our study should provide a database to further understand the physiological and pathological changes associated with the developing placenta in both uncomplicated and complicated pregnancies.
Supplementary Material
Supplemental Figure S1A. Clusters of genes in categories that are over-represented in 1st trimester versus term placentae that passed FDR<0.1. Overexpressed genes are shown in red. Main categories with common theme are labeled with horizontal bar (X axis). Number in parentheses next to category name refers to the group of genes labeled with a vertical line and the same number on the Y axis. Individual genes within each functional category and functional theme can be identified using Supplemental Figure S1B. List of GO categories shown with numbers on X axis are presented in Supplemental Figure S1C.
Supplemental Figure S1B shows list of genes used to build panel in S1A. Number for each gene corresponds to the number on Y axis in S1A.
Supplemental figure S1C shows list of categories corresponding to the X axis in S1A. Number for each GO category corresponds to the number on X axis in S1A.
Supplemental Figure S2A. Clusters of genes that are under-expressed in 1st trimester versus term placentae that passed FDR<0.1. Under-expressed genes are shown in green. Gene categories are shown under the cluster. All labels are similar to those shown in S1A.
Supplemental Figure S2B shows list of genes used to build a cluster.
Supplemental Figure S3. Cluster of genes that are under-expressed in 2nd trimester versus term placentae that passed FDR<0.1. Under-expressed genes are shown in green. List of genes and list of categories are shown in Y and X axis respectively.
List of genes that are differentially expressed at least 2-fold in the 1st trimester versus term placentae
List of genes that are differentially expressed at least 2-fold in the 2st trimester versus term placentae.
Acknowledgements
This study was supported by NICHD HD047892, P50 HD044404, NIEHS P30ES007033. This work was supported in part by the UW NIEHS sponsored Center for Ecogenetics and Environmental Health, Grant # NIEHS P30ES007033. T.N. is supported by fellowship of Astellas Foundation for Research on Metabolic Disorders.
Footnotes
Obstetrics-Fetal Pharmacology Research Unit
University of Washington Specialized Center of Research
References
- 1.Cross JC. Placental function in development and disease. Reproduction, Fertility, & Development. 2006;18:71–76. doi: 10.1071/rd05121. [DOI] [PubMed] [Google Scholar]
- 2.Pardi G, Cetin I. Human fetal growth and organ development: 50 years of discoveries. American Journal of Obstetrics & Gynecology. 2006;194:1088–1099. doi: 10.1016/j.ajog.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 3.Unadkat JD, Dahlin A, Vijay S. Placental drug transporters. Current Drug Metabolism. 2004;5:125–131. doi: 10.2174/1389200043489171. [DOI] [PubMed] [Google Scholar]
- 4.Carney EW, Scialli AR, Watson RE, DeSesso JM. Mechanisms regulating toxicant disposition to the embryo during early pregnancy: an interspecies comparison. Birth Defects Research Part C, Embryo Today: Reviews. 2004;72:345–360. doi: 10.1002/bdrc.20027. [DOI] [PubMed] [Google Scholar]
- 5.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 6.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biology. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athanassiou M, Hu Y, Jing L, Houle B, Zarbl H, Mikheev AM. Stabilization and reactivation of the p53 tumor suppressor protein in nontumorigenic revertants of HeLa cervical cancer cells. Cell Growth & Differentiation. 1999;10:729–737. [PubMed] [Google Scholar]
- 9.Ma FY, Anderson GM, Gunn TD, Goffin V, Grattan DR, Bunn SJ. Prolactin specifically activates signal transducer and activator of transcription 5b in neuroendocrine dopaminergic neurons. Endocrinology. 2005;146:5112–5119. doi: 10.1210/en.2005-0770. [DOI] [PubMed] [Google Scholar]
- 10.Biola A, Lefebvre P, Perrin-Wolff M, Sturm M, Bertoglio J, Pallardy M. Interleukin-2 inhibits glucocorticoid receptor transcriptional activity through a mechanism involving STAT5 (signal transducer and activator of transcription 5) but not AP-1. Molecular Endocrinology. 2001;15:1062–1076. doi: 10.1210/mend.15.7.0657. [DOI] [PubMed] [Google Scholar]
- 11.Takeda T, Kurachi H, Yamamoto T, Homma H, Morishige K, Miyake A, Murata Y. Participation of JAK, STAT and unknown proteins in human placental lactogen-induced signaling: a unique signaling pathway different from prolactin and growth hormone. Journal of Endocrinology. 1997;153:R1–3. doi: 10.1677/joe.0.153r001. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Sancho JM, Aguilera O, Garcia JM, Pendas-Franco N, Pena C, Cal S, Garcia de Herreros A, Bonilla F, Munoz A. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–1103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- 13.Eberhart CG, Argani P. Wnt signaling in human development: beta-catenin nuclear translocation in fetal lung, kidney, placenta, capillaries, adrenal, and cartilage. Pediatric & Developmental Pathology. 2001;4:351–357. doi: 10.1007/s10024001-0037-y. [DOI] [PubMed] [Google Scholar]
- 14.Adams RH, Porras A, Alonso G, Jones M, Vintersten K, Panelli S, Valladares A, Perez L, Klein R, Nebreda AR. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Molecular Cell. 2000;6:109–116. [PubMed] [Google Scholar]
- 15.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 16.Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- 17.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nature Reviews Genetics. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 18.Chan CC, Lao TT, Cheung AN. Apoptotic and proliferative activities in first trimester placentae. Placenta. 1999;20:223–227. doi: 10.1053/plac.1998.0375. see comment. [DOI] [PubMed] [Google Scholar]
- 19.Gheorghe C, Mohan S, Longo LD. Gene expression patterns in the developing murine placenta. Journal of the Society for Gynecologic Investigation. 2006;13:256–262. doi: 10.1016/j.jsgi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, McMaster M, Woo K, Janatpour M, Perry J, Karpanen T, Alitalo K, Damsky C, Fisher SJ. Vascular endothelial growth factor ligands and receptors that regulate human cytotrophoblast survival are dysregulated in severe preeclampsia and hemolysis, elevated liver enzymes, and low platelets syndrome. Am J Pathol. 2002;160:1405–1423. doi: 10.1016/S0002-9440(10)62567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibuya M, Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Experimental Cell Research. 2006;312:549–560. doi: 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Leach L, Babawale MO, Anderson M, Lammiman M. Vasculogenesis, angiogenesis and the molecular organisation of endothelial junctions in the early human placenta. Journal of Vascular Research. 2002;39:246–259. doi: 10.1159/000063690. [DOI] [PubMed] [Google Scholar]
- 23.Moon RT. Wnt/beta-catenin pathway. Science's Stke [Electronic Resource]: Signal Transduction Knowledge Environment. 2005;2005:cm1. [Google Scholar]
- 24.Pinto D, Clevers H. Wnt, stem cells and cancer in the intestine. Biology of the Cell. 2005;97:185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- 25.Goodwin AM, D'Amore PA. Wnt signaling in the vasculature. Angiogenesis. 2002;5:1–9. doi: 10.1023/a:1021563510866. [DOI] [PubMed] [Google Scholar]
- 26.Pollheimer J, Loregger T, Sonderegger S, Saleh L, Bauer S, Bilban M, Czerwenka K, Husslein P, Knofler M. Activation of the canonical wingless/T-cell factor signaling pathway promotes invasive differentiation of human trophoblast. American Journal of Pathology. 2006;168:1134–1147. doi: 10.2353/ajpath.2006.050686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clevers H. Wnt/β-Catenin Signaling in Development and Disease. Cell. 2006;127:469. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Brown LM, Lacey HA, Baker PN, Crocker IP. E-cadherin in the assessment of aberrant placental cytotrophoblast turnover in pregnancies complicated by pre-eclampsia. Histochem Cell Biol. 2005;124:499–506. doi: 10.1007/s00418-005-0051-7. [DOI] [PubMed] [Google Scholar]
- 29.Hemberger M, Cross JC, Ropers HH, Lehrach H, Fundele R, Himmelbauer H. UniGene cDNA array-based monitoring of transcriptome changes during mouse placental development. Proc Natl Acad Sci U S A. 2001;98:13126–13131. doi: 10.1073/pnas.231396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, Feng KT, Bernlohr DA, McDonagh S, Pereira L, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–1079. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1A. Clusters of genes in categories that are over-represented in 1st trimester versus term placentae that passed FDR<0.1. Overexpressed genes are shown in red. Main categories with common theme are labeled with horizontal bar (X axis). Number in parentheses next to category name refers to the group of genes labeled with a vertical line and the same number on the Y axis. Individual genes within each functional category and functional theme can be identified using Supplemental Figure S1B. List of GO categories shown with numbers on X axis are presented in Supplemental Figure S1C.
Supplemental Figure S1B shows list of genes used to build panel in S1A. Number for each gene corresponds to the number on Y axis in S1A.
Supplemental figure S1C shows list of categories corresponding to the X axis in S1A. Number for each GO category corresponds to the number on X axis in S1A.
Supplemental Figure S2A. Clusters of genes that are under-expressed in 1st trimester versus term placentae that passed FDR<0.1. Under-expressed genes are shown in green. Gene categories are shown under the cluster. All labels are similar to those shown in S1A.
Supplemental Figure S2B shows list of genes used to build a cluster.
Supplemental Figure S3. Cluster of genes that are under-expressed in 2nd trimester versus term placentae that passed FDR<0.1. Under-expressed genes are shown in green. List of genes and list of categories are shown in Y and X axis respectively.
List of genes that are differentially expressed at least 2-fold in the 1st trimester versus term placentae
List of genes that are differentially expressed at least 2-fold in the 2st trimester versus term placentae.