Abstract
Heat shock genes are highly evolutionarily conserved and are expressed to varying degrees in all organisms in response to stress. Heat shock 70 (hsp70) genes have been well characterized in a number of organisms, most notably Drosophila melanogaster, but not as yet for any of the major arthropod-borne viral mosquito vectors. To identify hsp70 genes in the yellow fever mosquito, Aedes aegypti (Diptera: Culicidae), basic local alignment searches of the Ae. aegypti genome were performed using D. melanogaster Hsp70 protein sequences as query. Two clusters of six previously unannotated AaHsp70 genes were identified and found to be organized into three pairs of nearly identical open reading frames, which mapped to two genomic scaffolds. Consistent with a designation as heat shock genes, no detectable level of expression of AaHsp70 genes was observed under normal rearing conditions (28°C), with robust expression observed with a heat shock of 37–39°C. Northern analysis showed heat-inducible expression of putative AaHsp70 genes at all life stages and in all tissues tested in a time- and temperature-dependent manner. Monitoring of AaHsp70 gene expression levels in field-caught Ae. aegypti may serve as a general marker for stress. In addition, promoter sequences from AaHsp70 genes may be used to control the expression of transgenes in an inducible manner.
Keywords: heat shock protein 70, Aedes aegypti, heat shock
Aedes aegypti is a significant vector of disease agents, capable of transmitting the viruses that cause yellow fever, dengue fever, and the more severe dengue hemorrhagic fever (Tatem et al. 2006). According to the WHO (2008), 2.5 billion people are at risk from dengue, with 50 million dengue infections worldwide each year. In 2007 in the Americas alone, there were 890,000 dengue infections, 26,000 of which were dengue hemorrhagic fever cases (WHO 2008).
Aedes aegypti is one of the most highly studied arthropods, not only for its importance as a vector of disease but also because of its ease to rear in the laboratory (Severson et al. 2004). With sequence data available for both Ae. aegypti and Anopheles gambiae, comparative genomics is an important tool for understanding vector biology. Comparative analysis with Drosophila melanogaster will also lead to better understanding of each of the mosquito genomes (Waterhouse et al. 2008).
Heat shock protein 70 (Hsp70) genes are members of a highly conserved gene family, with proteins related to D. melanogaster hsp70 found in prokaryotes and higher and lower eukaryotes (Craig 1985, Mukhopadhyay et al. 2003). The chaperone proteins encoded by these genes are responsible for preventing the cytotoxic aggregation of denatured proteins, as well as for the refolding of denatured proteins (Bettencourt and Feder 2002). Expression of hsp70 genes is induced by various stresses including heat, anoxia, and exposure to ethanol and chemical treatments including sodium arsenite, cadmium chloride, and sodium salicylate (Craig 1985, Tanguay 1988, Li and Werb 1982, Liu et al. 1994). D. melanogaster hsp70 genes are well characterized and serve as a reference for comparative genomics studies of hsp70 genes of other organisms (Severson et al. 2004, Waterhouse et al. 2008).
DmHsp70 promoter sequences are commonly used to drive the expression of exogenous gene products, to induce transposition events, and also for gene function studies in D. melanogaster (Golic and Lindquist 1989, Wimmer 2003) and heterologous systems such as B. mori (Uhlirova et al. 2002, Dai et al. 2007). The DmHsp70 promoter has also been shown to drive transgene expression in cultured mosquito cells (Zhao and Eggleston 1999). However, for some applications, an endogenously derived promoter may prove to be more useful than a heterologous promoter, particularly with respect to genetically modified mosquito strategies where limiting the amount of non-native DNA is critical.
Aedes aegypti is the best-characterized species within the subfamily Culicinae (Nene et al. 2007). A joint sequencing effort between the Broad Institute and The Institute for Genomic Research (TIGR) resulted in eight-folds coverage of the ≈1.3-Gb Liver-pool genome (Lobo et al. 2007, Nene et al. 2007). Before the completion of the Ae. aegypti genome sequencing project, a putative AaHsp70 gene was used by Fulton et al. (2001) as a marker for single-strand conformation polymorphism (SSCP) analysis based on an unpublished submission to GenBank. Isoe et al. (2007) also identified a putative AaHsp70 gene. This gene, however, was found not to be heat inducible (Isoe et al. 2007), suggesting that it may be a heat shock cognate instead. Heat shock cognates are closely related to heat shock proteins but are constitutively expressed and are not part of the stress response pathway. Last, a gene identified as hsp70 was annotated on genomic scaffold 1.116 and mapped to the p-arm of chromosome 3 (Nene et al. 2007). However, uncertainty remains as to the nature and accuracy of these preliminary annotations. In this paper, we definitively identify and describe the expression pattern of novel hsp70 genes from Ae. aegypti. We also present evidence that all three previous reports of AaHsp70 genes were actually descriptions of a heat shock cognate gene orthologous to D. melanogaster Hsc70-4.
Materials and Methods
Mosquito Strains and Rearing
Aedes aegypti (Liverpool strain) mosquitoes were maintained at 28°C and 80% RH with a photoperiod of 16 h of light and 8 h of darkness. Larvae were fed pulverized fish food and reared ≈300 per pan in 4 liters of reverse osmosis (RO) purified water until pupation. Adult mosquitoes were maintained on sucrose and were blood fed using artificial membrane feeders and defibrinated sheep blood (Colorado Serum Company, Denver, CO).
Amplification and Cloning of AaHsp70 Genes
AaHsp70 genes were amplified from Liverpool strain genomic DNA using Platinum Pfx polymerase chain reaction (PCR; Invitrogen, Carlsbad, CA) (2× Pfx amplification buffer, 0.3 mM dNTPs, 1 mM MgSO4, 0.3 μM primers, 1× enhancer solution, 1 U Platinum Pfx DNA polymerase). Genomic DNA was digested with either SacII or BglII to separate inverted hsp70 gene pairs. All attempts to amplify hsp70 genes from undigested mosquito DNA were unsuccessful, presumably because the nearly identical open reading frames and 5′ regions would be expected to form hairpin structures during the amplification process. Gene regions for putative AaHsp70Ba, -Bb, -Ca, and -Cb were amplified using SacII digested genomic DNA from male Ae. aegypti (94°C, 5 min; 94°C, 30 s, 60°C, 1 min, 68°C, 3 min, 35 cycles; 68°C, 10 min), and putative AaHsp70Aa and -Ab were amplified using BglII digested genomic DNA from male Ae. aegypti. Amplified AaHsp70 genes were cloned using KpnI restriction sites added to the primers used for the original amplification. Primers used to amplify AaHsp70 gene regions are listed in Table 1. Plasmids containing open reading frames from each gene were sequenced using ABI BigDye Terminators v 3.0 (Foster City, CA).
Table 1.
Oligonucelotide primers used to amplify and clone AaHsp70 paralogs
| Primersa | Sequence (5′ to 3′) | Expected length (bp) |
|---|---|---|
| Aa_F | ATCAAGATTGTCGACCAACTAGAAGGACC | 2,398 |
| Aa_R | TTTCTACTATATAAGCGCCCGGTTCG | |
| Ab_F | TCGTTTCCACTATATAAGCGCCCAGC | 2,346 |
| Ab_R | CTGAGTTCGGTATGTTTGAGAATGAGAATG | |
| Ba_F | CGCGTCTTCCAACATTCCTTACTGAACCTA | 2,374 |
| Ba_R | CCACTCACACAAAGCAATGAAATATGCGAC | |
| Bb_F | CGAATCACAAGCAAGAGCAATAAAGCGCC | 2,463 |
| Bb_R | GGCAATTTCGTAGCTCTATTGACATGTCCAG | |
| Ca_F | GGTAGCAGTCGAGTAAGCCAAGACAACGAA | 2,334 |
| Ca_R | AACAAGCTCAACATAATAGGAACATTAACAAACTTCC | |
| Cb_F | CCTGGCGTTTATATATAGGACCGATTCGAGC | 2,374 |
| Cb_R | AAGCATTAGCTTGAGTCGTCAATCTTTAAGATTACTG |
Cloning primers contained the sequence ttttggtacc at the 5′ end of each of the above oligonucleotides to introduce KpnI restriction sites.
Mosquito Heat Shock Regimens
Groups of adult female Ae. aegypti mosquitoes (n = 35–40) were heat shocked in a prewarmed oven ≈3–5 d after eclosion. Each heat shock lasted 1 h at a predetermined temperature ranging from 35 to 41°C in 2°C increments, with ≈80% humidity (35 ± 0.3, 37 ± 0.6, 39 ± 0.8, and 41 ± 0.4°C). Mosquitoes were permitted to rest at 28°C for varying amounts of time ranging from 30 min to 24 h after the heat shock regimen. Once the predetermined resting period was over, mosquitoes were harvested into microcentrifuge tubes and snap frozen in liquid nitrogen to preserve RNA. Third- and fourth-instar larvae and pupae were heat shocked using a 37°C water bath for 1 h and were permitted to rest for 30 min before being snap frozen. Groups of adult males were heat shocked for 1 h at 39°C and allowed to rest for 30 min before being snap frozen. The temperature during heat shock was monitored and verified using a HOBO data logger (Onset Computer, Bourne, MA).
Northern Analysis and Rapid Amplification of cDNA Ends
Total RNA (5 μg) from each experimental group was electrophoresed in a 1.2% agarose, 1× MOPS (0.023 M 3-morpholinopropane sulfonic acid, 0.3 mM NaOAc, 0.2 mM EDTA), and 2% formaldehyde gel at 90 V. RNA was blotted onto a positively charged nylon membrane (Immobilon-NY+; Milllipore, Concord, MA). Blots were prehybridized at 65°C in prewarmed Church’s buffer (0.25 M sodium phosphate buffer, 1 mM EDTA, 7% SDS) using a Fisher Isotemp hybridization oven (Fisher, Pittsburgh, PA). Random primed probes were labeled with [α-32P]dATP (specific activity 3000 Ci/mmol) using the Amersham Megaprime DNA Labeling System (GE Healthcare, Buckinghamshire, United Kingdom). The specific activity of the probes was determined using a Beckman-Coulter LS6500 Multi-purpose Scintillation Counter. Probes were purified using illustra NICK columns (GE Healthcare) and added to prewarmed Church’s buffer to hybridize overnight at 65°C. Blots were washed twice with 2× saline-sodium citrate (SSC) and 0.1% sodium dodecyl sulfate (SDS) for 20 min each at 65°C and twice with 0.2× SSC (0.03 M sodium chloride, 0.003 M sodium citrate) and 0.1% SDS for 20 min each at 65°C.
DNA templates to be radiolabeled as probes for Northern analysis were generated from putative 3′-untranslated regions (UTRs) of AaHsp70 gene paralogs and from sequence common to all 12 putative AaHsp70 genes. The common probe was amplified using One Step RT-PCR (50°C, 30 min, 95°C, 15 min; 94°C, 30 s, 60°C, 30 s, 72°C, 1 min, 35 cycles; 72°C, 10 min) using primers (5′-TTGGTTGATGTGGCTCCACTCTCATTGG-3′ and 5′-TTGTGCTCGAACTCGTCCTTCTCG-3′) and optional Q solution (Qiagen, Valencia, CA). To generate DNA fragments specific for pairs of AaHsp70 paralogs, a portion of each putative 3′ UTR was amplified using One Step RT-PCR and the following primers: AaHsp70Aa (5′-AAGTTGACTAAATTGAGTTGAGATACGAGACTGAATGAG-3′ and 5′-TTGTTATAGTTTATTTTCGTGAAAACATTCTACTTATGATTAC-3′), AaHsp70Ab (5′-AGGAGAAGTGAATGAGACTGAATGTTTTAGTAGAG-3′ and 5′-AACCTTATTCTCTAAGGCTTATGTCAGCAATTCC-3), AaHsp70Ba (5′-AATTGGGTTGACAAACGAGACTGAATGAG-3′ and 5′-TACAACGATAATAATTGCAAATACGATCTACGAATCC-3′), AaHsp70Bb (5′-TTGAGGAAGTCGACTAAAGCGAATGGAGAGG-3′ and 5′-TTCGTAAAAACAAGCTGTACACATTAAATAACTTTCTAAC-3′), AaHsp70Ca (5′-TTGAGGAAGTCGACTAAAGTGAATGGAGCG-3′ and 5′-AACAAGCTCAACATAATAGGAACATTAACAAACTTCC-3′), and AaHsp70Cb (5′-TTGAGGAAGTGGACTAAGTATATCAAGGCATTTAAACCC-3′ and 5′-AAGCATTAGCTTGAGTCGTCAATCTTTAAGATTACTG-3′). Gene fragments were cloned using the same oligonucleotide primers with added KpnI recognition sites in the same manner as the open reading frames. To generate cDNA for use in rapid amplification of cDNA ends (RACE) reactions, the Promega PolyAT-tract mRNA Isolation System was used to purify total RNA extracted from 60 female Liverpool Ae. aegypti. To perform 5′ and 3′ RACE, the Clontech (Mountain View, CA) SMART RACE amplification kit was used with gene-specific primers: for 5′ RACE (5′-CGATTTCCTTGGTCGTTGGCGATGATTTCC-3′, 5′-CGATTTCCCTGGTCGTTGGCGATGATTTCC-3′, 5′-CGATTTCCCTGGTCGTTGGCAATGATTTCC-3′) and for 3′RACE AaHsp70Aa (5′-TGCGGTACAAGCTGCCATCCTCAGTGGAGAC-3′), AaHsp70Ab (5′-CGATCATGACTCGATTGCATCAGGGTGG-3′), AaHsp70Ba (5′-ACGAAAAGCAACGCGAACGTGTCTCTGCC-3′), AaHsp70Bb (5′-CGATCATGACTCGTTTGCATCAAGGTGGAG-3′), AaHsp70Ca (5′-TTCGAGCACAAGATGCAAGAGCTGAGTCG-3′), and AaHsp70Cb (5′-ATCAGCTGGCCAGCAAGGAGGAAATGGACC-3′).
Phylogenetic Analysis
AaHsp70 gene sequences have been deposited in GenBank and are available using accession numbers FJ177309-14 and are listed in Table 2. Other sequences used for phylogenetic analysis include Anopheles gambiae Hsp70 genes AGAP004582 and AGAP012891, as well as AgHsc70B, ENSANGP00000017398; and D. melanogaster genes Hsp70Aa, CG31366; Hsp70Ab, CG18743; Hsp70Ba, CG31449; Hsp70Bb, CG31359; Hsp70Bbb, CG5834; Hsp70Bc, CG6489; and Hsc70-4, CG4264 (Sim et al. 2007). ClustalW parameters used to align Hsp70 protein sequences were pairwise gap opening penalty of 10, pairwise gap extension penalty of 0.1, multiple gap opening penalty of 10, and multiple gap extension penalty of 0.2 with negative matrix. A linearized neighbor-joining tree was produced using MEGA 4.1 (Tamura et al. 2007).
Table 2.
tBLASTn search with the D. melanogaster Hsp70Aa gene product yields 6–12 Ae. aegypti orthologs
| Gene | Supercontig |
Predicted protein |
Best Dma | Accession no. | ||
|---|---|---|---|---|---|---|
| No. | (Position) | No. AA | MW (kD) | |||
| AaHsp70Aa | 1.680 | (378,927–380,840) | 638 | 70.3 | Hsp70Bb | FJ177309 |
| AaHsp70Ab | 1.680 | (383,454–385,367) | 638 | 70.3 | Hsp70Bc | FJ177310 |
| AaHsp70Ba | 1.680 | (402,097–404,010) | 638 | 70.3 | Hsp70Bc | FJ177311 |
| AaHsp70Bb | 1.680 | (406,760–408,673) | 638 | 70.0 | Hsp70B | FJ177312 |
| AaHsp70Ca | 1.680 | (416,064–417,977) | 637 | 70.0 | Hsp70B | FJ177313 |
| AaHsp70Cb | 1.680 | (419,055–420,962) | 636 | 70.0 | Hsp70B | FJ177314 |
| AaHsp70Aa′ | 1.824 | (45,244–47,157) | 638 | 70.2 | Hsp70B | n/a |
| AaHsp70Ab′ | 1.824 | (49,384–51,297) | 638 | 70.3 | Hsp70B | n/a |
| AaHsp70Ba′b | 1.824 | (61,070–63,023) | n/a | n/a | n/a | n/a |
| AaHsp70Bb′ | 1.824 | (66,155–68,064) | 638c | 70.4 | Hsp70Bc | n/a |
| AaHsp70Ca′ | 1.824 | (86,660–88,569) | 638 | 70.1 | Hsp70Bc | n/a |
| AaHsp70Cb′ | 1.824 | (89,650–91,384) | 636 | 70.1 | Hsp70Bc/Bb | n/a |
| AaHsc70–2 | 1.389 | (987,893–989,673) | 587 | 64.4 | Hsc70-2 | n/a |
| AaHsc70–4 | 1.28 | (547,680–549,359) | 593 | 65.2 | Hsc70-4 | n/a |
| AaHsc70–4 | 1.116 | (1,035,564–1,037,399) | 651 | 71.1 | Hsc70-4 | n/a |
| AaHsp68 | 1.752 | (334,685–336,619) | 624 | 68.8 | Hsp68 | n/a |
| AaHsc-3a | 1.191 | (572,103–574,019) | 655 | 72.3 | Hsc70-3 | n/a |
| AaHsc-3b | 1.191 | (574,474–576,459) | 662 | 73.4 | Hsc70-3 | n/a |
Best match in D. melanogaster after BLASTp search of Flybase (FB2008_10).
Not available because of major gaps in the sequence assembly.
Two distinct mutations (nonsense and 1-bp deletion) interrupt the coding sequence of this gene; the data are presented as if the ORF were intact.
AA, amino acid; MW, molecular weight; n/a, not available.
Results
AaHsp70 Gene Organization
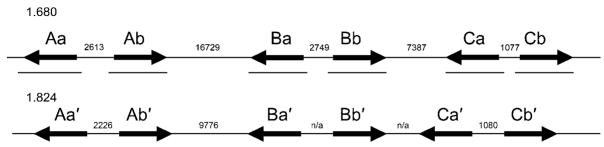
To identify hsp70 genes in Ae. aegypti, we performed tBLASTn searches of the Ae. aegypti genomic scaffolds using the D. melanogaster Hsp70Aa protein sequence. Eighteen putative genes were identified with an expected cut-off value of 0 (Table 2). The predicted protein sequences of 17 of the 18 genes (one sequence contained major gaps that prevented in silico translation) were used to back-query the D. melanogaster gene set, and the best match for each is reported in Table 2. Of the 18 genes, 12 were a best match for D. melanogaster hsp70. Six of the putative AaHsp70 genes mapped to genomic supercontig 1.680 (Table 2; Fig. 1), located on chromosome 1 (Nene et al. 2007), with a second cluster of six putative AaHsp70 genes on the unmapped supercontig 1.824 (Table 2; Fig. 1).
Fig. 1.
Genome organization and relationship of AaHsp70 genes. Arrangement of AaHsp70 genes in Ae. aegypti. AaHsp70 genes are indicated by solid arrows. Lines under each gene indicate areas reamplified, cloned, and sequenced in this project. Distances between genes are labeled in base pairs. n/a, not available due to gaps in sequence assembly.
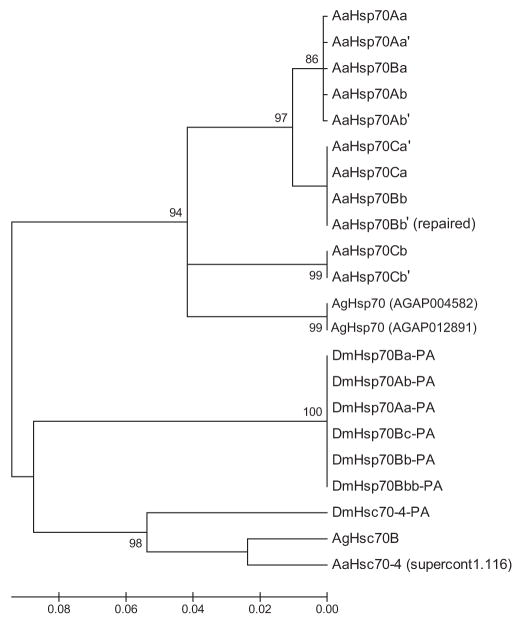
Putative AaHsp70 protein sequences were compared with An. gambiae (AgHsp70) and D. melanogaster (DmHsp70) genes and cognates using ClustalW and a neighbor-joining tree was produced (Fig. 2). The putative AaHsp70 genes formed an independent set of clades from An. gambiae and D. melanogaster hsp70 genes, consistent with the notion that hsp70 genes have undergone several independent duplication events in each of these organisms. To determine whether the sequences previously annotated as AaHsp70 (Fulton et al. 2001, Isoe et al. 2007) corresponded to the same gene sequences we describe here, or to a novel set of genes, we performed BLASTn searches using GenBank deposited nucleotide sequences as a query against the Ae. aegypti genomic supercontigs (Nene et al. 2007). Both DQ453756 (Isoe et al. 2007) and AI658418 (Fulton et al. 2001) were found to map to genomic supercontig 1.116 (positions 1031943–1032601 and 1032458–1035831, respectively). An open reading frame was detected in this region and the predicted protein sequence was used as a query to search the An. gambiae and D. melanogaster genomes using tBLASTn. As shown in Table 2 and Fig. 2, the heat shock–like gene identified on supercontig 1.116 is most closely related to AgHsc70B and DmHsc70–4, suggesting that the most appropriate annotation for this gene is AaHsc70–4.
Fig. 2.
Alignment of Ae. aegypti, An. gambiae, and D. melanogaster hsp70–predicted proteins. Gene sequences listed in the Materials and Methods section were aligned using MEGA 4.1 and bootstrap analysis was performed using the neighbor-joining method with 1,000 replications. Bootstrap support (>80) is listed on the node of each branch, where applicable. AaHsp70Bb′ (repaired) indicates the predicted protein after removing two deleterious mutations.
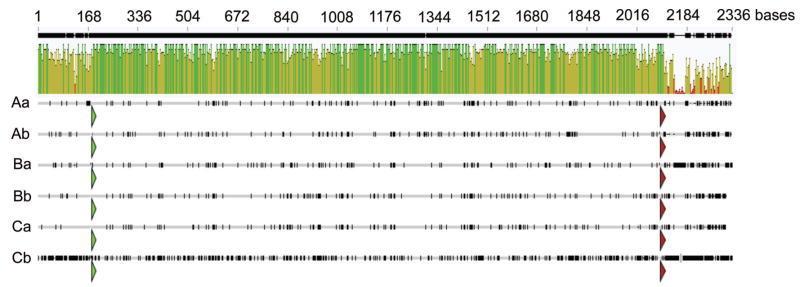
As the two clusters of six AaHsp70 genes were near identical to each other, we focused the rest of our analysis on the cluster located on scaffold 1.680, where the sequence assembly was of higher quality with fewer gaps. To document the exact start and stop of transcription of AaHsp70 transcripts, we performed RACE. Ninety-five percent of 5′ RACE clones (35/37 clones) supported a consistent start of transcription, which was found to be 182 bases upstream of the start of translation for AaHsp70Aa and AaHsp70Cb, 181 bases upstream for AaHsp70Ba, AaHsp70Bb, and AaHsp70Ca, and 180 bases upstream for AaHsp70Ab. Multiple 3′ RACE products were sequenced for each AaHsp70 paralog, with the longest most abundant clone assumed to represent the end of transcription. The 3′-UTRs were found to be far more variable in length than the 5′ region, with AaHsp70Ab (14/21 clones) being the shortest (135 nt 3′ UTR), followed by AaHsp70Ca (4/7 clones, 145 nt), AaHsp70Aa (5/8 clones, 150 nt), AaHsp70Bb (6/11 clones, 150 nt), AaHsp70Ba (3/4 clones, 208 nt), and AaHsp70Cb (9/9 clones, 282 nt). Each of the six putative open reading frames was nearly identical in length, with AaHsp70Aa, -Ab, -Ba, and -Bb at 1,914 nt; AaHsp70Ca at 1,911 nt; and AaHsp70Cb at 1,908 nt. To better resolve the within-cluster phylogenetic relationship of AaHsp70 genes, Geneious software (Biomatters, Aukland, New Zealand) was used to produce a ClustalW alignment of putative AaHsp70 gene transcripts (Fig. 3). Open reading frames of AaHsp70 genes were found to be 77–96% identical at the nucleotide level, with AaHsp70Cb being the most divergent. The 5′ untranslated regions of putative AaHsp70 genes were found to share 88% nucleotide identity, whereas the 3′-UTRs share only 38% identity. Pairwise comparisons between putative AaHsp70 genes showed that AaHsp70Bb and AaHsp70Ca were most similar, with >95% identity at the nucleotide level (Table 3). No other strong pairwise relationship was evident as AaHsp70Aa, AaHsp70Ab, and AaHsp70Ba were essentially equally similar to each other (93.5%), whereas AaHsp70Cb was equivalently divergent from the other five genes (Table 3). This is in agreement with the protein alignment presented in Fig. 1, where three AaHsp70 clades were also identified (Aa, Ab, and Ba; Bb and Ca; and Cb).
Fig. 3.
Sequence comparison of AaHsp70 genes. Complete transcripts for six AaHsp70 genes were aligned using ClustalW (Geneious, Biomatters, Aukland, New Zealand) to produce an identity graph. The start of each open reading frame is marked with a green arrow, and the end with a red arrow. Dark marks on lines representing each AaHsp70 gene indicate base changes from the consensus AaHsp70 sequence.
Table 3.
Nucleotide identity matrix of AaHsp70 open reading frames on supercontig1.680
| Aa | Ab | Ba | Bb | Ca | Cb | |
|---|---|---|---|---|---|---|
| AaHsp70Aa | 1.000 | 0.937 | 0.935 | 0.910 | 0.914 | 0.774 |
| AaHsp70Ab | 1.000 | 0.935 | 0.899 | 0.904 | 0.775 | |
| AaHsp70Ba | 1.000 | 0.908 | 0.909 | 0.783 | ||
| AaHsp70Bb | 1.000 | 0.956 | 0.782 | |||
| AaHsp70Ca | 1.000 | 0.781 | ||||
| AaHsp70Cb | 1.000 |
Expression of AaHsp70 Genes
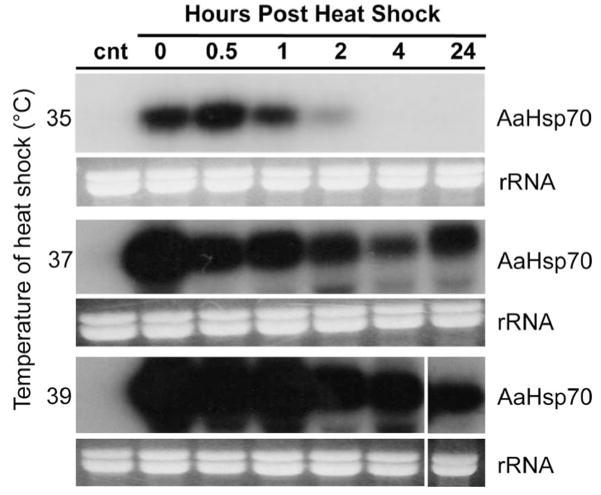
To confirm our bioinformatic annotation of this cluster as true heat shock genes, we next sought to determine whether mRNA expression could be induced through exposure to heat shock. Female Ae. aegypti were exposed to heat shock regimens with temperatures ranging from 35 to 41°C in 2°C increments for 1 h. Total RNA was harvested at various times after exposure to heat shock and was subject to Northern analysis using a probe capable of hybridizing with putative AaHsp70 genes but not the closely related heat shock cognates (Fig. 4). Expression of AaHsp70 mRNAs was induced to various extents in all heat shock regimens, whereas no expression was detected in control mosquitoes not subjected to heat shock (Fig. 4). The highest levels of expression were observed after exposure to 39°C, with only modest induction occurring at 35°C and heat shock transcripts detected only up to 2 h after heat shock. After heat shock at 37°C, AaHsp70 transcripts were detected up to 24 h, although expression did not match that seen at 39°C. AaHsp70 expression at 41°C could not be documented because all mosquitoes died during the 1-h heat shock regimen.
Fig. 4.
Expression levels of AaHsp70 genes increase with heat shock temperature. Female Ae. aegypti were heat shocked at temperatures from 35 to 39°C, and total RNA was hybridized to a 32P-labeled probe common to all AaHsp70 genes. All blots were hybridized simultaneously to ensure equivalent exposure and probe normalization (specific activity = 5.4 × 107 CPM). Control mosquitoes (cnt) were not heat shocked. Ethidium bromide–stained rRNA loading controls are shown below each blot.
Multiple copies of genes with the same function can indicate inactive evolutionary remnants (Ingolia et al. 1980). Thus, it was possible that not all of the AaHsp70 genes we identified were still active or responded to heat shock identically. Indeed, at least one of the AaHsp70 genes (Bb′) contained deleterious frame-shifts. To document the expression levels of each putative AaHsp70 gene, we repeated our heat shock regimen with female Ae. aegypti heat shocked at 39°C for 1 h. After a rest period of 0–4 h, total RNA was extracted and Northern analysis was performed using six paralog-specific probes derived from the unique 3′ UTR of each gene. Of the six putative AaHsp70genes we examined, at least five showed detectable expression by this method, with only AaHsp70Ab failing to produce a detectable hybridization signal (Fig. 5). The highest levels of expression were observed for AaHsp70Aa, AaHsp70Ba, and AaHsp70Ca transcripts, with weak expression seen for AaHsp70Cb (Fig. 5).
Fig. 5.
Northern analysis of individual AaHsp70 paralogs. Northern analysis was performed using a 32P-labeled probe derived from the 3′ untranslated regions of AaHsp70 paralogs. Exposure times were standardized based on specific activity of each probe (7.3 × 107–3.0 × 108 CPM). Mosquitoes were heat shocked at 39°C, and control mosquitoes (cnt) were not heat shocked. Ethidium bromide–stained rRNA loading controls are shown below each blot.
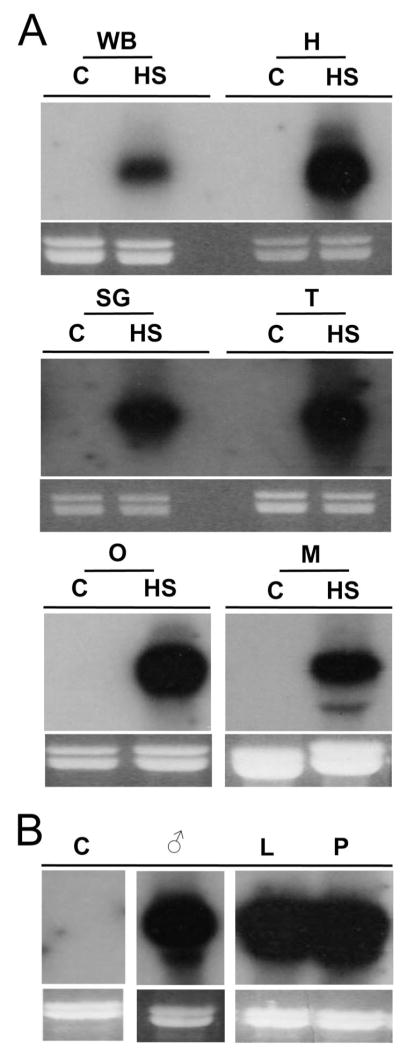
In D. melanogaster, hsp70 genes are expressed ubiquitously throughout the body of the fly after heat shock. To document any tissue specificity of putative AaHsp70 genes, RNA from heat-shocked Ae. aegypti heads, salivary glands, thoraxes lacking salivary glands, ovaries, midguts, as well as from heat-shocked larvae, pupae, and males was subject to Northern analysis using a probe capable of hybridizing with AaHsp70 genes but not the closely related cognates. As expected, robust expression was observed after heat shock in all tissues tested (Fig. 6A), in all life stages, and in male mosquitoes (Fig. 6B).
Fig. 6.
Tissue and life stage expression of AaHsp70 genes. Expression in specific tissues (A) and life stages (B) was analyzed by Northern analysis using a 32P-labeled probe designed to hybridize to all AaHsp70 genes. Tissues were dissections from female Ae. aegypti that had been heat shocked at 39°C for 1 h or kept at 28°C as controls (C). Tissues include whole body (WB), head (H), salivary glands (SG), thorax without salivary glands (T), ovaries (O), and midguts (M). RNA from males (¤), third and fourth instar larvae (L), and pupae (P) heat shocked at 39°C for 1 h was used for the life stage study. Ethidium bromide–stained rRNA loading controls are shown below each blot.
Discussion
The basic arrangement of hsp70 genes in Ae. aegypti closely resembles that of D. melanogaster (Gong and Golic 2004) and An. albimanus (Benedict et al. 1993), with a pair of nearly identical open reading frames organized as an inverted pair. We have successfully identified two clusters of three such pairs in close proximity in the Ae. aegypti genome. Although the presence of these two clusters of six AaHsp70 genes would indicate that this mosquito has up to 12 hsp70 paralogs, the fragmented nature of the genomic assembly does not allow us to rule out the possibility that these two clusters are allelic variants. Phylogenetic analysis of AaHsp70 genes compared with hsp70 genes from D. melanogaster and An. gambiae showed a pattern of divergence in which both Ae. aegypti and An. gambiae genes diverged from D. melanogaster before duplication of the ancestral inverted pair within each species, as predicted by Benedict et al. (1993). DmHsp70 genes are grouped at two loci on the right arm of chromosome 3 with the first inverted pair separated by ≈1.7 Kb, followed by three or four genes at the second locus organized with one gene proximal to the centromere separated by ≈40 Kb from two or three tandem genes of inverted polarity (Craig 1985, Gong and Golic 2004). It is believed that a duplication of the evolutionarily ancient, highly stable two gene cluster lead to the current organization of DmHsp70 genes, with tandem duplications leading to the fifth and sixth genes allowing D. melanogaster greater thermotolerance and niche expansion (Bettencourt and Feder 2001). It is possible that multiple copies of AaHsp70 may also contribute to the distribution and heat tolerance of Ae. aegypti. DmHsp70 genes are separated by large amounts of repetitive DNA resulting from transposable S-elements, and these are thought to have played a role in the duplication of heat shock genes from two to four in Drosophila (Bettencourt and Feder 2001, Evgen’ev et al. 2004, Gong and Golic 2004). The Ae. aegypti genome is highly repetitive, with ≈70% of the genome being transposable elements/repetitive DNA (Nene et al. 2007). At least two duplication events of the ancestral hsp70 gene two gene pair seem to have occurred in Ae. aegypti after divergence from both D. melanogaster and An. gambiae, and these may have occurred by similar mechanisms and for similar reasons. Unlike Drosophila, however, whose two pairs of heat shock genes are separated by >500 kb (Evgen’ev et al. 2004), and An. albimanus, whose two pairs are separated by ≈20 cM (Benedict et al. 1993), all three pairs of AaHsp70 genes were spaced within a span of 42 kb. Because the scaffold 1.824 is unmapped, the spacing between the two clusters of AaHsp70 genes remains unknown. In addition, unlike hsp70 genes described in An. albimanus, Ae. aegypti hsp70 genes were not found to be more closely related within each inverted pair compared with between pairs (Benedict et al. 1993). It is possible that the differences we observed between the AaHsp70 genes are the result of gene inactivation followed by drift rather than being a strict footprint of inheritance. Ultimately, we cannot draw firm conclusions at this point as to how the ancestral two-gene pair evolved into the six gene clusters we observed. Future studies of hsp70 genes from other members of the genus Aedes would likely provide additional insight.
The putative Ae. aegypti heat shock cognate identified as AaHsp70 by Isoe et al. (2007) was mapped to supercontig 1.116 on chromosome 3 (Nene et al. 2007). However, when we compared this sequence to the D. melanogaster genome, the best match is actually a heat shock cognate, a closely related gene that is constitutively expressed and not heat inducible. As shown in Fig. 2, the cognate identified by Isoe et al. (2007) forms a separate clade with cognates from both D. melanogaster and An. gambiae, further supporting the idea that this gene may be a heat shock cognate. Gene annotations are often the result of predictions based on homology before complete genome sequencing. However, examples such as this underscore the importance of confirming such predictions before experimentation.
To balance the benefits of hsp70 gene expression under stress conditions with detrimental effects of expression under normal conditions, DmHsp70 genes have been found to be self-regulating (Craig 1985, Lindquist 1986). DmHsp70 genes are targeted for degradation by signals in the 3′-UTRs, and under normal conditions, hsp70 transcripts are rapidly degraded (Petersen and Lindquist 1989, Feder et al. 1992). In cultured D. melanogaster cells, induced expression of DmHsp70 genes under normal conditions was deleterious to cells and resulted in a decreased growth rate (Solomon et al. 1991, Feder et al. 1992). Extreme temperatures induce long-lasting synthesis and very stable DmHsp70 mRNA, whereas at lower temperatures, expression proceeds until heat shock proteins have accumulated to a level proportional to the severity of heat shock before further transcription is repressed (Lindquist 1986, Nover 1991). AaHsp70 genes were found to exhibit a similar expression pattern, since under extreme heat shock conditions (39°C) high levels of expression are initiated rapidly and transcript was detected up to 24 h after heat shock, whereas expression of hsp70 transcripts was less robust and short lived under a more modest heat shock of 35°C (Fig. 4).
Induced mRNA expression levels were found to vary among the AaHsp70 paralogs we examined. Interestingly, the first gene of each inverted pair seemed to be expressed preferentially over the second (see Figs. 1 and 5), with AaHsp70Aa > AaHsp70Ab, AaHsp70Ba > AaHsp70Bb and AaHsp70Ca > AaHsp70Cb. Also, this expression seemed to be proportional, so that the highest/lowest level of expression was observed from the AaHsp70Aa/Ab pair and the next highest/lowest expression from AaHsp70Ca/Cb, followed by AaHsp70Ba/Bb, which had almost equivalent expression between the two genes. The reasons for this are currently unknown, but may be related to the positioning of replication complexes during transcriptional pausing. In D. melanogaster, RNA polymerase II is constantly bound upstream of the start of transcription to maintain an open chromatin configuration (Karpov et al. 1984, Craig 1985) because heat shock genes must be available for near immediate action under stress conditions. This configuration is likely to occur in Ae. aegypti as well and is consistent with the observation of high levels of AaHsp70 expression detected immediately after 37°C and 39°C heat shocks. Although we could not detect AaHsp70 Ab expression by Northern analysis, RACE transcripts were recovered for this gene, indicating some level of transcription.
AaHsp70 genes are expressed in all life stages and in all tissues tested in response to heat shock. Therefore, isolating heat inducible AaHsp70 promoter elements would be valuable for transgenesis and gene function studies, particularly when it is important to minimize the presence of exogenous sequences. Further studies of AaHsp70 regulatory regions would make such studies possible in Ae. aegypti. Hsp70 gene expression also serves as a marker for stress, and monitoring levels of AaHsp70 gene expression in wild populations could serve as a physiological indicator at the population level.
Acknowledgments
We thank M. Anderson, J. Overcash, M. Wiley, and B. Traver and other members of the Myles/Adelman laboratories for technical assistance. This project was supported by NIAID Grant R21AI071208-01 and by Virginia Tech startup funds to Z.N.A.
References Cited
- Benedict MQ, Cockburn AF, Seawright JA. The Hsp70 heat-shock gene family of the mosquito Anopheles albimanus. Insect Mol Biol. 1993;2:93–102. doi: 10.1111/j.1365-2583.1993.tb00130.x. [DOI] [PubMed] [Google Scholar]
- Bettencourt BR, Feder ME. Hsp70 duplication in the Drosophila melanogaster species group: how and when did two become five? Molec Biol Evol. 2001;18:1272–1282. doi: 10.1093/oxfordjournals.molbev.a003912. [DOI] [PubMed] [Google Scholar]
- Bettencourt BR, Feder ME. Rapid concerted evolution via gene conversion at the Drosophila hsp70 genes. J Molec Evol. 2002;54:569–586. doi: 10.1007/s00239-001-0044-7. [DOI] [PubMed] [Google Scholar]
- Craig EA. The heat shock response. CRC Crit Rev Biochem. 1985;18:239–280. doi: 10.3109/10409238509085135. [DOI] [PubMed] [Google Scholar]
- Dai H, Jiang R, Wang J, Xu G, Cao M, Wang Z, Fei J. Development of a heat shock inducible and inheritable RNAi system in silkworm. Biomolec Engl. 2007;24:625–630. doi: 10.1016/j.bioeng.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Evgen’ev MB, Zatsepina OG, Garbuz D, Lerman DN, Velikodvorskaya V, Zelentsova E, Feder ME. Evolution and arrangement of the hsp70 gene cluster in two closely related species of the virilis group of Drosophila. Chromosoma. 2004;113:223–232. doi: 10.1007/s00412-004-0312-6. [DOI] [PubMed] [Google Scholar]
- Feder JH, Rossi JM, Solomon J, Solomon N, Lindquist S. The consequences of expressing Hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- Fulton RE, Salasek ML, DuTeau NM, Black WCT. SSCP analysis of cDNA markers provides a dense linkage map of the Aedes aegypti genome. Genetics. 2001;158:715–726. doi: 10.1093/genetics/158.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The Flp recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Genomic deletions of the Drosphila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia TD, Craig EA, McCarthy BJ. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell. 1980;21:669–679. doi: 10.1016/0092-8674(80)90430-4. [DOI] [PubMed] [Google Scholar]
- Isoe J, Kunz S, Manhart C, Wells MA, Miesfeld RL. Regulated expression of microinjected DNA in adult Aedes aegypti mosquitoes. Insect Mol Biol. 2007;16:83–92. doi: 10.1111/j.1365-2583.2006.00704.x. [DOI] [PubMed] [Google Scholar]
- Karpov VL, Preobrazhenskaya OV, Mirzabekov AD. Chromatin structure of Hsp-70 genes, activated by heat-shock–selective removal of histones from the coding region and their absence from the 5′ region. Cell. 1984;36:423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Li GC, Werb Z. Correlation between synthesis of heat-shock proteins and development of thermotolerance in Chinese-hamster fibroblasts. Proc Natl Acad Sci USA. 1982;79:3218–3222. doi: 10.1073/pnas.79.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Liu RY, Corry PM, Lee YJ. Regulation of chemical stress-induced hsp70 gene expression in murine L929 cells. J Cell Sci. 1994;107:2209–2214. doi: 10.1242/jcs.107.8.2209. [DOI] [PubMed] [Google Scholar]
- Lobo NF, Campbell KS, Thaner D, Debruyn B, Koo H, Gelbart WM, Loftus BJ, Severson DW, Collins FH. Analysis of 14 BAC sequences from the Aedes aegypti genome: a benchmark for genome annotation and assembly. Genome Biol. 2007;8:R88. doi: 10.1186/gb-2007-8-5-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay I, Saxena DK, Chowdhuri DK. Hazardous effects of effluent from the chrome plating industry: 70 kDa heat shock protein expression as a marker of cellular damage in transgenic Drosophila melanogaster (hsp70-lacZ) Environ Health Perspect. 2003;111:1926–1932. doi: 10.1289/ehp.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi ZY, Megy K, Grabherr M, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L. Heat shock response. CRC: Boca Raton, FL; 1991. [Google Scholar]
- Petersen RB, Lindquist S. Regulation of Hsp70 synthesis by messenger-RNA degradation. Cell Regul. 1989;1:135–149. doi: 10.1091/mbc.1.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson DW, Knudson DL, Soares MB, Loftus BJ. Aedesaegypti genomics. Insect Biochem Mol Biol. 2004;34:715–721. doi: 10.1016/j.ibmb.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Sim C, Hong YS, Tsetsarkin KA, Vanlandingham DL, Higgs S, Collins FH. Anophelesgambiae heat shock protein cognate 70B impedes o’nyong-nyong virus replication. BMC Genom. 2007;8:231–242. doi: 10.1186/1471-2164-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Rossi JM, Golic K, McGarry T, Lindquist S. Changes in Hsp70 alter thermotolerance and heat-shock regulation in Drosophila. New Biologist. 1991;3:1106–1120. [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molec Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanguay RM. Transcriptional activation of heat-shock genes in eukaryotes. Biochem Cell Biol Biochim Biol Cell. 1988;66:584–593. doi: 10.1139/o88-069. [DOI] [PubMed] [Google Scholar]
- Tatem AJ, Hay SI, Rogers DJ. Global traffic and disease vector dispersal. Proc Natl Acad Sci USA. 2006;103:6242–6247. doi: 10.1073/pnas.0508391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlirova M, Asahina M, Riddiford LM, Jindra M. Heat-inducible transgenic expression in the silk-moth Bombyx mori. Dev Genes Evol. 2002;212:145–151. doi: 10.1007/s00427-002-0221-8. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Wyder S, Zdobnov EM. The Aedes aegypti genome: a comparative perspective. Insect Mol Biol. 2008;17:1–8. doi: 10.1111/j.1365-2583.2008.00772.x. [DOI] [PubMed] [Google Scholar]
- [WHO] World Health Organization. Dengue and dengue haemorrhagic fever, fact sheets. World Health Organization; Geneva, Switzerland: 2008. [Google Scholar]
- Wimmer EA. Innovations: applications of insect transgenesis. Nat Rev Genet. 2003;4:225–232. doi: 10.1038/nrg1021. [DOI] [PubMed] [Google Scholar]
- Zhao YG, Eggleston P. Comparative analysis of promoters for transient gene expression in cultured mosquito cells. Insect Mol Biol. 1999;8:31–38. doi: 10.1046/j.1365-2583.1999.810031.x. [DOI] [PubMed] [Google Scholar]