SUMMARY
Cell type-specific expression of epithelial and mesenchymal isoforms of Fibroblast Growth Factor Receptor 2 (FGFR2) is achieved through tight regulation of mutually exclusive exons IIIb and IIIc, respectively. Using a novel application of cell-based cDNA expression screening we identified two paralogous epithelial cell type-specific RNA binding proteins that are essential regulators of FGFR2 splicing. Ectopic expression of either protein in cells that express FGFR2-IIIc caused a switch in endogenous FGFR2 splicing to the epithelial isoform. Conversely, knockdown of both factors in cells that express FGFR2-IIIb by RNA interference caused a switch from the epithelial to mesenchymal isoform. These factors also regulate splicing of CD44, p120-Catenin (CTNND1), and hMena (ENAH); three transcripts that undergo changes in splicing during the epithelial to mesenchymal transition (EMT). These studies suggest that Epithelial Splicing Regulatory Proteins 1 and 2 (ESRP1 and ESRP2) are coordinators of an epithelial cell type-specific splicing program.
INTRODUCTION
Alternative splicing generates multiple mRNAs from a single gene transcript, greatly expanding proteomic diversity in complex organisms. Regulation of splicing is achieved by auxiliary cis-elements that bind regulatory proteins that enhance or silence splicing of adjacent exons (Black, 2003; Matlin et al., 2005). Most known splicing regulators are RNA binding proteins (RBPs), such as the SR and hnRNP family of proteins, that are expressed fairly ubiquitously, albeit with some differences in expression between tissues (Hanamura et al., 1998). Modulation of these proteins’ activity and subcellular localization by post-translational modifications contributes to cell type-specific splicing decisions (Allemand et al., 2005; Stamm, 2008). Post-transcriptional regulation of the mRNAs encoding RBPs by microRNAs can also influence cell type-specific splicing decisions (Boutz et al., 2007; Makeyev et al., 2007). Furthermore, alternatively spliced exons are subject to combinatorial control by numerous splicing regulators with both negative and positive effects on splicing (Black, 2003; Smith and Valcarcel, 2000). A very limited number of cell type-specific mammalian splicing factors have been identified that are cell type-specific. Nonetheless, the identification of additional splicing regulatory proteins with distinct cell type-specific differences in expression remains an elusive goal.
An alternative splicing event with well-defined cell type-specificity and functional consequences is the choice between mutually exclusive exons IIIb and IIIc of fibroblast growth factor receptor 2 (FGFR2) (Figure 1A). The FGFR2-IIIb splice variant is exclusive to epithelial cells, while FGFR2-IIIc is mesenchymal, and the resulting receptors have distinct differences in ligand binding specificity (Orr-Urtreger et al., 1993; Zhang et al., 2006). The compartment specific expression of these FGFR2 splice variants and their ligands is essential for regulation of cell proliferation and differentiation during development (Eswarakumar et al., 2005). Previous studies identified a number of auxiliary cis-elements and RBPs that regulate FGFR2 exon IIIb and exon IIIc splicing (Hovhannisyan and Carstens, 2007; Mauger et al., 2008) and references therein). To identify additional splicing factors that promote FGFR2-IIIb expression we carried out a genome-wide, high-throughput cDNA overexpression screen. Among the previously uncharacterized splicing regulators identified using this approach were epithelial cell type-specific splicing regulators.
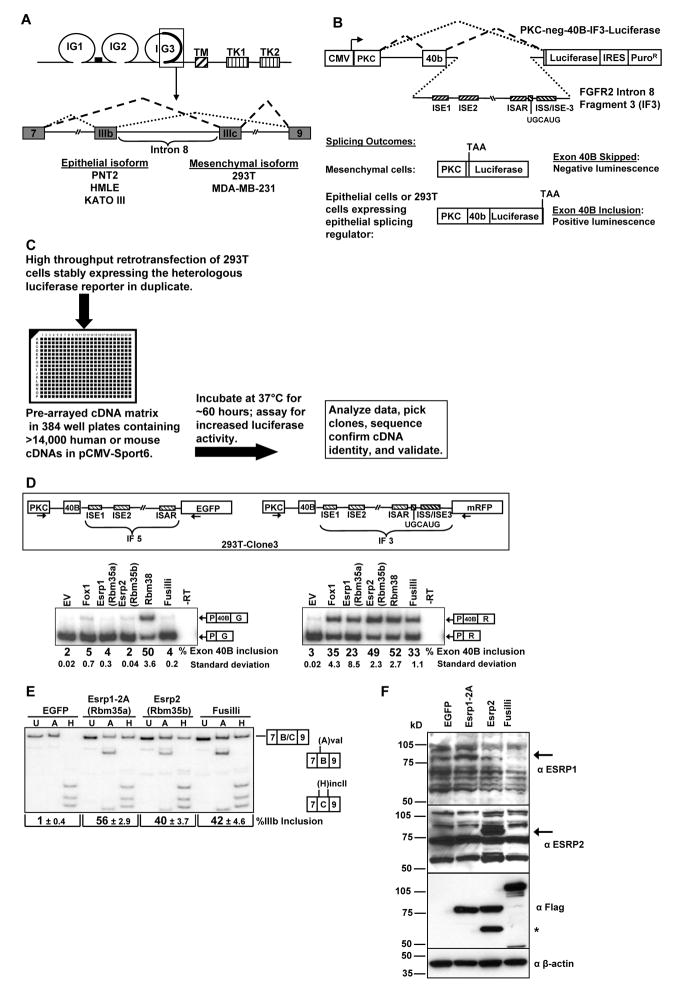
Figure 1. Identification of FGFR2 splicing regulators Rbm35a (Esrp1) and Rbm35b (Esrp2) in a high throughput cDNA expression screen.
(A) Schematic of the FGFR2 protein (top) and the pre-mRNA in the region encoding exons IIIb and IIIc. IG=immunoglobulin-like domains. TM=transmembrane domain. TK=tyrosine kinase domains.
(B) Schematic of the reporter minigene and screening strategy. An FGFR2 intron 8 fragment (IF3) required for IIIb inclusion in epithelial cells is positioned downstream of a heterologous exon. Auxiliary intronic cis-elements are indicated by hatched boxes.
(C) Flowchart summarizing the cell-based screen.
(D) Schematic of the minigenes stably expressed in 293T-clone 3 (top). Cells were transiently transfected with empty vector (EV) or cDNAs for Fox-1, Rbm35a (Esrp1), Rbm35b (Esrp2), Rbm38, or Fusilli and exon inclusion determined by RT-PCR. Average percentages of exon 40B inclusion with standard deviations compiled from three experiments are indicated below a representative gel. The Rbm35a, Rbm35b, and Rbm 38 cDNAs represented here are the MGC clones from the screening collection.
(E) 293T cells were transduced with pMXs-based retroviruses containing cDNAs for Rbm35a (Esrp1), Rbm35b (Esrp2), Fusilli, or EGFP. The Rbm35a cDNA used here is a full length clone containing alternative exons 14 and 15 (2A). FGFR2 splice variant analysis was determined by an RT-PCR protocol in which products are digested with AvaI (A) or HincII (H) which specifically cut exon IIIb and exon IIIc containing products, respectively. U indicates undigested PCR products. Percent exon IIIb inclusion is calculated as Exon IIIb Product (lane H)/(Exon IIIb + Exon IIIc (lane A)) product. The average percentage of IIIb inclusion with standard deviations was compiled from three experiments.
(F) Expression of Esrp1, Esrp2, and Fusilli protein in cells from (E) as determined by immunoblotting with anti-FLAG and anti- ESRP1 (RBM35A) and anti-ESRP2 (RBM35B) antibodies. β-actin is used as a loading control. Arrows indicate the respective protein bands. The asterix represents a variant of Esrp2 suspected to result from use of an alternative translational start site.
RESULTS
Identification of splicing regulatory proteins using a high throughput cDNA expression screen
We used luciferase-based reporter minigenes to carry out a high throughput cDNA expression screen for factors that promote the epithelial pattern of FGFR2 splicing. A heterologous minigene, PKC-neg-40B-IF3-Luc consists of a 40 nt exon (40B) whose inclusion in spliced transcripts is required for translation of the luciferase coding sequence (Figure 1B). This exon is included in ~3% of spliced transcripts in stably transfected cells. Insertion of an FGFR2 intron 8 fragment (Intron Fragment 3: IF3) confers epithelial cell type-specific splicing enhancement to the heterologous exon and a corresponding increase in luciferase activity ((Hovhannisyan et al., 2006) and data not shown). As 293T cells express the mesenchymal FGFR2-IIIc isoform we generated a 293T cell clone that stably expresses the PKC-neg-40B-IF3-Luc minigene and employed a reverse transfection approach to screen ~15,000 cDNAs within the Mammalian Gene Collection (MGC) library for factors that promote exon 40B inclusion (Figure 1C). Using a median six-fold increase in luciferase activity across two duplicate wells as the cutoff we identified a total of 28 cDNAs in the screen, corresponding to 22 unique genes (Table 1). Four of these hits were false positives, but the remaining 18 cDNAs enhanced luciferase activity and increased exon 40b splicing (Figure S1A, B). Satisfyingly, 15 of the 18 validated hits encoded RBPs, including eight known mammalian splicing regulators.
Table 1.
cDNAs that promoted a ≥6-fold change in luciferase activity in the screen.
| Gene Symbola | FoldChange in Screen | MGC Clone Number | RNA Binding Domain | Validated Change in Splicing |
|---|---|---|---|---|
| Zgpat (1)b | 30.4 | BC021513.1 | CCCH Zinc finger, G-Patch | Yes |
| Rbm38 | 26.6 | BC006687.1 | RRM | Yes |
| Esrp1(Rbm35a) | 25.5 | BC031468.1 | RRM | Yes |
| Zgpat (2)b | 14.1 | BC027218.1 | CCCH Zinc finger, G-Patch | Yes |
| SFRS3b,c | 13.5 | BC000914.1 | RRM | Yes |
| Cugbp2c | 12.5 | BC026856.1 | RRM | Yes |
| SEPT9 | 12.0 | BC054004.1 | Yes | |
| Vgll4 | 11.4 | BC048841.1 | RRM | No |
| BOLL | 11.0 | BC033674.1 | RRM | Yes |
| Sfrs3b,c | 10.6 | BC071196.1 | RRM | Yesd |
| Esrp2 (Rbm35b) | 10.6 | BC031444.1 | RRM | Yes |
| Khdrbs1c | 10.5 | BC002051.1 | KH Domain | Yes |
| RBM4b,c | 9.5 | BC032735.1 | RRM | Yes |
| Rcc1 | 9.2 | BC019807.1 | Yes | |
| Prkacab | 8.6 | BC054834.1 | Yes | |
| Brunol4c | 8.4 | BC048405.1 | RRM | Yes |
| Ssb | 8.4 | BC003820.1 | RRM | Yes |
| Rbm4bb,c | 8.1 | BC019488.1 | RRM | Yes |
| BRUNOL5c | 7.8 | BC028101.1 | RRM | Yes |
| RNPS1 | 7.7 | BC108316.1 | RRM | Yes |
| Zcchc10 | 7.4 | BC025078.1 | CCHC Zinc finger | Yes |
| SNRP70c | 6.8 | BC001315.1 | RRM | Yes |
| PRKACAb | 6.8 | BC039846.1 | Yes | |
| ZGPATb | 6.4 | BC032612.1 | CCCH Zinc finger, G-Patch | Yes |
| WNT1 | 6.3 | BC005449.1 | No | |
| Pspc1 | 6.1 | BC026772.1 | RRM | No |
| Zbtb7c | 6.1 | BC018187.1 | C2H2 Zinc finger | No |
| Zgpat (2)b | 6.0 | BC027218.1 | CCCH Zinc finger, G-Patch | Yes |
All caps denotes a human cDNA, lower case represents mouse cDNAs.
Genes represented by multiple, independent cDNAs or transfections.
Known regulators of mammalian splicing.
Not tested directly, but inferred from the results obtained with human SFRS3.
Two paralogous splicing factors identified in the screen, Rbm35a and Rbm35b, require the FGFR2 auxiliary cis-element ISE/ISS-3 to modulate splicing of the reporter minigene
Subsequent validations showed that most of the factors identified in the screen did not require FGFR2 intron 8 sequence elements to enhance exon 40B splicing (Figure S1B). Among the seven cDNAs that demonstrated a dependence on FGFR2 intron 8 sequences, the two with the most robust enhancement of splicing were two RNA Recognition Motif (RRM) containing proteins, Rbm35b and Rbm38. A paralog of Rbm35b, Rbm35a, was also a hit in the screen, and we therefore directed further investigation into these three gene products. Rbm38 (RNPC1) has not been implicated in mammalian splicing, although a C. elegans ortholog, SUP-12, regulates splicing of the worm FGFR Egl-15 (Kuroyanagi et al., 2007). Rbm35a and Rbm35b are also previously uncharacterized mammalian splicing factors, but are also orthologous with a C. elegans splicing regulator, sym-2 (Barberan-Soler and Zahler, 2008). In previous work we characterized an auxiliary cis-element, ISE/ISS-3 (Intronic Splicing Enhancer/Intronic Splicing Silencer-3) that functions specifically in epithelial cell types to enhance splicing of the upstream exon IIIb and silence the downstream exon IIIc (Hovhannisyan and Carstens, 2005). ISE/ISS-3 is located in intron 8 downstream of a UGCAUG motif that is a binding site for the Fox family of splicing regulators. Binding of Fox-2 to this element has been shown to play an important role in FGFR2 splicing regulation (Baraniak et al., 2006). In order to determine whether Rbm35a, Rbm35b, or Rbm38 require ISE/ISS-3 and/or the UGCAUG motif, we transfected these cDNAs in a 293T cell clone containing a previously described mRFP-based heterologous minigene that also contains the full FGFR2 Intron Fragment 3 (IF3) (Newman et al., 2006). The same cell clone also stably expresses a control EGFP-based minigene in which ISE/ISS-3 and the UGCAUG motif have been deleted (Intron Fragment 5: IF5). As predicted, transient transfection of a cDNA for Fox-1 enhanced exon inclusion only in the minigene that contained its binding site (Figure 1D). Rbm38 promoted a similar degree of exon 40B splicing from both minigenes, suggesting that it did not bind to or require ISE/ISS-3 or the UGCAUG motif and most likely interacts with upstream FGFR2 sequence elements (Figure 1D). However, the ability of Rbm35a and Rbm35b to promote splicing was dependent upon the presence of ISE/ISS-3 and/or the UGCAUG motif to enhance exon 40B splicing. A cDNA encoding the D. Melanogaster ortholog for Rbm35a and Rbm35b, Fusilli, achieved a similar splicing outcome.
Ectopic expression of Rbm35a or Rbm35b causes a switch in endogenous FGFR2 splicing from the mesenchymal to epithelial isoform
Transduction of 293T cells with retroviral vectors encoding FLAG-tagged cDNAs for Rbm35a or Rbm35b induced a substantial switch in splicing of endogenous FGFR2 transcripts from exon IIIc to exon IIIb splicing (Figure 1E). Immunoblotting with anti-FLAG, anti-RBM35A, and anti-RBM35B antibodies confirmed that this switch corresponded with expression of proteins of the size predicted for full length Rbm35a and Rbm35b (Figure 1F). Transduction with virus expressing a cDNA for D. melanogaster Fusilli also caused a switch in endogenous FGFR2 splicing. The MGC Rbm35a cDNA clone used in the previous transient transfection (Figure 1D) is truncated near the 5′ end. In Figure 1E and subsequent studies we used full length Rbm35a cDNAs, including two different splice variants, that displayed greater splicing activity than either the truncated clone of Rbm35a or Rbm35b (Figure S2).
Rbm35a and Rbm35b are epithelial cell type-specific splicing regulatory proteins (ESRPs)
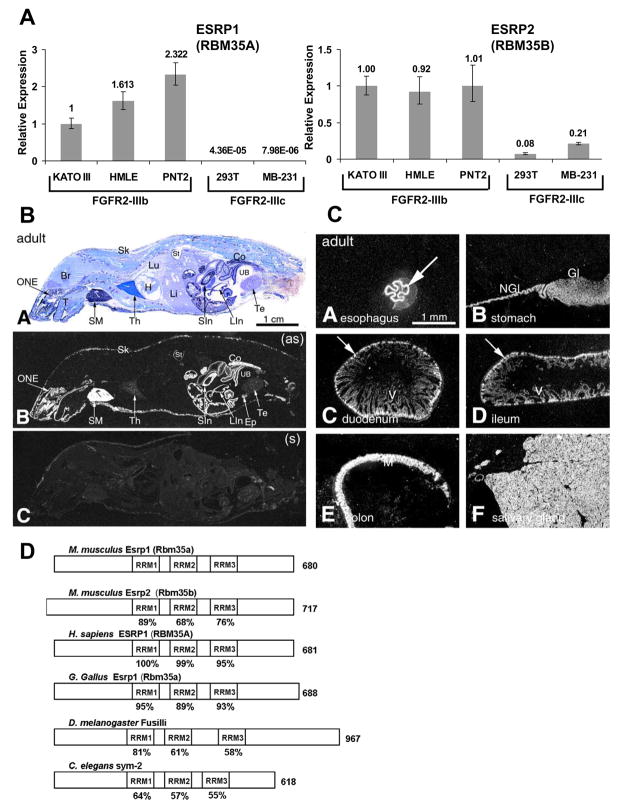
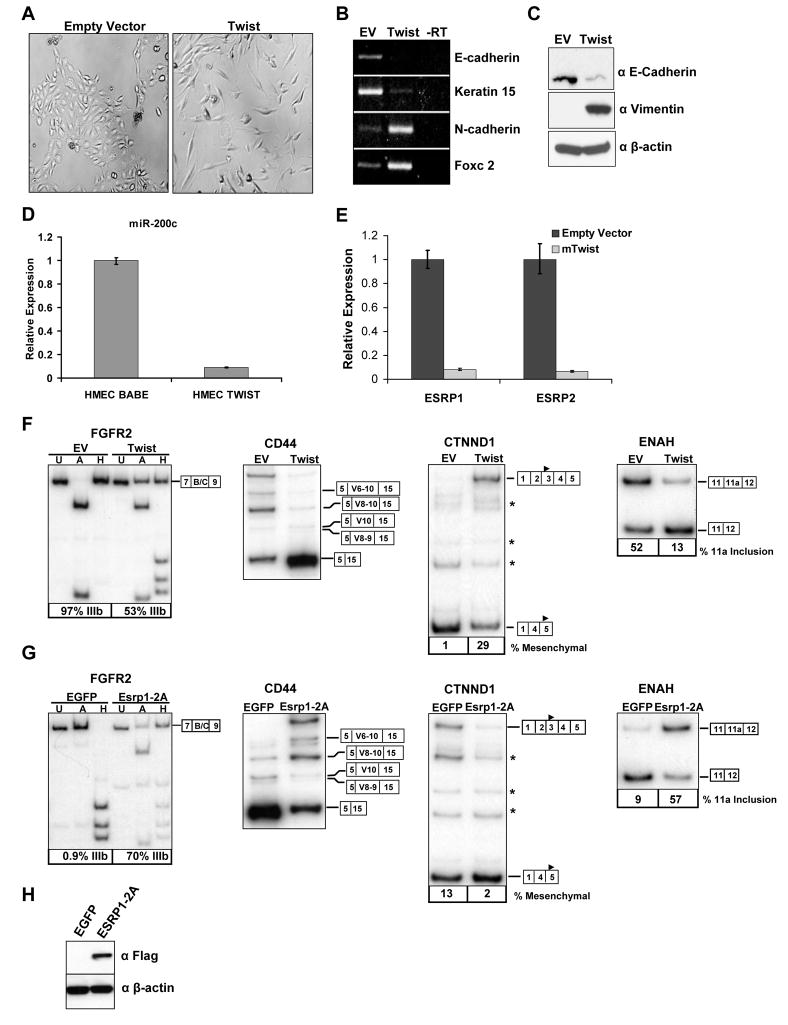
Analysis of RBM35A and RBM35B mRNA expression in a panel of cell lines available in the lab demonstrated that expression of both genes correlated with the epithelial FGFR2-IIIb isoform, with very low to undetectable expression in FGFR2-IIIc expressing cell lines (Figure 2A and Figure S3). Furthermore, microarray data from the NCI60 panel of cell lines revealed that cell lines classified as “epithelial” based on the E-cadherin/vimentin protein ratio expressed substantially higher levels of RBM35A and/or RBM35B than cells classified as “mesenchymal” (Figure S4) (Park et al., 2008). A number of these “epithelial” cell lines have been confirmed to express FGFR2-IIIb in the data presented here as well as in a previous study (Figure S3B, G) (Cha et al., 2008). Epithelial specific expression of Rbm35a within brain sections was previously shown by in situ hybridization analysis in mice (McKee et al., 2005). To more comprehensively examine Rbm35a mRNA expression, we carried out in situ hybridization analysis of whole P1 and adult mouse tissue sections as well as panels of tissues with defined epithelial cell layers. These studies revealed distinct epithelial-specific expression in diverse tissues and organs with particularly notable levels of expression in skin and gastrointestinal epithelia (Figure 2B, C and Figures S5–8). Evidence that Rbm35b expression is also epithelial-specific in mouse embryos is available in an online in situ hybridization database (Visel et al., 2004).
Figure 2. Rbm35a and Rbm35b expression is epithelial cell type-specific.
(A) Quantitative RT-PCR analysis of ESRP1 and ESRP2 expression in a set of human cell lines used in this study. Mean expression values from triplicate assays ± SD are shown relative to KATO III.
(B) In situ hybridization analysis showing epithelial-specific expression in diverse mouse tissues and organs. Panel A = Whole-body section of an adult mouse with cresyl violet staining. Panel B = X-ray film autoradiography detection of Rmb35a mRNA, seen as bright labeling. High-level labeling is observed in the olfactory neuroepithelium, salivary glands, stomach, small and large intestine and skin. Abbreviations: Br –brain; Co – colon; Ep – epididymis; H – heart; Li – liver; LIn – large intestine; SIn – intestine; Lu – lung; ONE –olfactory neuroepithelium; Sk – skin; SM – submaxillary gland; St – stomach; T – tongue; Te – testis; Th –thymus; UB – urinary bladder; (as) – antisense; (s) – sense (Panel C).
(C) In situ hybridization analysis in the adult GI tract. Arrows indicate epithelial expression in defined epithelial layers. Gl – glandular region of the stomach wall; M – mucosa; NGl – non-glandular region of the stomach wall; V – villi.
(D) Schematic presentation of ESRP orthologues with the three RRM domains boxed and length in amino acids on the right. The percent identity of the RRM domains relative to mouse Esrp1 is indicated below the boxes.
The mammalian ESRPs and their orthologs in chicken, D. melanogaster, and C. elegans contain three RNA Recognition Motifs (RRMs) and display significant phylogenetic sequence conservation within these domains, particularly RRM1 (Figure 2D and Figure S9). Our demonstration that D. Melanogaster Fusilli can regulate endogenous FGFR2 splicing provides the first evidence that this fly protein is a splicing factor. Furthermore, its ability to substitute for the mammalian protein in human cells suggests that these orthologs bind similar RNA target sequences and interact with conserved components of multiprotein splicing regulatory complexes. Epithelial-specific expression of Fusilli in the stomodeum and proctodeum was previously noted by in situ hybridization analysis of fly embryos (Wakabayashi-Ito et al., 2001). Collectively, these data provide strong evidence that these factors are evolutionarily conserved epithelial cell type-specific splicing proteins. We therefore propose the names Epithelial Splicing Regulatory Proteins 1 and 2 (ESRP1 and ESRP2) to replace the generic gene symbols RBM35A and RBM35B, respectively.
The ESRPs are required for expression of FGFR2-IIIb
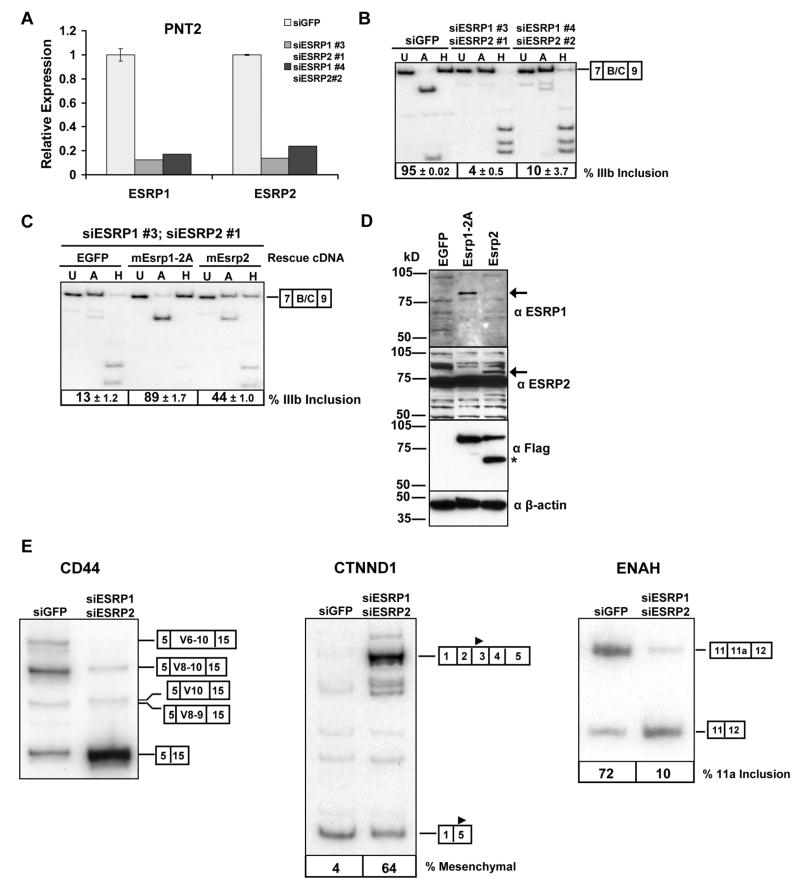
To determine whether expression of ESRP1 or ESRP2 is required for expression of epithelial FGFR2-IIIb we performed RNA interference (RNAi) to deplete these gene products in the human prostatic epithelial cell line PNT2, in which we could achieve effective knockdown (Figure 3A). Two separate combinations of effective siRNAs against ESRP1 and ESRP2 caused a significant switch from exon IIIb to exon IIIc splicing in the endogenous FGFR2 transcript (Figure 3B). To further establish that this switch was due to the loss of ESRP1 and ESRP2 expression, we carried out a “rescue experiment” with mouse cDNAs for Esrp1 and Esrp2 that were immune to knockdown by these human siRNAs. PNT2 cells were transduced with an EGFP control or vectors containing the cDNAs encoding FLAG-tagged mouse Esrp1 or Esrp2. The cells were subsequently transfected with siRNAs against human ESRP1 and ESRP2 and endogenous FGFR2 isoform analysis was performed. In contrast to the controls, cells transduced with RNAi resistant cDNAs for Esrp1 maintained predominant FGFR2-IIIb expression (Figure 3C). However, the cDNA for Esrp2 only partially preserved FGFR2-IIIb expression when both endogenous factors were depleted. Independent knockdown of ESRP1 in PNT2 cells caused a partial switch towards FGFR2-IIIc splicing whereas knockdown of ESRP2 alone caused no change in FGFR2 splicing (data not shown). Thus, at least in these cells, it appears that the preservation of the FGFR2-IIIb splicing pathway is more dependent on ESRP1 than ESRP2. Expression of the FLAG-tagged Esrp1 and Esrp2 protein in these experiments was verified by immunoblotting with anti-FLAG antibodies as well as antibodies against each protein (Figure 3D). These data thus provide fairly conclusive evidence that the expression of at least one of these epithelial cell type-specific protein paralogs is required for FGFR2-IIIb expression.
Figure 3. ESRP1 and/or ESRP2 is necessary for expression of FGFR2-IIIb and the regulation of CD44, CTNND1, and ENAH splicing.
(A) Quantitative RT-PCR showing efficient knockdown of ESRP1 and ESRP2 in PNT2 cells using two different combinations of siRNAs targeting each transcript compared with control siRNAs against EGFP. Mean expression values from triplicate assays ± SD are shown relative to the EGFP control.
(B) ESRP1 and ESRP2 knockdown by RNAi causes a switch in endogenous FGFR2 from exon IIIb to exon IIIc splicing. Average percentages of exon IIIb inclusion with standard deviations from three transfections are indicated below a representative RT-PCR.
(C) PNT2 cells were transduced with cDNAs for EGFP, mouse Esrp1, or Esrp2, and subjected to siRNAs against human ESRP1 and ESRP2. FGFR2 splice variant analysis from three experiments is shown along with a representative RT-PCR gel.
(D) Expression of Esrp1, Esrp2, and Fusilli protein in cells from (C) as determined by immunoblotting with anti-FLAG and anti- ESRP1 (RBM35A) and anti-ESRP2 (RBM35B) antibodies. β-actin is used as a loading control. Arrows indicate the respective protein bands. The asterix is as described in the legend to Figure 1F.
(E) Depletion of ESRP1 and ESRP2 also causes a switch in splicing of CD44, p120-catenin (CTNND1), and ENAH splicing. The splice variants are indicated by boxes at the right of each gel. The solid triangle in exon 3 or exon 5 of CTNND1 indicates the different translational start sites used in the respective isoforms.
The ESRPs regulate splicing of CD44, p120-Catenin (CTNND1) and hMena (ENAH)
The profound switch in endogenous FGFR2 splicing from the epithelial to mesenchymal isoform upon depletion of ESRP1 and ESRP2 suggested that these cell type-specific factors might regulate additional epithelial-specific transcript variants. Inclusion of several “variable” exons of CD44 transcripts, including exons 8–10 (V8–V10), has been shown to be epithelial-specific (Ponta et al., 2003). Depletion of ESRP1 and ESRP2 in PNT2 cells resulted in a significant decrease in the inclusion of CD44 exons 8–10 and increase in the standard isoform (CD44s) in which all of the variable exons are skipped (Figure 3E). Delta catenin (CTNND1), also known as p120-Catenin, expresses mesenchymal specific splice variants that contain alternative exons 2 and 3 (Keirsebilck et al., 1998). Skipping of these exons in epithelial cells, results in a shorter protein isoform that initiates translation in exon 5. Expression of the mesenchymal p120-Catenin isoform is induced during the epithelial to mesenchymal transition (EMT) (Ohkubo and Ozawa, 2004). Knockdown of ESRP1 and ESRP2 in PNT2 cells also induced expression of the mesenchymal isoform of p120-Catenin (Figure 3E). ENAH contains an alternative exon, 11a, that is predominantly included in epithelial cell lines and skipped in mesenchymal cells (Pino et al., 2008). Knockdown of ESRP1 and ESRP2 led to a significant decrease in ENAH exon 11a inclusion (Figure 3E). These three examples of additional targets of the ESRPs suggest that they regulate a larger number of epithelial vs. mesenchymal splice variants. Furthermore, they illustrate examples of regulated targets in which they can promote epithelial-specific exons (CD44, ENAH), silence mesenchymal exons (CTNND1), or both (FGFR2).
The ISE/ISS-3 auxiliary cis-element is a target binding site for ESRP1 and ESRP2
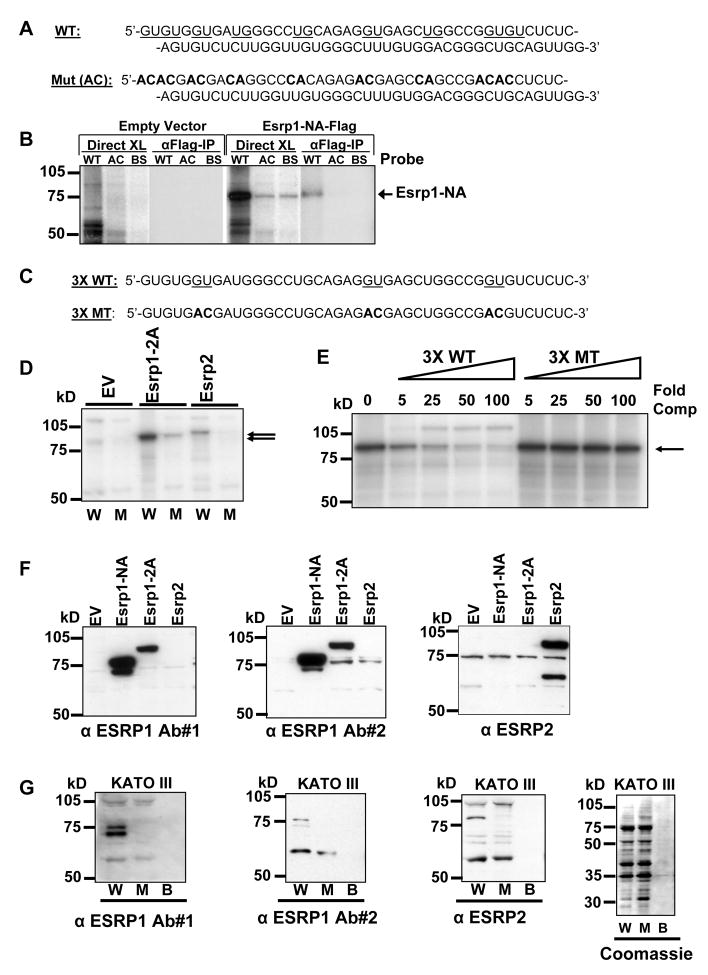
To investigate whether these proteins bound specifically to ISE/ISS-3 we performed UV crosslinking experiments with radiolabelled ISE/ISS-3 RNAs and previously defined functional mutants and control RNAs (Hovhannisyan et al., 2006). The RNAs were incubated with nuclear extracts from 293T cells transiently transfected with cDNAs encoding FLAG-tagged Esrp1 or empty vector. In Esrp1 transfected cells a band of the size predicted for the FLAG-tagged Esrp1 was crosslinked to the wild-type ISE/ISS-3 (WT), but not a mutant in which GU or UG dinucleotides in the 5′ half of the element were replaced with AC (AC), or an unrelated RNA (BS)(Figure 4A, B). Immunoprecipitation with anti-FLAG antibodies confirmed the identity of the crosslinked Esrp1 protein. An RNA sequence containing three tandem copies of the 5′ end of ISE/ISS-3 (3X WT) is nearly equivalent in function to the full length ISE/ISS-3 and three more discrete GU to AC mutations (3X MT) abrogate splicing activity (Figure 4C)(Hovhannisyan and Carstens, 2007). Crosslinking of overexpressed Esrp1 and Esrp2 was observed with the wild-type, but not the functional mutant of this sequence, suggesting that they bind specific GU-rich sequence motifs (Figure 4D). To verify that this difference in crosslinking was due to differential binding, we also carried out competition experiments using 3X WT and 3X MT unlabelled “cold” competitor RNAs. These experiments show that crosslinking of ESRP1 to the wild type sequence is effectively competed by the wild type, but not the mutant, competitor (Figure 4E). We validated that the overexpressed cDNAs produce Esrp1 and Esrp2 protein by immunoblotting of these extracts with anti-ESRP1 and anti-ESRP2 antibodies (Figure 4F). To determine whether endogenous Esrp1 and Esrp2 protein binds to this sequence we carried out RNA affinity pulldown experiments using FGFR2-IIIb expressing KATO III cells, for which preparation of bulk nuclear extracts is feasible. Proteins that bound to beads containing the 3X WT or 3X MT RNAs, or beads alone were analyzed by SDS-PAGE gels and immunoblotting using anti-ESRP1 and anti-ESRP2 antibodies. Immunoreactive bands of the expected size for ESRP1 and ESRP2 were identified in pulldowns from the wild-type, but not the mutant 3X ISE/ISS-3 sequence, or to beads alone (Figure 4G). Coomassie staining showed that similar amounts of total protein bound to both RNA columns. In the case of ESRP1 the same protein was recognized by two different antibodies, providing additional evidence that this protein represents endogenous ESRP1. We had difficulty unambiguously identifying endogenous ESRP1 or ESRP2 by direct Western analysis with these antibodies. These results therefore also allowed us to verify ESRP1 and ESRP2 protein expression in an epithelial cell line. It is noteworthy that both of these proteins eluded detection in mass spectrometry analysis of pulldowns from KATO III nuclear extracts using this same assay. We suspect that promiscuous binding by more abundant RNA binding proteins that bind this sequence masked the identification of peptides corresponding to the ESRPs.
Figure 4. ESRP1 and ESRP2 bind to GU-rich sequences in ISE/ISS-3.
(A) At top, the sequence of ISE/ISS-3 (WT) is shown and the sequences that were mutated to AC or CA are underlined. The resulting mutant sequence (Mut (AC)) is shown at bottom.
(B) Rbm35a binds specifically to ISE/ISS-3 (WT), but not to the mutated sequence (AC) or unrelated RNA (BS). Results of direct crosslinking (Direct XL) and crosslinking followed by IP of the FLAG-tagged proteins are shown.
(C) At top the sequence of the wild-type 5′ half (43 nt) of ISE/ISS-3 present as 3 tandem copies in the 3X WT sequence. A more discreet set of 3 GU to AC mutations is underlined. The resulting sequence present as three tandem copies in the 3X MT mutant sequence is shown at bottom.
(D) Esrp1 and Esrp2 bind three tandem copies of the 5′ half of ISE/ISS-3 in a sequence-specific manner. UV crosslinking was performed using nuclear extracts from 293T cells transiently transfected with FLAG tagged cDNAs for the Esrp1-2A, Esrp2, or empty vector (EV). Arrows indicate the crosslinked bands. W=3X WT sequence. M=3X MT sequence. kD=Sizes of molecular weight markers.
(E) UV crosslinking competition using nuclear extracts from 293T cells transiently transfected with Esrp1-2A. Extracts were incubated with radiolabelled 3X WT RNAs and unlabelled 3X WT or 3X MT competitor RNAs with the molar fold-excess of competitor (Fold comp) indicated.
(F) Nuclear extracts from 293T cells transiently transfected with cDNAs for two splice variants of Esrp1, Esrp2 and empty vectors immunoblotted with antibodies against endogenous Esrp1 and Esrp2. Two different antibodies against Esrp1 specifically detect the overexpressed protein.
(G) RNA pull-down assays from KATO III cell nuclear extracts show sequence-specific binding of endogenous ESRP1 and ESRP2 to the 3X WT (W), but not 3X mutant (M). Proteins eluted from beads containing the indicated RNAs or beads alone (B) were immunoblotted with antibodies against ESRP1 or ESRP2. A Coomassie stain showing total protein content in the RNA pulldown assay is also shown.
Downregulation of the ESRPs coincides with loss of epithelial splicing during the EMT and ectopic expression of ESRP1 in mesenchymal cells restores an epithelial splicing program
A switch from FGFR2-IIIb to FGFR2-IIIc has been shown during the epithelial to mesenchymal transition (EMT), a developmental process associated in pathophysiological conditions with fibrosis and cancer metastasis (Thiery and Sleeman, 2006). Changes in CD44 and CTNND1 splicing also occur in models of EMT and the same changes in splicing occur upon knockdown of the ESRPs. We therefore investigated whether ESRP expression is lost in a human mammary epithelial cell line, HMLE, during the induction of an EMT by the transcription factor Twist (Yang et al., 2004). Analysis of FGFR2 splicing following Twist-induced EMT showed a partial switch from FGFR2-IIIb to FGFR2-IIIc that coincided with a morphological EMT, decrease in epithelial markers, and increase in mesenchymal markers (Figure 5A, B, C, and D). These changes were associated with downregulation of the mRNAs for ESRP1 and ESRP2 (Figure 5E). We also noted changes in CD44, CTNND1, and ENAH splicing that were similar to those seen after knockdown of ESRP1 and ESRP2 in PNT2 cells (Figure 5F). Induction of an EMT in the same cell line upon knockdown of E-Cadherin by RNAi is also associated with downregulation of both ESRP1 and ESRP2 (Onder et al., 2008). Using published microarray data from this experiment, we examined ESRP expression during the EMT along with that of previously published epithelial and mesenchymal markers using gene cluster analysis. We also performed a similar analysis using microarray data from a breast cancer model in which MCF10F cells selected for invasive properties in vitro underwent an EMT. A switch from epithelial to mesenchymal CD44 splicing was previously observed in this model of the EMT (Huang et al., 2007). In both analyses ESRP1 and ESRP2 clustered with epithelial markers and correlated inversely with expression of mesenchymal markers during the EMT (Figure S10). Furthermore, ESRP1 and ESRP2 mRNA expression correlated highly with that of E-Cadherin in the microarray analyses of the NCI60 panel of cell lines (Table S1). These results suggest that a decrease in ESRP expression may be a general feature of the EMT and that the ESRPs are a component of an epithelial gene signature.
Figure 5. Expression of ESRP1 and ESRP2 is abrogated during the epithelial to mesenchymal transition (EMT).
(A) The morphologies of the HMLE cells expressing either the empty pBabe-Puro or pBabe-Puro-Twist by phase contrast microscopy.
(B) Expression of epithelial genes E-cadherin and Keratin 15 and mesenchymal genes N-cadherin and Foxc2 by RT-PCR of RNAs from HMLE cells expressing either the control vector (EV) or Twist. Mean expression values from triplicate assays ± SD are shown relative to control vector.
(C) Immunoblots confirming Twist induced downregulation of E-Cadherin protein and upregulation of the mesenchymal Vimentin protein. β-actin is included as a loading control.
(D) Downregulation of ESRP1 and ESRP2 mRNA during Twist induced EMT determined by qRT-PCR.
(E) Downregualtion of the epithelial miR-200c during Twist induced EMT as determined by qRT-PCR.
(F) Twist induced EMT causes a change in splicing of FGFR2, CD44, CTNND1, and ENAH. Asterixes indicate additional known minor splice variants.
(G) Ectopic expression of Esrp1 in mesenchymal MDA-MB-231 cells causes opposite changes in splicing of FGFR2, CD44, CTNND1, and ENAH to those that occur in the EMT.
(H) Expression of FLAG-tagged Esrp1 in MDA-MB-231 cells as determined by immunoblotting with anti-FLAG antibodies.
The loss of ESRP expression during the EMT suggested that ectopic expression of the ESRPs in mesenchymal cells would restore an epithelial splicing program. To test this hypothesis we transduced the mesenchymal MDA-MB-231 cell line with virus encoding FLAG-tagged ESRP1 or control EGFP virus. Examination of FGFR2, CD44, CTNND1, and ENAH splicing demonstrated that the expression of ESRP1 caused a switch towards the epithelial splicing pathway for each of these transcripts (Figure 5G). Similar changes in splicing were also observed with ectopic expression of ESRP1 in the mesenchymal MDA-MB-435 cell line (Figure S11).
DISCUSSION
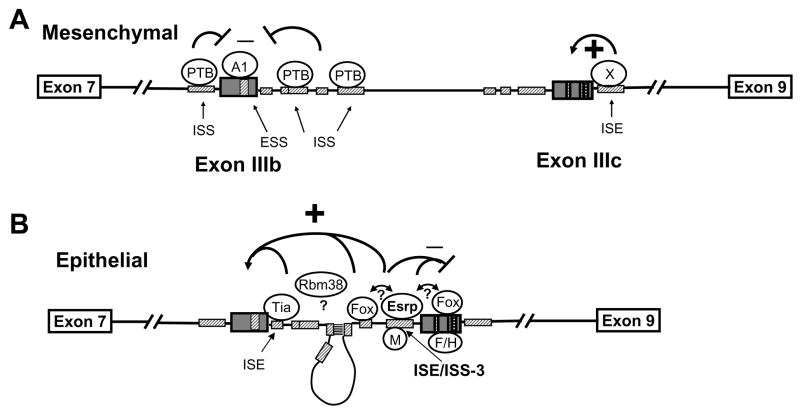
We identified two paralogous epithelial cell type-specific splicing proteins, ESRP1 and ESRP2, that are required for the expression of epithelial FGFR2-IIIb. We also determined that ISE/ISS-3 is an ESRP binding site and mutations that abolish the function of this element abrogate binding. This cis-element was previously shown to enhance splicing of the upstream exon IIIb and silence the downstream exon IIIc and our studies indicate that the ESRPs participate in both of these functions. Fox-2 has also been shown to both enhance exon IIIb and silence exon IIIc splicing and the proximity of a Fox binding site to ISE/ISS-3 suggests that cooperative interactions between these proteins may be required for both functions. Several proteins, including hnRNP M, hnRNP F, and hnRNP H silence exon IIIc but do not directly affect exon IIIb splicing (Hovhannisyan and Carstens, 2007; Mauger et al., 2008). It is noteworthy that hnRNP F/H are close homologs of the ESRPs and both function in FGFR2 splicing regulation (Figure S9). However, while hnRNP F and H bind to G triplets in exon IIIc, mutations in ISE/ISS-3 that abrogate ESRP binding do not directly affect G triplets, suggesting that they have somewhat different binding specificity. Although its binding site in FGFR2 is not defined, co-transfection studies suggest that RBM38 enhances exon IIIb splicing in the absence of effects on exon IIIc splicing (data not shown). Because knockdown of both ESRP1 and ESRP2 in an epithelial cell line caused a switch in endogenous FGFR2 splicing towards the mesenchymal pathway, we propose that in the absence of these proteins, the combinatorial balance of other splicing regulators favors the mesenchymal FGFR2 splicing pathway. The ESRP proteins, however, most likely collaborate with other, more ubiquitously expressed splicing regulators to remodel the RNP complexes assembled on FGFR2 transcripts in epithelial cells and tip the balance towards exon IIIb inclusion and exon IIIc repression (Figure 6).
Figure 6. Model for the mechanism of FGFR2 cell type-specific splicing.
(A) Combinatorial control by ubiquitous regulatory proteins favors exon IIIb silencing and exon IIIc inclusion in mesenchymal cells. Crosshatched boxes indicate known FGFR2 auxiliary cis-elements. PTB=polypyrimidine tract binding protein. A1=hnRNPA1. X= Unknown factor(s) that bind an ISE downstream of exon IIIc.
(B) In epithelial cells the ESRPs collaborate with other regulatory proteins to activate exon IIIb splicing and silence exon IIIc splicing. Potential interactions between these proteins are indicated by double arrows and question marks. M=hnRNP M. Tia=Tia1 or TiaR. Fox=Fox family members. F/H=hnRNP F or H. A base pairing interaction between two complementary sequences in the intron (ISE2 and ISAR) likely serves to position ESRP1 more closely to exon IIIb where it can act in conjunction with other factors (such as RBM38 and Tia1) to activate splicing (Muh et al., 2002). Note that proteins other than the ESRPs shown binding to FGFR2 transcripts only in mesenchymal or epithelial cells are expressed in both cell types and may be bound to the cognate elements in either cell type.
Although a very limited number of cell type-specific splicing regulators have been identified in mammals, there are almost certain to be numerous additional splicing factors with cell type-specific expression that will be found among the large number of uncharacterized RBPs (David and Manley, 2008). To date, the best characterized mammalian cell type- or tissue-specific splicing factors such as Nova, nPTB, Fox-1/2, Muscleblind (MBNL), and CELF family members regulate neural or muscle-specific splicing events (Li et al., 2007; Matlin et al., 2005). Studies of the neural-specific Nova proteins revealed that they co-regulate splicing of numerous targets encoding proteins that function at neuronal synapses (Ule et al., 2005). Studies of the neural and muscle-enriched Fox proteins have similarly shown that their targets are enriched for gene products that function at neuromuscular junctions and that are implicated in neurological and muscular diseases (Zhang et al., 2008). Such examples show that identification of co-regulated splicing events that comprise these splicing regulatory networks can potentially identify proteins that are functionally linked with roles in common cellular and developmental processes (Keene, 2007). The limited examples of co-regulated targets of the ESRPs we have identified here suggest that they similarly co-regulate the splicing of numerous transcripts whose different splice variants have distinct roles in epithelial and mesenchymal cells that contribute to the unique functions and characteristics of each cell type.
Epithelial- and mesenchymal- specific isoforms that are regulated by the ESRPs are likely to participate in epithelial-mesenchymal crosstalk during early vertebrate development and to have important roles in epithelial to mesenchymal transitions during development as well as in disease processes such as cancer metastasis and tissue fibrosis. Interesting examples of coordination among the ESRP targets described here are consistent with these hypotheses. For example, expression of epithelial FGFR2-IIIb in the apical ectodermal ridge (AER) is required for limb induction in response to mesenchyme-derived FGF-10 (Xu et al., 1998). Expression of the epithelial variant of CD44 in the AER was shown to facilitate a reciprocal interaction of AER-derived FGF-8 with receptors in the underlying mesenchyme that most likely include FGFR2-IIIc as well as FGFR1 (Sherman et al., 1998). p120-Catenin (CTNND1) associates with E-cadherin at the plasma membrane and promotes cell-cell adhesion, but paradoxically can also promote cell motility and invasion in cells that have lost E-cadherin expression (Yanagisawa et al., 2008). These seemingly contradictory functions of p120-catenin appear to be due to the different activities of the splice variants that predominate in epithelial vs. mesenchymal cells as the mesenchymal p120-catenin isoforms, but not the epithelial isoforms, promote cell motility and invasion in mesenchymal cells (such as MDA-MB-231) that have lost E-cadherin expression (Yanagisawa et al., 2008). Although there is some controversy regarding the relationship between the EMT and tumor metastasis there are numerous demonstrations that the EMT is one mechanism that can contribute to the metastatic process (Yang and Weinberg, 2008). Our studies showing coordinated changes in splicing that accompany the EMT suggest that the differential functions of these epithelial and mesenchymal splice variants may contribute to metastasis. Enhanced cell motility and invasive capacity mediated by the mesenchymal isoform of p120-catenin is just one example of a splicing change during the EMT that might contribute to metastasis. However, whether these changes in splicing are required for the EMT to occur will require further investigation.
Given the limitations of biochemical approaches, a number of groups have developed fluorescence-based splicing reporter minigenes to establish genetic screens for splicing regulators ((Newman et al., 2006) and references therin). An application of this approach in C. elegans led to the identification of the previously noted SUP-12, whose mammalian ortholog, RNPC1 (RBM38) was not known to be a splicing regulator prior to our study (Kuroyanagi et al., 2007). Mammalian cell-based screens were successful in identification of two factors that regulate Tau exon 10 inclusion through expression cloning with a pooled cDNA library (Kar et al., 2006; Wu et al., 2006). Cell-based screening approaches also recently uncovered hnRNP L-like (hnRNP LL) as a splicing regulator of CD45 in activated T cells (Oberdoerffer et al., 2008; Topp et al., 2008). Further application of genome-wide, high throughput screening approaches has the potential to further expand the number of known splicing regulators. Parallel overexpression of multiple cDNAs at the same time using reverse transfection in a gain-of-function approach has now been scaled up to permit genome-wide screens in high throughput format; an approach that we applied successfully in this study (Luesch, 2006). While the relative merits and liabilities of an RNAi screen versus a cDNA expression screen can be debated, functionally redundant proteins such as ESRP1 and ESRP2 may elude detection in an arrayed RNAi screen. In addition to the ESRPs, we also discovered several additional previously uncharacterized mammalian splicing regulators including RBM38 that also merit further investigation. These studies thus illustrate the potential of mammalian genetic screens in high throughput format to complete a more comprehensive catalog of mammalian splicing regulators.
EXPERIMENTAL PROCEDURES
Plasmids
Minigene PKC-neg-40B-IF3-Luc-Puro was derived from PKC-neg-40B-IF3-EGFP-Puro (Newman et al., 2006) by substitution of a firefly luciferase CDS from pGL4 for EGFP. Additional details of plasmid construction are provided in Supplementary Data.
Cell culture and transfection
293T cells used in the screen and validations were maintained in DMEM with 5–10% FBS and transfected with Transit 293 (Mirus). Details on additional cell lines used are described in the Supplementary Data.
Viral production and transduction
Retroviral transductions were performed using the 293T-N16 packaging cell line. Cells were transfected in 6 cm dishes with 5.7 μg of the retroviral expression vector and 0.3 μg of pCMV-VSV-G using Transit 293. After 16–20 hours the media was replaced with fresh DMEM with 5% FBS and virus was harvested after an additional 24 hours. Target cells were transduced with a 50/50 mix of viral supernatant and growth media. Selection was carried out using 2 μg/ml puromycin or 40 μg/ml blasticidin.
RT-PCR
RNA was isolated using Trizol (Invitrogen). Reverse transcription, PCR, and minigene splicing analysis were performed essentially as described (Newman et al., 2006). The identity of all splice variants was confirmed by DNA sequencing.
Real-time qRT-PCR
1μg of total RNA was used to generate cDNA with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Analysis was performed in triplicate using TaqMan Gene Expression Assays for human RBM 35A and RBM35B, normalized to 18s rRNA (Applied Biosystems). The ddCt method of relative quantification was used on a 7500 Fast Real-time PCR machine with SDS software (Applied Biosystems).
For microRNA qRT-PCR, 7.5ng of RNA was used to generate cDNA using the microRNA Reverse Transcription Kit. MicroRNA expression levels were measured in quadruplicate using the microRNA Taqman Assays for human miR-200c (Applied Biosystems), normalized to RNU48 (snoRNA, C/D box 48).
RNA interference
PNT2 cells were seeded in 12 well plates and transfected with siRNAs twice over a period of 48 hours using Lipofectamine 2000. RNA was extracted 48 hours after the second siRNA transfection. Qiagen siRNAs and Ambion siRNAs were used at final concentrations of 40nM and 10nM, respectively. For the RNAi rescue experiments, PNT2 cells were transduced with pMXs-IRES-blast2 vectors and selected in Blasticidin for 5–7 days prior to transfection with siRNAs.
Antibodies and immunoblotting
Total cell extracts were harvested in RIPA buffer. Immunoblotting was performed as described (Hovhannisyan and Carstens, 2007). Full details on the antibodies are in the Supplementary Data.
UV-crosslinking and immunoprecipitation
The constructs and protocol for UV crosslinking and immunoprecipitation were previously described, but with further modifications as described in the Supplementary Data (Hovhannisyan and Carstens, 2007).
Supplementary Material
Acknowledgments
We thank Kunio Inoue, S. Ananth Karumanchi, Toshio Kitamura, Reini F. Luco, Tom Misteli, Marcus Peter, and Robert Weinberg for reagents, Tom Jongens, Steve Liebhaber, and Rebecca Wells for critical review of the manuscript, Jeanne Geskes for assistance with the high throughput screen, John Tobias for assistance with microarray analysis, Jose Russo for use of microarray data, and Sendurai Mani, Scott Dessain, and Michael Beers for helpful discussions. C.C.W is supported by Genetics Training Grant NIGMS T32-GM08216. This research was supported by the Pennsylvania Department of Health and NINDS R01 NS054794 (J.B.H), DOD grant PC 991539 (R.P.C), NIH grant R01 CA093769 (R.P.C), and a Penn Genome Frontiers Institute Seed Grant (R.P.C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci U S A. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S, Zahler AM. Alternative splicing regulation during C. elegans development: splicing factors as regulated targets. PLoS Genet. 2008;4:e1000001. doi: 10.1371/journal.pgen.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JY, Lambert QT, Reuther GW, Der CJ. Involvement of fibroblast growth factor receptor 2 isoform switching in mammary oncogenesis. Mol Cancer Res. 2008;6:435–445. doi: 10.1158/1541-7786.MCR-07-0187. [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. Rna. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan RH, Carstens RP. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol Cell Biol. 2005;25:250–263. doi: 10.1128/MCB.25.1.250-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan RH, Carstens RP. Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem. 2007;282:36265–36274. doi: 10.1074/jbc.M704188200. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan RH, Warzecha CC, Carstens RP. Characterization of sequences and mechanisms through which ISE/ISS-3 regulates FGFR2 splicing. Nucleic Acids Res. 2006;34:373–385. doi: 10.1093/nar/gkj407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fernandez SV, Goodwin S, Russo PA, Russo IH, Sutter TR, Russo J. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17beta-estradiol. Cancer Res. 2007;67:11147–11157. doi: 10.1158/0008-5472.CAN-07-1371. [DOI] [PubMed] [Google Scholar]
- Kar A, Havlioglu N, Tarn WY, Wu JY. RBM4 interacts with an intronic element and stimulates tau exon 10 inclusion. J Biol Chem. 2006;281:24479–24488. doi: 10.1074/jbc.M603971200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keirsebilck A, Bonne S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol. 2007;27:8612–8621. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Luesch H. Towards high-throughput characterization of small molecule mechanisms of action. Mol Biosyst. 2006;2:609–620. doi: 10.1039/b609384a. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh SJ, Hovhannisyan RH, Carstens RP. A Non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2) J Biol Chem. 2002;277:50143–50154. doi: 10.1074/jbc.M207409200. [DOI] [PubMed] [Google Scholar]
- Newman EA, Muh SJ, Hovhannisyan RH, Warzecha CC, Jones RB, McKeehan WL, Carstens RP. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. Rna. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci. 2004;117:1675–1685. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino MS, Balsamo M, Di Modugno F, Mottolese M, Alessio M, Melucci E, Milella M, McConkey DJ, Philippar U, Gertler FB, et al. Human Mena+11a isoform serves as a marker of epithelial phenotype and sensitivity to epidermal growth factor receptor inhibition in human pancreatic cancer cell lines. Clin Cancer Res. 2008;14:4943–4950. doi: 10.1158/1078-0432.CCR-08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Sherman L, Wainwright D, Ponta H, Herrlich P. A splice variant of CD44 expressed in the apical ectodermal ridge presents fibroblast growth factors to limb mesenchyme and is required for limb outgrowth. Genes Dev. 1998;12:1058–1071. doi: 10.1101/gad.12.7.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. Rna. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi-Ito N, Belvin MP, Bluestein DA, Anderson KV. fusilli, an essential gene with a maternal role in Drosophila embryonic dorsal-ventral patterning. Dev Biol. 2001;229:44–54. doi: 10.1006/dbio.2000.9954. [DOI] [PubMed] [Google Scholar]
- Wu JY, Kar A, Kuo D, Yu B, Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol Cell Biol. 2006;26:6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem. 2008;283:18344–18354. doi: 10.1074/jbc.M801192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.