Abstract
Regulators of G-protein signaling (RGS) proteins enhance the intrinsic GTPase activity of G protein α (Gα) subunits and are vital for proper signaling kinetics downstream of G protein–coupled receptors (GPCRs). R7 subfamily RGS proteins specifically and obligately dimerize with the atypical G protein β5 (Gβ5) subunit through an internal G protein γ (Gγ)-subunit–like (GGL) domain. Here we present the 1.95-Å crystal structure of the Gβ5–RGS9 complex, which is essential for normal visual and neuronal signal transduction. This structure reveals a canonical RGS domain that is functionally integrated within a molecular complex that is poised for integration of multiple steps during G-protein activation and deactivation.
Multifarious hormones, neurotransmitters, growth factors and other extracellular stimuli produce their physiological effects by activating G protein–coupled receptors (GPCRs) and their associated heterotrimeric G proteins1. RGS proteins attenuate heterotrimeric G-protein signaling by enhancing the intrinsic GTPase activity of Gα subunits and are vital for proper signal transduction kinetics2–4. The R7 (class C) subfamily of RGS proteins encompasses the four mammalian proteins RGS6, RGS7, RGS9 and RGS11 (ref. 4), which are all highly expressed in the central nervous system5,6. RGS9 is the best-characterized R7-RGS protein and is expressed in two isoforms: RGS9-1 mediates the rate-limiting step during response recovery of rod phototransduction7, whereas RGS9-2 is required for proper signal transduction downstream of certain opioid and dopamine receptors8–10. Humans lacking functional RGS9-1 are temporarily visually impaired by sudden changes in light levels11. Mice without RGS9-2 develop exacerbated dependence and withdrawal to morphine and exhibit dyskinesias induced by the stimulation of D2-dopamine receptors that are similar to the side effects observed in patients treated for psychoses and Parkinson’s disease10.
Members of the R7 subfamily of RGS proteins contain functional domains in addition to the characteristic RGS domain, which provides catalytic GTPase activating function. A Dishevelled/Egl-10/Pleckstrin homology (DEP) domain participates in proper subcellular localization of these RGS proteins through interaction with the recently discovered membrane-targeting proteins RGS9 anchor protein (R9AP) and R7 binding protein (R7BP)12–14. A G protein γ (Gγ)-like (GGL) domain, which shares sequence homology with the Gγ subunits of heterotrimeric G proteins, mediates obligate heterodimerization with the most divergent G protein β (Gβ) subunit, Gβ5 (ref. 15). Although it has been suggested that Gβ5–R7-RGS complexes might support nucleotide exchange on G protein α (Gα) subunits in conjunction with agonist-stimulated GPCRs16, the function of Gβ5 in these complexes remains unknown.
To better understand the functional implication of Gβ5–R7-RGS complexes in mediating the activation and deactivation of G protein–mediated signaling cascades, we present here the high-resolution crystal structure of Gβ5–RGS9. In this structure, Gβ5 scaffolds the various domains of RGS9 and contains a highly conserved surface that is consistent with its ability to bind Gα subunits but that is occluded by RGS9. The structure highlights features that dictate the exclusive pairing of Gβ5 with R7-RGS proteins and suggests a mechanism for filtering the interactions between activated Gα proteins and the RGS domain that uses other portions of Gβ5–RGS9.
RESULTS
Overall structure of Gβ5–RGS9
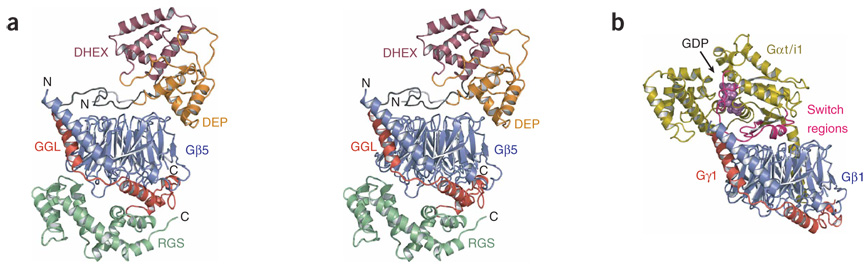
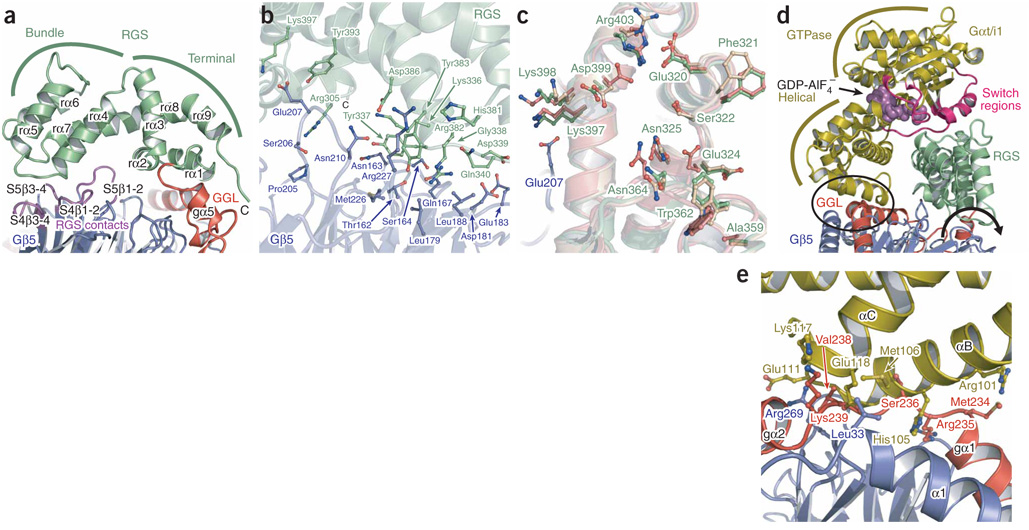
We solved the crystal structure of mouse Gβ5 in complex with a fragment of mouse RGS9 (residues 1–422) that encompasses all domains (DEP, GGL and RGS) common to both the retinal and neuronal forms of RGS9 (Fig. 1). Gβ5 recapitulates the toroidal β-propeller common to domains of WD repeats and is sandwiched between the N- and C-terminal lobes of RGS9 (Fig. 1a). Three distinct RGS9 interfaces bury more than 25% of the total solvent-accessible surface area of Gβ5 and involve 96 out of 353 Gβ5 amino acids (Supplementary Figs. 1 and 2 online). First, the N-terminal lobe of RGS9 packs against the ‘top’ surface of Gβ5, which is analogous to the surface used by Gα subunits to bind conventional Gβγ dimers17,18 (Fig. 1). Second, similar to what is seen in Gγ subunits, the centrally located GGL domain of RGS9 intertwines with the N-terminal helical extension of Gβ5 before wrapping around the side of the β-propeller. Finally, the RGS domain packs against the bottom face of Gβ5. The congruent Cα atoms from both independent dimers in the asymmetric unit superimpose with a root mean square (r.m.s) deviation of 1.0 Å, supporting the biological relevance of the observed domain packing.
Figure 1.
Structure of Gβ5–RGS9. (a) Stereo ribbon diagram of Gβ5 (blue) in dimeric complex with RGS9 including its N-terminal (light gray), Dishevelled/Egl-10/Pleckstrin homology (DEP; orange), DEP helical extension (DHEX; maroon), G protein γ (Gγ)-subunit–like (GGL; red), regulators of G-protein signaling (RGS; green) and DHEX-GGL linker (dark gray) regions. (b) Ribbon representation of the transducin Gαβγ heterotrimer17 (PDB 1GOT), with the Gβ subunit oriented similar to Gβ5 in a. Gα, gold; Gα-switch regions, magenta; Gβ, blue; Gγ, red; GDP, purple spheres. Structure figures were generated with PyMOL software (http://pymol.sourceforge.net).
The DEP–DHEX domains
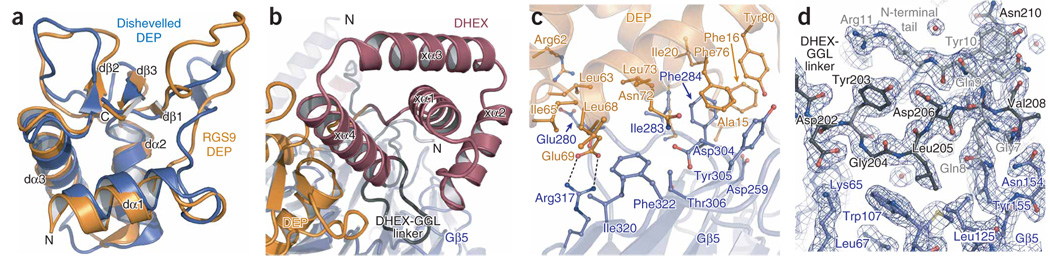
The N-terminal lobe of RGS9 consists of two extended regions flanking a DEP domain adjacent to a novel fold, here termed a DEP helical extension (DHEX). The DEP domain forms a core tri-helical bundle capped by an antiparallel β-sheet with two divergent loops (Fig. 2a and Supplementary Fig. 1). The core regions of the RGS9 and Dishevelled19 DEP domains are similar and superpose with a 1.3-Å r.m.s. deviation (Fig. 2a). The RGS9 dα1-dβ1 loop shows alternate conformations in the two complexes within the crystal asymmetric unit and is likely to be inherently flexible. The DHEX domain, previously referred to as the interdomain12 or R7 homology domain20, has no close structural homologs and instead forms a ‘helix-wrap’ composed of a central helix encircled by three antiparallel α-helices (Fig. 2b) (see the discussion of the Dali search in the Methods section). The fourth DHEX helix (xα4) interfaces with the β-sheet and variable loops of the DEP domain to form a single DEP-DHEX superdomain that packs against the extended flanking regions (the N-terminal portion of RGS9 and the DHEX-GGL linker; Fig. 1a and Fig 2b). Whereas the DHEX domain makes no direct contacts with Gβ5, the DEP domain, the N-terminal region of RGS9 and the DHEX-GGL linker provide an extensive interface that buries 2600 Å2 of surface area between the two proteins (Fig. 2c,d). Most of the surface of Gβ5 that contacts the N-terminal region of RGS9 and the DHEX-GGL linker is congruent with the surface of Gβ1 that contacts the switch II region of Gα-GDP subunits17,18 (Fig. 2d and Fig 3, and Supplementary Figs. 1 and 2 online). In contrast, Gβ5 and the DEP domain interact through a hydrophobic core surrounded by a ring of electrostatic interactions, and this interface does not include analogous regions of Gβ subunits that are needed to engage Gα-GDP (Fig. 2c, and Supplementary Figs. 1 and 2). Consistent with this extensive interface, the isolated N-terminal lobe functions in trans with separately expressed GGL-RGS fragments for the Caenorhabditis elegans R7-RGS proteins, which are known as EGL-10 and EAT-16 (ref. 21).
Figure 2.
RGS9 N-terminal lobe. (a) Ribbon diagram of superposed RGS9 (orange) and Dishevelled (blue) Dishevelled/Egl-10/Pleckstrin homology (DEP) domains. RGS9-DEP domain secondary structure elements are labeled. (b) RGS9-DEP helical extension (RGS9-DHEX; maroon) domain with secondary structure elements labeled. (c) Specific contact residues between the RGS9-DEP domain (orange) and Gβ5 (blue) are represented as balls and sticks. (d) 2Fo – Fc electron density map of the RGS9 DHEX-Gγ–like (GGL) linker (dark gray) and N-terminal (light gray) elements with neighboring Gβ5 residues (blue) contoured at 1σ (dark-blue mesh).
Figure 3.
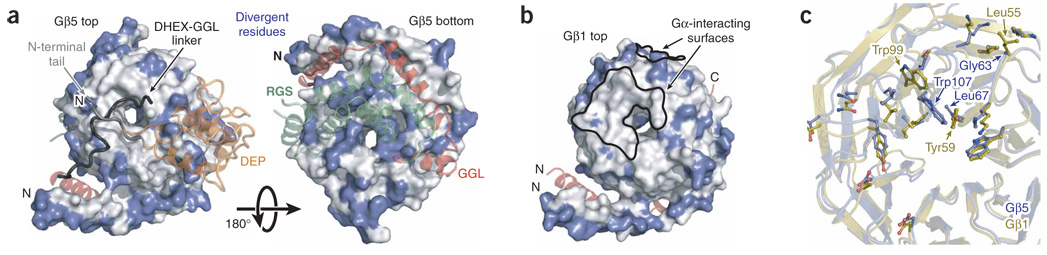
Conserved Gα binding interface on Gβ5. (a) Surface representation of the N-terminal lobe (top) and regulators of G-protein signaling (RGS) domain (bottom) binding faces of Gβ5. Divergent residues as indicated in Supplementary Figure 1 are blue. A transparent ribbon representation of RGS9 is shown and colored according to Figure 1a. The DEP helical extension (DHEX) domain and RGS domain elements that have no Gβ5 contacts are deleted for clarity. (b) Surface representation of Gβ1 (ref. 17) oriented similarly to the top face of Gβ5 in a. The residues that align with divergent Gβ5 residues are colored blue for comparison with the figure in a, and the surfaces of Gβ1 that interface with Gα subunits are indicated with black outlines. (c) Superposed Gβ5 (blue) and Gβ1 (gold) structures with Gαt-binding residues of Gβ1 (refs. 17,23) and structurally equivalent Gβ5 residues depicted as balls and sticks.
The N-terminal lobe of R7-RGS proteins is also important for subcellular localization through interaction with the membrane-targeting proteins R9AP and R7BP12–14. The DEP domain is necessary, but not sufficient, to bind these anchoring proteins14,22, and recent studies also implicate the DHEX domain20. The apposition of the DEP and DHEX domains observed in the Gβ5–RGS9 structure (Fig. 1a and Fig 2b) supports a model whereby anchoring proteins bind to a surface that includes both domains.
The Gβ5 propeller
The structure revealed here illustrates that Gβ5 and Gβ1 fold into essentially identical seven-bladed β-propellers (β-sheets S1–S7) with equivalent N-terminal helical extensions23 (r.m.s. deviation of 1.1 Å; Fig. 1a,b). The most substantial structural difference between Gβ5 and Gβ1 involves the insertion of a 310 helix in the loop between β-strands 3 and 4 of β-sheet S3 (Supplementary Fig. 1).
Gβ1, Gβ2, Gβ3 and Gβ4 share 80–90% sequence identity, whereas Gβ5 shows approximately 50% sequence conservation with Gβ1–4 (ref. 24; Supplementary Fig. 1). Not surprisingly, most of the functionally divergent residues within Gβ5 cluster at the interfaces with the DEP and RGS domains (Fig. 3a). As previously noted for other Gβ subunits23, surface-exposed residues within the N-terminal helix are also notably nonconserved (Fig. 3a). Conversely, the surface of Gβ5 that interacts with the GGL domain is highly conserved in other Gβ subunits, probably reflecting a common origin of GGL and Gγ domains and their role in heterodimerization with Gβ subunits.
The discovery that R7-RGS proteins interact with Gβ5 subunits naturally led to the hypothesis that these complexes share structural and functional commonalities with conventional Gβγ dimers25, and the structure of Gβ5–RGS9 indicates that the main determinants of Gα binding are retained in Gβ5 (refs. 17,26) (Fig. 3b,c). Only two residues of Gβ5 (Gly63 and Leu67) differ from analogous residues (Leu55 and Tyr59) of Gβ1 that interact with Gα-GDP subunits, and these differences are unlikely to affect the interaction with Gα-GDP. Indeed, Leu55 and Tyr59 of Gβ1 can be mutated to alanine without a substantial loss of Gα binding or receptor-catalyzed nucleotide exchange26. Trp99 of Gβ1 is necessary for high-affinity binding of Gα-GDP subunits26, and the analogous residue in Gβ1 (Trp107) adopts a distinct rotameric conformation that is probably influenced by its interaction with the N-terminal lobe of RGS9. Nevertheless, despite these minor differences, it has not been possible to reconstitute heterotrimers consisting of Gβ5, various R7-RGS proteins and Gα-GDP subunits25,27, presumably because the N-terminal lobe of R7-RGS proteins ‘cap’ the top of Gβ5, which is needed to engage Gα-GDP subunits (Fig. 1a and Fig 3).
The GGL domain
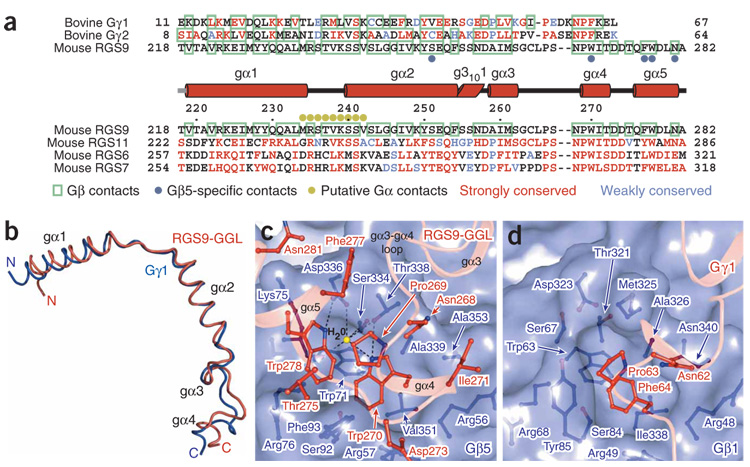
The interface of Gβ5 with the GGL domain is the most extensive of the three interfacial regions. Contacts occur between 54 Gβ5 residues and 39 RGS9 residues, which together bury 4,700 Å2 of solventaccessible surface area (Fig. 4a, and Supplementary Figs. 1 and 2). Our Gβ5–RGS9 crystal structure clearly demonstrates that the RGS9-GGL domain is structurally identical to conventional Gγ subunits17, and the GGL domain superposes on equivalent atoms from the transducin Gγ structure with an r.m.s. deviation of 1.1 Å (Fig. 1 and Fig 4b). Whereas the pattern of interactions between Gβ5 and the GGL domain largely mirror those observed between Gβ and Gγ subunits (Fig. 4a, and Supplementary Fig. 1 and 2), R7-RGS proteins show stringent specificity for Gβ5 (refs. 25,28). Conversely, Gγ subunits do not form stable complexes with Gβ5 but instead specifically bind Gβ1–4 (refs. 28,29). Comparison of the Gβ5–RGS9 structure with other Gβγ complexes suggests that the different Gβ specificity of the GGL domain versus Gγ results from the cumulative effect of several individually minor alterations. For example, RGS9-Ser251 forms a hydrogen bond with Gβ5-Tyr248 that cannot be recapitulated with either the equivalently positioned Gγ hydrophobic residues or the phenylalanine of Gβ1–4, respectively (Supplementary Figs. 1 and 2). More importantly, RGS9-Trp270 forms many contacts as it inserts into a deep hydrophobic cleft within Gβ5 (Fig. 4c, and Supplementary Fig. 2), and its replacement with the analogous phenylalanine found in Gγ subunits would be likely to reduce specificity of GGL domains for Gβ5 (Fig. 4c,d). The small Gβ5 residues Thr338 and Ala353 better accommodate Trp270 than would the larger Gβ1–4 methionine and asparagine residues. Gβ5-Thr338 also contributes to a unique hydrogen bond with RGS9-Trp270, and both residues participate in a water-mediated hydrogen bond network with several neighboring residues (Fig. 4c). The role of RGS9-Trp270 in conferring specificity of binding to Gβ5 is supported by previous mutational and binding studies using other R7-RGS proteins28. A cluster of hydrophobic residues centered on RGS9-Trp270 is extended by residues from a novel GGL domain helix (gα5), which folds back against the shorter and conformationally altered GGL domain gα3–gα4 loop (Fig. 4c). This gα5 helix provides additional specific interactions with Gβ5 and also provides a bridge to the RGS domain.
Figure 4.
Gγ-subunit–like (GGL) domains and Gγ subunits are structurally equivalent. (a) Structure-based sequence alignments of the RGS9-GGL domain with Gγ subunits and other mammalian R7-RGS-GGL domains. Residue numbers indicated above the R7-RGS–GGL domain sequences are for mouse RGS9. The helical elements of the RGS9-GGL domain are indicated above the R7-RGS-GGL domain alignments as cylinders. Residues contacting Gβ subunits17,18 are boxed in green. The residues that are identical or strongly conserved with RGS9-GGL residues are colored red, and residues that are weakly conserved are colored blue. RGS9-GGL domain residues supporting Gβ5-specific contacts are indicated with blue dots. These are defined as contacts that could not be reproduced in a complex with other Gβ subunits owing to chemical or steric conflicts. Residues proposed to contact an RGS-domain–bound Gα subunit are labeled with gold dots. (b) Backbone trace of the superposed RGS9-GGL (red) domain and Gγ1 (ref. 17; dark-blue) structures. GGL domain secondary structure elements are labeled. (c) RGS9-W270 binding pocket shown as a translucent surface with individual Gβ5 (blue) and GGL domain (red) residues shown as balls and sticks. The GGL domain backbone is represented as a transparent ribbon. A hydrogen-bonded water molecule (yellow sphere) is depicted. (d) Gγ1-F64 (red) and Gβ1 (blue) binding interface, depicted similarly to the figure in c.
The RGS domain
The structure of the RGS domain is not altered by incorporation into the Gβ5–RGS9 heterodimer and superimposes well with the structure of the isolated RGS domain of bovine RGS9 (ref. 30) (r.m.s. deviation = 0.9 Å). The structures of this and other RGS domains are similarly composed of terminal and bundle subdomains31 (Fig. 5a). Coalescence of the bundle subdomain (rα4–rα7) brings the termini of the RGS domain in apposition to form the terminal subdomain (rα1–rα3 and rα8–rα9). The interface between the RGS domain and Gβ5 is the smallest of the three Gβ5–RGS9 interfaces, burying only 1,450 Å2 of solvent-accessible surface area (Fig. 5a,b, and Supplementary Figs. 1 and 2). Most of the direct contacts occur between two RGS domain loops (rα4–rα5 and rα6–rα7) and the loops within β-sheets S4 and S5 of Gβ5 (Fig. 5a,b). Loops between S4β1–S4β2 and S5β1–S5β2 of Gβ5 are longer than their counterparts in other Gβ subunits and contribute to the RGS9–Gβ5 interface (Fig. 5a,b, and Supplementary Figs. 1 and 2). RGS9-Arg305 is the only residue within the RGS terminal subdomain that directly contacts Gβ5 (Fig. 5b). However, several residues in this subdomain also show interdomain interactions with residues of the GGL domain gα2–g3101 elements and the gα5 helix, which further stabilize the Gβ5–RGS domain interface (Supplementary Fig. 1).
Figure 5.
The RGS9-RGS domain interfaces with Gβ5 and activated Gα subunits. (a) Ribbon diagram of the RGS9-RGS domain (green) interface with the Gγ-subunit–like (GGL) domain (red) and Gβ5 (blue). Gβ5 loop residues contacting the RGS domain are purple. Bundle and terminal RGS subdomains are indicated with green lines. (b) Transparent ribbon diagram of the RGS domain (green) and Gβ5 (blue) with interfacial contact residues depicted as balls and sticks. (c) Superimposed structures from the isolated RGS9-RGS domain (wheat) and RGS9-RGS domains in complexes with Gαt/i (coral) and Gβ5 (green) are depicted as transparent ribbon diagrams along with the Gβ5 (blue) structure. The Gαt/i contact residues from all three RGS domain structures and the Gβ5 residue (Glu207) that shares a common RGS domain contact (Lys397) with Gαt/i30 are depicted as balls and sticks. The isolated and Gαt/i-bound RGS9-RGS domain coordinates PDB accession numbers are 1FQI and 1FQJ, respectively. (d) A model of Gαt/i (ref. 30; gold) docked onto the RGS domain (green) of Gβ5–RGS9 is depicted and colored similarly to Figure 1. The GTPase and all-helical subdomains of the Gα subunit are indicated with gold lines. A region where the Gα subunit clashes with the GGL domain and Gβ5 is circled. The direction of a proposed reorientation of the RGS domain is indicated with a curved arrow. The orientation of this image is rotated clockwise ~90° around the y axis with respect to the image in a. (e) Magnified and ~90° rotated view of the encircled region of d, with sterically clashing residues shown as balls and sticks.
Lys397 within the RGS domain of RGS9 is the only Gβ5 contact residue that also interacts with Gα; however, this residue is positioned similarly in both complexes and could participate in electrostatic interactions with both proteins concomitantly30 (Fig. 5b,c, and Supplementary Figs. 1 and 2). Other Gα binding residues generally have the same orientation as analogous residues in the structure of the isolated RGS9-RGS domain, particularly Glu324, Asn325, Ala359, Trp362, Asn364 and Asp399, which participate in enhancing Gα-GTPase activity30 (Fig. 5c). We conclude that Gβ5 association does not directly influence the functional architecture of the RGS9-RGS domain.
The loop-to-loop nature of the Gβ5–RGS domain interface potentially allows flexibility, although conservation of the contacts and relative orientations between the RGS domain and Gβ5 in both dimers of the crystal asymmetric unit support the functional relevance of the observed interface. To ascertain whether this orientation would allow binding of Gα-GTP and GTPase acceleration, we superimposed the structure of the isolated RGS domain of bovine RGS9 bound to Gαt/i1-GDP-AlF4−(ref. 30) onto the structure of Gβ5–RGS9 (Fig. 5d). Notably, the docked Gα molecule fits well, with the exception of several residues in the αB–αC region of the all-helical domain that clash with the GGL domain and Gβ5 (Fig. 5d,e). Clashes in these elements could explain why Gα subunits have decreased affinity for Gβ5–R7-RGS complexes compared with isolated RGS domains from R7-RGS proteins32. Slight rotation of the RGS domain around its interface with Gβ5 (Fig. 5d) could allow activated Gα to bind Gβ5–RGS9 without steric hindrance. An alternative, and possibly complementary, mechanism is that side chains from the αB–αC region of Gα and the RGS9-GGL domain could adopt rotomer orientations that produce specific interactions between the two proteins (Fig. 4a and Fig 5e). Such an interface would provide an attractive mechanism for orchestrating the Gα subunit binding selectivity of R7-RGS proteins. Confirmation of the precise nature and functional role of this proposed interface awaits future experiments.
DISCUSSION
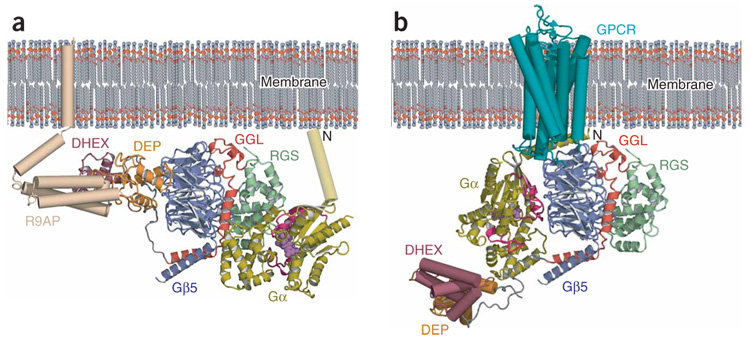
Several pieces of evidence suggest an overall orientation of Gβ5–RGS9 relative to membranes (Fig. 6a). For instance, the N terminus of RGS-bound Gα must be directed toward the membrane to allow anchoring through its lipophilic post-translational modifications33. Also, the R7-RGS N-terminal lobe would be oriented in a way that supports its interaction with membrane-anchoring proteins. A model accounting for these factors reveals a cluster of positive charge from the RGS9 N-terminal lobe favorably oriented toward the negatively charged membrane surface (Supplementary Fig. 3 online).
Figure 6.
Membrane orientation of the Gβ5–RGS9 complex. (a) A model of the membrane-relative orientation and interprotein interactions of the Gβ5–RGS9 complex. Gβ5 and RGS9, colored as in Figure 1, with Gαt/i (ref. 30; gold) docked and colored as in Figure 5d. A modeled N-terminal helix for Gαt/i is depicted as a cylinder, and a wheat-colored R9AP cartoon is positioned near the RGS9-Dishevelled/Egl-10/Pleckstrin homology (DEP) and RGS9-DEP helical extension (DHEX) domains. (b) A model of the Gβ5–RGS9 complex, colored and positioned as in a, with the DEP and DHEX domains arbitrarily repositioned to allow docking of a Gαt structure17 on Gβ5. The Gα subunit is colored as in Figure 1b. A cartoon depiction of a G protein–coupled receptor (GPCR)52 (cyan) is positioned in the membrane.
The structure of Gβ5–RGS9 represents a conjunction of signaling modules necessary for both G-protein activation and deactivation. Extensive genetic studies of C. elegans describe a model of reciprocal activation and deactivation of two Gα subunits mediated by distinct Gβ5–R7 complexes operating downstream of individual GPCRs34,35. In this model, GPB-2 (the Gβ5 ortholog), in complex with the R7-RGS proteins EGL-10 or EAT-16, heterotrimerizes with GDP-bound EGL-30 (Gαq) or GOA-1 (Gαo), respectively34,35. Thismodel suggests that GPCR-promoted nucleotide exchange on the Gα subunit of one GPB-2–R7-RGS–Gα heterotrimer results in release of the GPB-2–R7-RGS dimer, which functions as a GTPase-activating protein to inhibit the opposing G-protein signaling pathway. Consistent with such a Gβ5–R7-RGS–Gα heterotrimer model and the conserved Gα binding interface on Gβ5 described earlier (Fig. 3b,c), mouse Gαt-GDP was shown to stoichiometrically immunoprecipitate with RGS9 and Gβ5 from retinal rod outer segments13. Although these studies did not characterize the interaction mechanism of Gαt, its confirmed GDP-loaded state suggests that binding was mediated not by the RGS domain but by another element of this complex.
The biochemical and genetic studies discussed above indicate that an uncapping mechanism may exist in vivo to disrupt interactions between Gβ5 and the R7-RGS N-terminal lobe, and thus allow Gβ5 interaction with Gα subunits. Binding of R9AP and R7BP to the N-terminal lobe of R7-RGS proteins20,22 could contribute to the release of contacts with the putative Gα-interacting surface of Gβ5. Additionally, interactions between the DEP domain and certain GPCRs could participate in uncapping the conserved Gα binding surface of Gβ5 (refs. 10,36). Several RGS9 residues in the DHEX-GGL linker (Asn210–Thr218) make no contacts with Gβ5 (Fig. 1a and Supplementary Fig. 1) and could function as a hinge upon which the RGS9 N-terminal lobe pivots away from Gβ5. This same region of RGS6 and RGS7 is 25 residues longer than that region in RGS9 or RGS11 (Supplementary Fig. 1), perhaps introducing functional variability in the magnitude of N-terminal lobe reorientation with respect to Gβ5. Interestingly, repositioning of the RGS9 N-terminal lobe in our membrane orientation model would allow a ground-state Gα subunit to bind to Gβ5 with a similar membrane orientation to that generally accepted for conventional Gαβγ heterotrimers17 (Fig. 6b). Although understanding of the full implications of this arrangement awaits further work, the crystal structure of Gβ5–RGS9 together with existing biochemical and cellular studies point to a role of Gβ5–R7 dimers interacting directly with specific GPCRs and Gα subunits to modulate visual and neuronal signal transduction.
METHODS
Expression and crystallization of Gβ5 and RGS9
The coding sequences of Mus musculus Gβ5 (full-length short isoform) and RGS9 (residues 1–422) were amplified by PCR as described previously25,37 and inserted into the NcoI and XhoI endonuclease restriction sites of pFastBacHT (Invitrogen) and a modified pFastBacHT vector in which the His6 tag was replaced with a glutathione S-transferase (GST) tag (pFastBacGST), respectively. Baculovirus isolates were prepared following the Bac-to-Bac method (Invitrogen).
Recombinant His6-Gβ5 and GST-RGS9 proteins were coexpressed in High-Five insect cells for 48 hours at 27 °C after virus infection. Insect cells were lysed in 20 mM Tris (pH 7.5), 300 mM NaCl, 10 mM imidazole (pH 7.5), 1 mM DTT, 10% (v/v) glycerol, 0.5% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and Complete protease inhibitor cocktail (Roche) using an Emulsi-Flex C5 homogenizer (Avestin). His6-Gβ5 and GST-RGS9 complex was purified using sequential His Trap HP and GSTrap FF (GE Healthcare) column chromatography steps, and both the His6 and GST tags were cleaved with TEV protease. The cleaved tags and TEV protease were removed by repeating both affinity chromatography steps. The final purified complex was dialyzed and concentrated to 6.5 mg ml−1 in 20 mM PIPES (pH 6.5), 150 mM NaCl, 2 mM DTT and 5% (v/v) glycerol for crystallization.
Crystals of Gβ5–RGS9 were grown at 18 °C in sitting drops over a reservoir containing 100 mM Bis-Tris (pH 6.3), 5% (v/v) MME-PEG 550 and 10 mM DTT. Crystals were cryoprotected by stepping up to 20% (v/v) MME-PEG 550 and 20% (v/v) glycerol in the drop buffer. Rhenium or platinum derivatives were prepared by soaking crystals for 20–24 h in reservoir buffer with 5% (v/v) glycerol and 5 mM K2ReCl6 or 1 mM K2Pt(NO2)4, respectively, followed by back-soaking and cryoprotection. Derivatives with iodide were prepared by soaking cryoprotected crystals for 30–60 s in a final cryoprotection buffer containing 1 M NaI. Crystals were screened for diffraction after derivatization and cryoprotection at the University of North Carolina structural X-ray facility (Chapel Hill, North Carolina, USA).
Structure determination and analysis
The Gβ5–RGS9 structure was solved by combining phases obtained using both multiple isomorphous replacement with anomalous scattering (MIRAS) and molecular replacement (MR) techniques. All X-ray diffraction data sets for native and derivative crystals were collected at 100 K on SER-CAT beamline 22-ID at the Advanced Photon Source at the Argonne National Laboratory in Argonne, Illinois, USA. The data were indexed, integrated and scaled using HKL2000 (ref. 38). Heavy atom binding positions for eight rhenium, one platinum and ten iodine atoms were located and refined with CCP4 (ref. 39) programs, including FFT40 and MLPHARE39. The phasing power values (acentric/centric) were 0.78/0.54, 2.33/1.27, 2.48/1.29, 2.46/1.28 and 0.69/0.50 for the K2ReCl6 peak, K2Pt(NO2)4 peak, inflection and remote and NaI data sets, respectively. Phases obtained from heavy atom substructures were improved by solvent flattening, histogram matching and noncrystallographic symmetry averaging using the program DM41. MR replacement was performed using Phaser42 with a first search ensemble containing Gβγ structures23,43 (PDB 1TBG, 2TRC) and a second search ensemble composed of RGS9-RGS domain structures30 (PDB 1FQI, 1FQJ, 1FQK). MIRAS and MR phase combination was performed using Sigma-A44. The model building and refinement were done with Coot45 and Refmac5 (ref. 46). Translation, libration and screw-rotation displacement (TLS) refinement was performed using different TLS groups for each of the two Gβ5–RGS9 dimers in the crystal asymmetric unit and for the various structural domains and elements of both RGS9 (residues 7–15, 16–104, 105–118, 119–192, 193–218, 219–273, 274–289 and 290–421) and Gβ5 (residues 9–36, 37–53 and 54–353). Data collection and structure refinement statistics are shown in Table 1. For the final refined structure, 91.9% and 8.1% of the residues were in the favored and allowed regions, respectively, of the Ramachandran plot. The mean B factor reported reflects the sum of both TLS-derived and residual B factor components. Electron density was visible for residues 7–421 from both RGS9 molecules (chains A and C) and for residues 9–353 and 1–353 of Gβ5 chains B and D, respectively. The increased N-terminal stability of the Gβ5 chain D results from crystal contacts and is not likely to reflect native structure.
Table 1.
Data collection and refinement statistics for multiple isomorphous replacement with anomalous scattering (MIRAS) structures
| Native | K2ReCl6 | K2Pt(NO2)4 | NaI | |||
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P212121 | P212121 | P212121 | P212121 | ||
| Cell dimensions a, b, c (Å) | 110.2, 119.0, 132.0 | 109.3, 120.8, 129.6 | 110.2, 119.1, 130.1 | 110.7, 118.9, 131.0 | ||
| Peak | Peak | Inflection | Remote | |||
| Wavelength | 1.0000 | 1.1766 | 1.0719 | 1.0722 | 1.0630 | 1.5400 |
| Resolution (Å) | 50–1.95 | 50–2.80 | 50–2.20 | 50–2.25 | 50–2.45 | 50–2.90 |
| (2.02–1.95)a | (2.90–2.80) | (2.28–2.20) | (2.33–2.25) | (2.54–2.45) | (3.00–2.90) | |
| Rsym | 0.075 (0.434) | 0.108 (0.518) | 0.075 (0.460) | 0.076 (0.588) | 0.080 (0.614) | 0.116 (0.470) |
| I / σI | 22.4 (3.3) | 20.5 (2.1) | 35.3 (1.9) | 33.3 (1.9) | 34.7 (1.8) | 21.6 (2.2) |
| Completeness (%) | 99.9 (99.7) | 95.0 (69.1) | 92.6 (60.2) | 92.4 (57.7) | 96.7 (74.0) | 90.9 (58.6) |
| Redundancy | 4.1 (4.1) | 8.9 (5.3) | 13.1 (7.8) | 12.8 (6.9) | 13.2 (7.0) | 9.3 (5.2) |
| Refinement | ||||||
| Resolution (Å) | 20.0–1.95 | |||||
| No. reflections | 119,175 | |||||
| Rwork/Rfree | 18.3/22.5 | |||||
| No. atoms | ||||||
| Protein | 12365 | |||||
| Water | 890 | |||||
| B-factors | ||||||
| Protein | 34.8 | |||||
| Water | 40.2 | |||||
| r.m.s. deviations | ||||||
| Bond lengths (Å) | 0.010 | |||||
| Bond angles (°) | 1.16 | |||||
A total of four crystals were used in solving the structure.
Values in parentheses are for the highest-resolution shell.
Ungapped superposition of the two Gβ5–RGS9 complexes in the crystal asymmetric unit was performed with the LSQKAB module of CCP4 (ref. 39) using Cα atoms from 415 RGS9 and 345 Gβ5 residues. Additional structure superpositions were performed with the SSM function of Coot45. Superposition of the DEP domains from RGS9 and Dishevelled (PDB 1FSH) was carried out using 71 out of 86 and 85 Cα atoms, respectively, with six gaps. Gβ5 and Gβ1 (PDB 1GOT) were superimposed using 330 out of 353 and 339 Cα atoms, respectively, with five gaps. Gγ transducin (PDB 1GOT) and the RGS9 GGL domain were superimposed using 50 out of 55 and 54 Cα atoms, respectively, with two gaps. The bovine (PDB 1FQI) and mouse RGS9-RGS domains were superimposed using 134 Cα atoms from each, without gaps. Sequence alignments were initially performed with ClustalX47 then hand edited based on comparison of superposed structures.
A Gαt/i docked model was prepared by superimposing the RGS domain from the Gβ5–RGS9 structure with an RGS9-RGS domain and Gαt/i complex structure30 (PDB 1FQJ). A fragment of PDEγ that was also present in this structure would have been positioned directly behind the model of Gαt/i as it is presented in Figure 6a, but was removed from the figure to minimize clutter. An N-terminal helix was positioned on the docked Gαt/i (Fig. 6a) by superposition with a Gαt structure containing this structural element17 (PDB 1GOT). Gαt was docked on the top surface of Gβ5 (Fig. 6b) by superposition of Gβ5 with Gβ1 from the heterotrimeric structure17 (PDB 1GOT). Buried solvent-accessible surface area was calculated with CCP4 program AreaIMol48. Specific residue contact distances were determined as atoms within 120% of their summed van der Waals radii for van der Waals contacts or atoms closer than 3.5 Å for hydrogen bonds, as measured using the CCP4-Molecular Graphics49 program. The PDB was searched for structures with similarity to the DHEX domain using DALI50, and the closest structural homolog had a z score of 4.5 and an r.m.s. deviation of 3.4 Å over 76 Cα atoms in ten fragments. Electrostatics calculations were performed, and Supplementary Fig. 3 was prepared using SPOCK51. Although electrostatic potential maps calculated at ±2 kT or ±5 kT were qualitatively similar, Supplementary Figure 3 was prepared using those from the ±2 kT calculations to highlight the basic patch near the N-terminal lobe of RGS9.
Supplementary Material
Note Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank L. Betts for suggestions in analyzing diffraction data. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. We thank the SER-CAT beamline staff for assistance in data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences. This research was funded by grants from the US National Institutes of Health (P01-GM65533 and R01-GM081881 to J.S. and T.K.H.), the American Cancer Society (PF-06-034-01-GMC to M.L.C) and the University of North Carolina Lineberger Comprehensive Cancer Center (M.L.C.).
Footnotes
Accession codes. Protein Data Bank: Atomic coordinates and structure factors for the Gβ5 and RGS9 complex have been deposited with the accession code 2PBI.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Gilman AG. Nobel Lecture. G proteins and regulation of adenylyl cyclase. Biosci. Rep. 1995;15:65–97. doi: 10.1007/BF01200143. [DOI] [PubMed] [Google Scholar]
- 2.Berman DM, Gilman AG. Mammalian RGS proteins: barbarians at the gate. J. Biol. Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 3.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 4.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 5.Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J. Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat. Rev. Drug Discov. 2002;1:187–197. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]
- 7.Krispel CM, et al. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Zachariou V, et al. Essential role for RGS9 in opiate action. Proc. Natl. Acad. Sci. USA. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman Z, et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- 10.Kovoor A, et al. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J. Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiguchi KM, et al. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- 12.Lishko PV, Martemyanov KA, Hopp JA, Arshavsky VY. Specific binding of RGS9-Gb5L to protein anchor in photoreceptor membranes greatly enhances its catalytic activity. J. Biol. Chem. 2002;277:24376–24381. doi: 10.1074/jbc.M203237200. [DOI] [PubMed] [Google Scholar]
- 13.Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc. Natl. Acad. Sci. USA. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a Novel Neuronal protein interacting with RGS proteins of the R7 family. J. Biol. Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- 15.Sondek J, Siderovski DP. Gγ-like (GGL) domains: new frontiers in G-protein signaling and β-propeller scaffolding. Biochem. Pharmacol. 2001;61:1329–1337. doi: 10.1016/s0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- 16.Jones MB, Siderovski DP, Hooks SB. The Gβγ dimer as a novel source of selectivity in G-protein signaling: GGL-ing at convention. Mol. Interv. 2004;4:200–214. doi: 10.1124/mi.4.4.4. [DOI] [PubMed] [Google Scholar]
- 17.Lambright DG, et al. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 18.Wall MA, Posner BA, Sprang SR. Structural basis of activity and subunit recognition in G protein heterotrimers. Structure. 1998;6:1169–1183. doi: 10.1016/s0969-2126(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 19.Wong HC, et al. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat. Struct. Biol. 2000;7:1178–1184. doi: 10.1038/82047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson GR, Semenov A, Song JH, Martemyanov KA. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9-2. J. Biol. Chem. 2006;282:4772–4781. doi: 10.1074/jbc.M610518200. [DOI] [PubMed] [Google Scholar]
- 21.Patikoglou GA, Koelle MR. An N-terminal region of Caenorhabditis elegans RGS proteins EGL-10 and EAT-16 directs inhibition of Gαo versus Gαq signaling. J. Biol. Chem. 2002;277:47004–47013. doi: 10.1074/jbc.M208186200. [DOI] [PubMed] [Google Scholar]
- 22.Martemyanov KA, et al. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J. Neurosci. 2003;23:10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a GA protein βγ dimer at 2.1Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 24.Watson AJ, Katz A, Simon MI. A fifth member of the mammalian G-protein β-subunit family. Expression in brain and activation of the β2 isotype of phospholipase C. J. Biol. Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- 25.Snow BE, et al. A G protein γ subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gβ5 subunits. Proc. Natl. Acad. Sci. USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ford CE, et al. Molecular basis for interactions of G protein βγ subunits with effectors. Science. 1998;280:1271–1274. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 27.Posner BA, Gilman AG, Harris BA. Regulators of G protein signaling 6 and 7. Purification of complexes with Gβ5 and assessment of their effects on G protein-mediated signaling pathways. J. Biol. Chem. 1999;274:31087–31093. doi: 10.1074/jbc.274.43.31087. [DOI] [PubMed] [Google Scholar]
- 28.Snow BE, Betts L, Mangion J, Sondek J, Siderovski DP. Fidelity of G protein β-subunit association by the G protein γ-subunit-like domains of RGS6, RGS7, and RGS11. Proc. Natl. Acad. Sci. USA. 1999;96:6489–6494. doi: 10.1073/pnas.96.11.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dingus J, et al. G Protein βγ dimer formation: Gβ and Gγ differentially determine efficiency of in vitro dimer formation. Biochemistry. 2005;44:11882–11890. doi: 10.1021/bi0504254. [DOI] [PubMed] [Google Scholar]
- 30.Slep KC, et al. Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 Å. Nature. 2001;409:1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 31.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4¯ -activated Giα1: stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 32.Skiba NP, et al. RGS9-G β 5 substrate selectivity in photoreceptors. Opposing effects of constituent domains yield high affinity of RGS interaction with the G protein-effector complex. J. Biol. Chem. 2001;276:37365–37372. doi: 10.1074/jbc.M106431200. [DOI] [PubMed] [Google Scholar]
- 33.Chen CA, Manning DR. Regulation of G proteins by covalent modification. Oncogene. 2001;20:1643–1652. doi: 10.1038/sj.onc.1204185. [DOI] [PubMed] [Google Scholar]
- 34.Hajdu-Cronin YM, Chen WJ, Patikoglou G, Koelle MR, Sternberg PW. Antagonism between Goα and Gqα in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 1999;13:1780–1793. doi: 10.1101/gad.13.14.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robatzek M, Niacaris T, Steger K, Avery L, Thomas JH. eat-11 encodes GPB-2, a Gβ5 ortholog that interacts with Goα and Gqα to regulate C. elegans behavior. Curr. Biol. 2001;11:288–293. doi: 10.1016/s0960-9822(01)00074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballon DR, et al. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 37.Hooks SB, et al. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J. Biol. Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 40.Ten Eyck LF. Crystallographic fast Fourier transforms. Acta Crystallogr. A. 1973;29:183–191. [Google Scholar]
- 41.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 43.Gaudet R, Bohm A, Sigler PB. Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 44.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A. 1986;42:140–149. [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 49.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 50.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 51.Christopher JA. The Spock Homepage. 1998 〈 http://quorum.tamu.edu/〉.
- 52.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note Supplementary information is available on the Nature Structural & Molecular Biology website.