Abstract
Background
In humans, the intestinal microbiota plays an important role in the maintenance of host health by providing energy, nutrients, and immunological protection. Applying current molecular methods is necessary to surmount the limitations of classical culturing techniques in order to obtain an accurate description of the microbiota composition.
Results
Here we report on the comparative assessment of human fecal microbiota from three age-groups: infants, adults and the elderly. We demonstrate that the human intestinal microbiota undergoes maturation from birth to adulthood and is further altered with ageing. The counts of major bacterial groups Clostridium leptum, Clostridium coccoides, Bacteroidetes, Bifidobacterium, Lactobacillus and Escherichia coli were assessed by quantitative PCR (qPCR). By comparing species diversity profiles, we observed age-related changes in the human fecal microbiota. The microbiota of infants was generally characterized by low levels of total bacteria. C. leptum and C. coccoides species were highly represented in the microbiota of infants, while elderly subjects exhibited high levels of E. coli and Bacteroidetes. We observed that the ratio of Firmicutes to Bacteroidetes evolves during different life stages. For infants, adults and elderly individuals we measured ratios of 0.4, 10.9 and 0.6, respectively.
Conclusion
In this work we have confirmed that qPCR is a powerful technique in studying the diverse and complex fecal microbiota. Our work demonstrates that the fecal microbiota composition evolves throughout life, from early childhood to old age.
Background
The composition of the intestinal microbiota plays a significant role in human immunology, nutrition and pathological processes [1]. Describing the complexity and ecology of the intestinal microbiota is important for defining its effects on overall human health. This level of understanding has been hindered by the limited sensitivity and inherent biases of culture-based techniques. Recently, the study of the gut microbiota has received renewed interest due to the development of molecular methods for more accurately assessing its composition and diversity, formerly thought to contain a mere 400–500 bacterial species [2]. Bacterial strains which are not cultivable under conventional methods have thus been identified [3]. This has markedly increased the information available concerning the complexity of the human bowel microbiota, from which over 1250 Operational Taxonomic Units have been identified corresponding to several dominant phyla [4,5].
Although the microbiota in adults has been extensively studied, investigation into structural changes and compositional evolution from infants to the elderly has only recently begun. Very little information is available pertaining to possible variations that occur with ageing. In healthy adults, 80% of the identified fecal microbiota can be classified into three dominant phyla: Bacteroidetes, Firmicutes and Actinobacteria [6]. In general terms the Firmicutes to Bacteroidetes ratio is regarded to be of significant relevance in human gut microbiota composition [7]. On a more refined level, however, the fecal microbiota is a highly complex and diverse bacterial ecosystem. Within this ecosystems exists a hierarchy of dominant (> 109 Colony Forming Units (CFU)/g)) anaerobic bacteria, represented by the genera Bacteroides, Eubacterium, Bifidobacterium, Peptostreptococcus, Ruminococcus, Clostridium and Propionibacterium, and sub-dominant (< 109 CFU/g), bacteria of the Enterobacteriaceae family, especially E. coli, and the genera Streptococcus, Enterococcus, Lactobacillus, Fusobacterium, Desulfovibrio and Methanobrevibacter [8].
Establishment of the intestinal microbiota has been shown to be a progressive process [9]. This process of increasing diversity is required for proper development and is important for overall health. The major functions attributed to the microbiota present in the gut begin to manifest at the end of the second year of life and comprise: i) nutrients absorption and food fermentation [10], ii) stimulation of the host immune system [11] and iii) barrier effects against pathogens [12]. Once climax composition is achieved near the end of adolescence, this ecosystem displays a high stability in healthy adults [13]. Although the intestinal microbiota is relatively stable throughout adult life, recent studies indicated that modifications occur in the composition in elderly individuals. For example, a reduction in the numbers of Bifidobacteria and Bacteroides has been observed, accompanied also by a decrease of Lactobacilli. A commensurate increase in the number of facultative anaerobes also highlights the variation between adults and elderly individuals [14-17]. Such variation was also observed by Ley et al. [7] when a correlation between body weight and gut microbial ecology was analysed. The microbiota in obese subjects shows an elevated proportion of Firmicutes and a reduced population of Bacteroides. Conversely, a decreased Firmicutes/Bacteroidetes ratio has been directly related to weight loss [7].
The work presented here aims to continue to expand our understanding of the intestinal flora including its establishment, composition, and evolution. To that end, we focused on the important ratio between Firmicutes and Bacteroidetes. We used a qPCR-based approach to enumerate changes in bacterial populations in the human intestine.
This work is part of the few culture-independent studies [18] which use adapted molecular approaches to analyze modulations of fecal microbiota related to ageing.
Results
Microbiota specificities related to age
Average bacterial counts for each human age-group are summarized in Table 1. In adults, the Bacteroidetes and Firmicutes are the most prevalent phyla present, the latter of which combines the values obtained for the dominant C. leptum and C. coccoides groups and the sub-dominant Lactobacillus group. The Bifidobacterium genus is present in eight to ten-fold lower numbers than the two major phyla. E. coli was found to be present at 7.7 log10 CFU/g, also consistent with its characteristic sub-dominant population in adults.
Table 1.
Composition of the human microbiota compared in three age groups
| TaqMan detection | SYBR-Green detection | |||||||
|---|---|---|---|---|---|---|---|---|
| Firmicutes | Firmicutes | Firmicutes | ||||||
| n | All-bacteria (a) | C. leptum group (b) | C. coccoides group (b) | Bacteroides/Prevotella group (b) | Bifidobacterium genus (b) | E. coli (b) | Lactobacillus/Leuconostoc/Pediococcus group (b) | |
| Infant | 21 | 10.7 ± 0.1 (A) | -3.2 ± 0.4 (A) | -3.2 ± 0.4 (A) | -1.5 ± 0.3 (A) | -0.6 ± 0.2 (A) | -1.5 ± 0.3 (A) | -3 ± 0.2 (A) |
| Adult | 21 | 11.5 ± 0.1 (B) | -0.7 ± 0.1 (B) | -1.2 ± 0.1 (B) | -1.5 ± 0.1 (AB) | -2.3 ± 0.3 (B) | -3.8 ± 0.1 (B) | -3.9 ± 0.3 (AB) |
| Elder | 20 | 11.4 ± 0.1 (B) | -1.1 ± 0.1 (C) | -1.8 ± 0.1 (A) | -1 ± 0.1 (A) | -2.3 ± 0.3 (B) | -2.4 ± 0.2 (C) | -4.2 ± 0.2 (B) |
n represents the number of samples in each group.
(a) All-bacteria results obtained by qPCR were expressed as the mean of the log10 value ± SEM.
(b) Results were expressed as the mean of the log10 value ± SEM of normalized data calculated as the log of targeted bacteria minus the log of All-bacteria number.
The non parametric Wilcoxon test was performed.
Data not sharing the same letter within a column are significantly diferrent at p < 0.05.
Quantification of samples from infants showed total bacterial counts to be nearly ten-fold lower in log10 values (10.7) than in adults and seniors (11.5 and 11.4, respectively). It is worth noting that while they constitute the major dominant groups in adults and elderly, C. leptum and C. coccoides groups are only observed at a sub-dominant level in infants. Bifidobacteria was clearly the most abundant group measured in infants. Owing to lower overall numbers of bacteria in infants, the Bifidobacterium genus represented a major fraction of the dominant bacterial species found in the infant fecal microbiota, far above Firmicutes and Bacteroidetes. Infants were also found to harbor an E. coli population at a level characteristic of a dominant group, 109 CFU/g, contrary to the level observed in adults.
Normalized quantitative PCR data
When normalized against all bacterial group counts, the qPCR data (Table 1) can be represented as a percentage of total bacterial counts. Statistical analysis of the data show that C. leptum, and C. coccoides levels are significantly lower in infants (-3.2 and -3.2 Δlog10 respectively) than in adults (-0.7 and -1.2 Δlog10 respectively), while Bacteroides levels are equivalent in each age group. Alternatively, Bifidobacterium levels are greater in infants (-0.6 Δlog10) than in adults (-2.3 Δlog10) and seniors (-2.3 Δlog10). Lactobacillus counts are greater in infants (-3 Δlog10) than in seniors (-4.2 Δlog10) with an equivalent value in adults (-3.9 Δlog10). Interestingly, E. coli levels exhibit a progression between the three age groups since the highest counts are found in infants (-1.5 Δlog10), then decrease in adults (-3.8 Δlog10), ultimately stabilizing at an intermediate level in seniors (-2.4 Δlog10).
Finally, analysis of each bacterial population revealed no significant differences for the elderly when compared with those for adults with the exception of C. leptum, C. coccoides and E. coli, which as in infants, showed counts characteristic of a dominant group.
Firmicutes/Bacteroidetes ratio
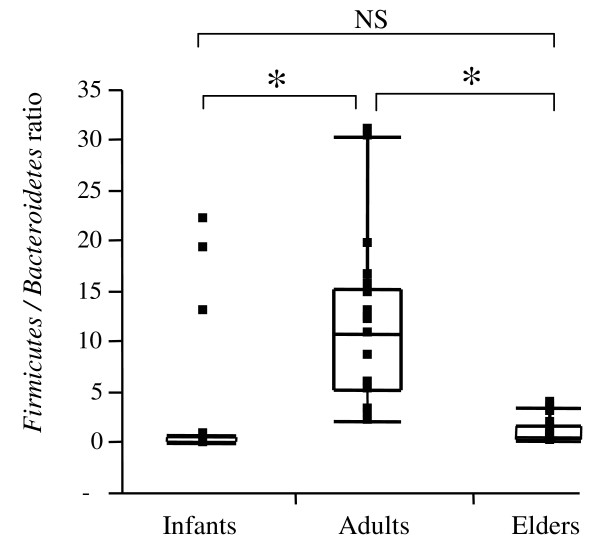
For the Firmicutes/Bacteroidetes ratio, we observed significant differences between infants and adults (0.4 and 10.9, respectively) and between adults and elderly (10.9 and 0.6, respectively) (Figure 1). Notably, no significant differences were found between infants and elderly.
Figure 1.
Box-and-Whisker plot of Firmicutes/Bacteroidetes ratios in the three age-groups. Horizontal lines represent the paired comparison. Boxes contain 50% of all values and whiskers represent the 25th and 75th percentiles. Significantly different (P < 0.05) ratios are indicated by *, while NS corresponds to non-significant differences.
Discussion
The microbiota of the large intestine plays an important role in host metabolism and maintenance of host health [19]. The accurate description of this bacterial community is an important question that has long remained a challenge owning to the limitations of culturing and isolation techniques. We have thus employed current molecular methods, i.e. quantitative PCR, to tackle this problem. Our work has allowed for a detailed description of the complex composition of the human intestinal microbiota which can serve as a basis to monitor gut microbiota changes in connection with diet or health.
Our results defining a standard adult profile, together with previous reports, showed that C. leptum, C. coccoides, Bacteroides and Bifidobacterium represent the four dominant groups of the adult fecal microbiota [8,20,21]. Sub-dominant groups are Lactobacilli Enterobacteriaceae, Desulfovibrio, Sporomusa, Atopobium as well as other bacterial groups including Clostridium clusters XI, XIVb, and XVIII [21,22].
Total bacterial counts overall were found to be significantly lower in infants than in adults and seniors. In infant fecal microbiota, we observed Bifidobacterium as the dominant group. This population dominance has been documented as a conserved feature during early gastrointestinal tract colonization [23]. Moreover, this observation is strongly related to diet, being enhanced by breast feeding [24,25]. In order to account for this observed effect, our infant panel consistent of 21 individuals aged 1 to 10 months, 14 of which were breast-fed, and 7 which were formula-fed. The older infants in our study received a more diverse diet. Significant higher numbers of Bifidobacterium were observed in infants versus adults and seniors. We conclude, therefore, that the high level of Bifidobacterium observed in our panel was not strictly correlated to breast feeding and could be considered as a broad signature of the infant microbiota during the first year of life.
This observation confirms previous reports indicating that the gastrointestinal tract is first colonized by facultative anaerobes, such as E. coli [23]. Strict anaerobes, such as Clostridium, colonize at later stages, as can be seen by the relatively low levels of C. leptum and C. coccoides in infants [23]. Our results are in agreement with these previous reports. We hypothesize that diet change must be considered among the primary causes for such a shift of microbiota between infants and adults.
In the case of elderly subjects, our qPCR results indicated a significant increase in the counts of E. coli when compared to adults. This data is consistent with other publications indicating that elderly subjects harbor a different E. coli microbiota profile compared to younger adults [26-28].
Concerning the microbiota of the elderly, a number of authors reported a reduction in the numbers and diversity of many protective commensal anaerobes, such as Bacteroides and Bifidobacteria. These reports also suggest a shift in the dominant bacterial species [17,19]. The Firmicutes to Bacteroidetes ratio was already shown to be of significant relevance in signaling human gut microbiota status [7]. This previous work focused on lean individuals and used 16S ribosomal RNA gene sequencing. Our measurements of the Firmicutes/Bacteroidetes ratio in adults obtained by our species-specific qPCR are in agreement with those obtained by Ley et al. [7]. Compared with young adults, the elderly have a different digestive physiology, characterized at a physiological level by a reduction in transit and of digestive secretions. These changes could explain the observed changes in the fecal microbiota associated with advancing age.
Conclusion
Our results illustrate a measurable progression of bacterial species colonizing the human intestinal tract during different stages of life. This progression is easily observed and quantified using qPCR to evaluate numbers of bacteria belonging to major dominant and subdominant groups of the human fecal microbiota. The Firmicutes/Bacteroidetes ratio undergoes an increase from birth to adulthood and is further altered with advanced age. This ratio appears applicable in highlighting variations between infants, adults and the elderly. It can be linked to overall changes in bacterial profiles at different stages of life.
Methods
Sample collection
Fecal samples from 21 adults (25 to 45 years old) were recovered from previous sampling [20]. Fresh fecal samples were obtained from 21 infants (3 weeks to 10 months old) and 20 elderly subjects (70 to 90 years old). Infants in the study group were currently feeding with either breast milk (n = 16) or formula (n = 7). None of the infant subjects had been exposed to antibiotics. Adult and elderly subjects consumed an unrestricted Western-type diet. All subjects from these two age classes were not under antibiotic treatment or taking any other drugs known to influence the fecal microbiota composition for at least three months prior to sampling. All subjects were free of known metabolic or gastrointestinal diseases. Whole stools were collected in sterile boxes and immediately stored at 4°C under anaerobic conditions using an Anaerocult® A (Merck, Nogent sur Marne, France). Samples were frozen within 4 hours at -20°C as 200 mg aliquots and stored for further analysis. Adults and elderly subjects were volunteers. Parents of infants gave written informed consent for this work. All procedures were approved by an ethics committee.
DNA extraction
DNA was extracted from the 200 mg aliquots of feces as described previously [29,30]. After the final precipitation with isopropanol, nucleic acids were centrifuged and pellets were suspended in 225 μl of phosphate buffer and 25 μl of potassium acetate. After the RNase treatment, DNA was recovered by centrifugation and pellet was suspended in TE buffer.
Real-time qPCR
Real-time qPCR was performed using an ABI 7000 Sequence Detection System apparatus with system software version 1.2.3 (Applied-Biosystems) [20,31]. Total numbers of bacteria were inferred from averaged standard curves as described by Lyons et al. [32].
TaqMan® qPCR was adapted to quantify total bacteria populations in addition to the dominant (<1% of faecal bacteria population) bacterial species C. coccoides, C. leptum, Bacteroides/Prevotella and Bifidobacterium. qPCR using SYBR-Green® was performed for the sub-dominant bacterial species Escherichia coli and for the Lactobacillus/Leuconostoc/Pediococcus group. Primers and probes used in this study were designed based on 16S rRNA sequences. A detailed description can be found in Furet et al [20] and Firmesse et al [31].
Normalization of quantitative PCR data
Normalization was done by subtracting the value obtained for the "all bacteria" group from the values for the other bacterial groups in our study [20].
Firmicutes/Bacteroidetes ratios
An estimation of the total amount of Firmicutes was obtained by adding bacterial values obtained from C. coccoides, C. leptum and Lactobacillus. For Firmicutes/Bacteroidetes ratios, calculations were obtained for each individual using CFU counts.
Statistics
The non-parametric Wilcoxon test was performed using JMP® software (Abacus Concepts, Berkeley, CA). The comparative results of Firmicutes/Bacteroidetes ratios were visualized as box-and-whisker plots showing: the median and the interquartile (midspread) range (boxes containing 50% of all values), the whiskers (representing the 25th and 75th percentiles) and the extreme data points. Statistical significance was accepted at P < 0.05.
Authors' contributions
DM, FL and JPF carried out all PCR experiments. OF performed statistical studies. HS and VDG helped to draft the manuscript with the assistance of all authors. JD and GC conceived and coordinated the study. All authors read and approved the manuscript.
Contributor Information
D Mariat, Email: denis.mariat@jouy.inra.fr.
O Firmesse, Email: o.firmesse@AFSSA.FR.
F Levenez, Email: florence.levenez@jouy.inra.fr.
VD Guimarăes, Email: valeria.guimaraes@cea.fr.
H Sokol, Email: sokol_harry@yahoo.fr.
J Doré, Email: joel.dore@jouy.inra.fr.
G Corthier, Email: gerard.corthier@jouy.inra.fr.
J-P Furet, Email: jean-pierre.furet@jouy.inra.fr.
Acknowledgements
We thank Dr Sean P Kennedy for critical reading of the manuscript.
References
- Blaut M, Collins MD, Welling GW, Dore J, Van Loo J, De Vos W. Molecular biological methods for studying the gut microbiota: the EU human gut flora project. Br J Nutr. 2002;87(Suppl 2):S203–11. doi: 10.1079/BJN/2002539. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Collier CT, Koike S, Mackie RI, Gaskins HR. Molecular ecological analysis of the gastrointestinal microbiota: a review. J Nutr. 2004;134:465–472. doi: 10.1093/jn/134.2.465. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Berstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Doré J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay C, Sutren M, Rochet V, Saunier K, Doré J, Rigottier-Gois L. Design and validation of 16S rDNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ Microbiol. 2005;7:933–946. doi: 10.1111/j.1462-2920.2005.00763.x. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh P, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Harmsen HJ, Raangs GC, He T, Degener JF, Welling GW. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol. 2002;68:2982–2990. doi: 10.1128/AEM.68.6.2982-2990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e117. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JL. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3(4):331–41. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébuterne X. Gut changes attributed to ageing: effects on intestinal microflora. Curr Opin Clin Nutr Metab Care. 2003;6:49–54. doi: 10.1097/00075197-200301000-00008. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Macfarlane GT. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol. 2002;51:448–454. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- Van Tongeren SP, Slaets JPJ, Harmsen HJM, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodmansey EJ. Intestinal bacteria and ageing. J Appl Microbiol. 2007;102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- Saunier K, Doré J. Gastrointestinal tract and the elderly: functional foods, gut microflora and healthy ageing. Dig Liver Dis. 2002;34(Suppl 2):S19–24. doi: 10.1016/S1590-8658(02)80158-X. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001;48:198–205. doi: 10.1136/gut.48.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet JP, Firmesse O, Gourmelon M, Bridonneau C, Tap J, Mondot S, Doré J, Corthier G. Comparative assessment of human and farm animal fecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- Rigottier-Gois L, Bourhis AGL, Gramet G, Rochet V, Doré J. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol Ecol. 2003;43:237–245. doi: 10.1111/j.1574-6941.2003.tb01063.x. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- Salminen S, Isolauri E. Intestinal colonization, microbiota and probiotics. J Pediatr. 2006;149:S115–S120. doi: 10.1016/j.jpeds.2006.06.062. [DOI] [Google Scholar]
- Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2006;72(4):2359–65. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal microflora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–7. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- Bartosch S, Fite A, Macfarlane GT, McMurdo ME. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol. 2004;70:3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Sakamoto M, Kitahara M, Benno Y. Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T-RFLP. Microbiol Immunol. 2003;47:557–570. doi: 10.1111/j.1348-0421.2003.tb03418.x. [DOI] [PubMed] [Google Scholar]
- He F, Ouwehand AC, Isolauri E, Hosoda M, Benno Y, Salminen S. Differences in composition and mucosal adhesion of bifidobacteria isolated from healthy adults and healthy seniors. Curr Microbiol. 2001;43:351–354. doi: 10.1007/s002840010315. [DOI] [PubMed] [Google Scholar]
- Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-unit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay C, Rigottier-Gois L, Holmstrom K, Rajilic M, Vaughan EE, De Vos WM, Collins MD, Thiel R, Namsolleck P, Blaut M, Doré J. Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol. 2005;71(7):4153–5. doi: 10.1128/AEM.71.7.4153-4155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmesse OA, Mogenet AJL, Bresson JLG, Corthier GJP, Furet JP. Lactobacillus rhamnosus R11 consumed in a food supplement survived human digestive transist without modifying microbiota equilibrium as assessed by real time Polymerase Chain Reaction. J Mol Microbiol Biotechnol. 2008;14:90–99. doi: 10.1159/000106087. [DOI] [PubMed] [Google Scholar]
- Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38(6):2362–5. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]