Abstract
Background
RAS genes acquire the most common somatic gain-of-function mutations in human cancer, and almost all of these mutations are located at codons 12, 13, 61, and 146.
Methods
We present a method for detecting these K-RAS hotspot mutations in 228 cases of colorectal cancer. The protocol is based on the multiplex amplification of exons 2, 3 and 4 in a single tube, followed by primer extension of the PCR products using various sizes of primers to detect base changes at codons 12, 13, 61 and 146. We compared the clinicopathological data of colorectal cancer patients with the K-RAS mutation status.
Results
K-RAS mutation occurred in 36% (83/228) of our colorectal cancer cases. Univariate analysis revealed a significant association between K-RAS mutation at codon 12 of exon 2 and poor 5-year survival (p = 0.023) and lymph node involvement (p = 0.048). Also, K-RAS mutation at codon 13 of exon 2 correlates with the size of the tumor (p = 0.03). Multivariate analysis adjusted for tumor size, histologic grade, and lymph node metastasis also indicated K-RAS mutations at codon 12 and 13 of exon 2 correlate significantly with overall survival (p = 0.002 and 0.025). No association was observed between codon 61 and 146 and clinicopathological features.
Conclusion
We demonstrated a simple and fast way to identify K-RAS mutation.
Background
The RAS genes encode a family of GTPases that act as signal switch molecules for many important cellular processes. Common in human cancers, activating ras mutations cause the deregulation of ras protein activity, which results in the loss of GTPase activity and the gain of oncogenic activity [1-7]. In fact, RAS genes are the genes that most commonly show somatic gain-of-function mutations in human cancers [8,9]. There are three cellular RAS genes, encoding the K-ras, H-ras and N-ras proteins, and all show activating mutations in human tumors. Of the three RAS genes, K-RAS is the most frequently mutated, which makes it an ideal target for cancer treatment [9,10]. Meanwhile, K-RAS mutation plays an important role of the response rate of anti-EGFR antibodies treatment for patients with metastatic colorectal cancer [11,12].
RAS point mutations are highly prevalent in human cancers and mostly occur in codons 12, 13 and 61. These mutations render the ras proteins insensitive to GTP-induced hydrolysis of GTP to GDP and lock them in the activated state [2,8,9]. Mutations in codons 10, 11, 15, 18, 19, and 22 have also been reported, but their biological significance is unclear [13-18]. Several methods have been developed for detecting K-RAS mutations [19-26], but these methods do not simultaneously recognize all base changes at codons 12, 13 or 61 of K-RAS. Somatic missense mutation at codon 146 were found in three independent studies of colorectal cancers from Hong Kong and the United States, and it was suggested that these mutations may make an equal or greater contribution to colorectal cancer than the codon 61 mutation [27-29]. Direct sequencing of exons 2 and 3 can recognize the base changes in codons 12, 13 and 61. But, codon 146 which is located at exon 4, is too far apart (19,771 base pairs) from exon 2 to be sequenced in a single reaction. In this study, we employed a method to simultaneously detect the base changes in codons 12, 13, 61 and 146 of K-RAS in a single tube and establish their clinical significance in colorectal cancer.
Results
K-RAS mutation detection assay
We designed primers adjacent to codons 12, 13, 61 and 146 of the K-RAS gene, as these are the most common mutations in cancer. For mutation analysis of codons 12 and 13, we used different-sized primers (sense strand) to recognize the change of the first base separately, and we designed antisense strand primers to detect the change of the second base of the codon to avoid interference between primers of the same direction. We did not design primers to detect change at the third base of codon 12 or 13 because these mutations do not result in the change of the amino acid. For the analysis of codon 61, we designed three different-sized primers (sense strand) to distinguish changes in the first, second, and third bases separately. The primers were made to be different in size either by adding different lengths of poly(dT) tails to the 5'-end or extending the primer sequence in order to allow for separation based on the differences in size.
First, we analyzed codon 12 mutations using a 20mer sense strand primer to detect the first base changes of codon 12 and a 29mer antisense strand primer to detect the second base changes of codon 12. To the tube with codon 12 primers, we added two more primers to detect the base changes at the first and second bases of codon 13 (41mer sense strand for the first base; 49mer antisense strand for the second base). The results suggested that the primers worked well in spite of the fact that the primers used to detect the first or the second base changes of codons 12 and 13 are of the same direction, suggesting that these primers do not interfere with each other. The extended products were analyzed on the Beckman Coulter automatic sequencer in a 17-minute run, and all the base changes of codons 12 and 13 were detected. These results indicated that the direction of the primer do not play an important role in detecting the base changes at a given codon. We then used different-sized primers of the same direction (sense strand) to recognize the first and the second base changes separately, and all the base changes of codons 12 and 13 were detected. We also used degenerate probes (containing a degenerate base at the last base of the probe to detect the second base change of codon 12 or 13) and they also worked well (data not shown). After this preliminary study using four probes to detect codon 12 and codon 13 mutations, we added five more probes to detect codon 61 and codon 146 mutations. This collection of probes defines our multiplex PCR and primer extension method for codons 12, 13, 61, and 146.
We then used our multiplex PCR and primer extension method to assay 228 cases of colorectal cancer. The results showed that 83 cases have K-RAS mutations in the hotspots that we screened. Of the 83 mutant cases, there were 58 cases of codon 12 mutations including 7 cases of GGT→TGT, 3 cases of GGT→AGT, 18 cases of GGT-GTT, 29 cases of GGT→GAT and 1 case of GGT→GCT. There are 21 cases of codon 13 mutations, including 1 case of GGC→CGC and 20 cases of GGC→GAC. There was one example showing a mutation of codon 61 (CAA→CAT), and there were three cases of codon 146 mutations, all of which were of the GCA→ACA mutation. The mutations in all of these cases were heterozygous.
Sensitivity of the K-RAS mutation assay
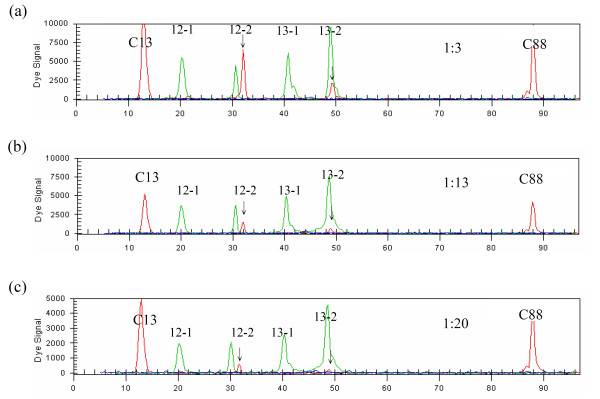
We next determined the minimal amount of DNA required for reliable mutation detection. We diluted DNA samples harbouring codon 12 and 13 mutations, and analyzed the samples as described above. For each of the mutations, we found that 10 ng of DNA was enough for the multiplex PCR reaction. We subsequently determined the lowest concentration of mutant DNA in a background of normal DNA needed for mutation detection. We mixed tumor DNA (heterozygous for the mutation at codons 12 and 13) with control DNA in different ratios (1:3, 1:13 and 1:20) (Fig. 1). The result showed that this method can detect K-RAS mutations in a DNA mixture containing as little as 5% mutant DNA.
Figure 1.
Determining the sensitivity of multiplex PCR and primer extension analysis of K-RAS mutation by serial dilution of the samples. Upper (a), middle (b) and lower (c) panels are 1: 3, 1: 13 and 1: 20 dilution, respectively.
Mutational analysis of K-RAS gene in colorectal cancers
We used direct sequencing to analyze the mutations of the K-RAS gene in the coding and intron-exon junction regions of exons 2, 3, and 4. As expected, the results showed hotspot K-RAS mutation in 83 of the 228 cases. The results of sequencing analysis were identical to the results of multiplex PCR with primer extension analysis. There were no compound heterozygous mutations or homozygous mutations in any of these cases, and the mutational frequencies of codons 12, 13, 61, and 146 were 25.4%, 9.2%, 0.4% and 1.3%, respectively. We did not find any other mutations, such as mutations at codons 10, 11, 15, 18, 19 or 22. We can therefore conclude that our method was very accurate in profiling the mutations in colorectal cancers, and will detect the majority, if not all, of the K-RAS mutations commonly seen in cancer.
Correlation between K-RAS mutations and clinicopathological data in colorectal cancers
We compared the clinicopathological data of colorectal cancer patients with the mutational status of K-RAS in their cancerous tissues. The univariate analysis revealed that there is a significant association between K-RAS mutation at codon 12 of exon 2 and poor 5-year survival. This mutation also serves as a good indicator of lymph node involvement in this disease (Table 1). Furthermore, K-RAS mutation at codon 13 of exon 2 correlates with the size of the tumor (>3 cm, p = 0.03). In the multivariate analysis which incorporated independent prognostic factors of tumor size, histologic grade, and lymph node metastasis, we found that K-RAS mutations at codon 12 and 13 of exon 2 were both significant with regard to overall survival (p = 0.002 and 0.025) (Table 2). Lymph node involvement also correlates significantly with overall survival (p = 0.001). No association was observed between codon 61 and 146 and clinicopathological features.
Table 1.
Correlation between clinicopathological features and K-RAS mutation of codon 12 1st base and codon 13 2nd base
| mutation of codon 12 1st base | mutation of codon 13 2nd base | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| no | yes | total | p-value | no | yes | total | p-value | ||
| Tumor size | <= 3 cm | 55 | 5 | 57 | 1 | 56 | 1 | 57 | 0.03 |
| >3 cm | 160 | 8 | 168 | 149 | 19 | 168 | |||
| Grade | Well | 4 | 0 | 4 | 0.749 | 4 | 0 | 4 | 0.416 |
| Moderate | 171 | 9 | 180 | 161 | 19 | 180 | |||
| Poor | 10 | 1 | 11 | 11 | 0 | 11 | |||
| Lymph node metastasis | - | 107 | 2 | 109 | 0.048 | 101 | 8 | 109 | 0.621 |
| + | 88 | 8 | 96 | 87 | 9 | 96 | |||
| Stage | I, II | 76 | 0 | 76 | 0.121 | 70 | 6 | 76 | 0.782 |
| III, IV | 76 | 4 | 80 | 72 | 8 | 80 | |||
| Survival | <= 5 years | 114 | 9 | 123 | 0.023 | 109 | 14 | 123 | 0.162 |
| >5 years | 104 | 1 | 105 | 99 | 6 | 105 | |||
p-value by Chi-square Test or Fisher's Exact Test when appropriated
Table 2.
Multivariate analysis (Cox regression) of independent prognostic factors in patients with colorectal cancer
| Harzard Ratio | 95% CI | p-value | ||
|---|---|---|---|---|
| Mutation Codon 12 1st base | - | 1.0 | 1.14 to 4.71 | 0.002 |
| + | 2.3 | |||
| Codon 13 2nd base | - | 1.0 | 1.09 to 3.58 | 0.025 |
| + | 2.0 | |||
| Tumor size | <= 3 cm | 1.0 | 0.60 to 1.48 | 0.803 |
| >3 cm | 0.9 | |||
| Grade | well | 1.0 | 0.26 to 2.69 | 0.762 |
| moderate/poor | 0.8 | |||
| Lymph node metastasis | - | 1.0 | 1.33 to 2.86 | 0.001 |
| + | 2.0 |
Discussion and Conclusion
K-RAS mutations are common in human cancers and play a very important role in various processes of cancer development, including cancer initiation, metastasis, prognosis and response for treatment [1-7,10,11,30-32]. Therefore, mutation detection of K-RAS is of clinical importance in cancer studies. In this study, we present a simple assay to detect K-RAS mutations in colorectal cancer. Several methods can be used to detect K-RAS mutations, including PCR and direct sequencing, PCR-RFLP and direct sequencing, PCR-SSCP and direct sequencing, PCR and probe hybridization, and allele-specific competitive blocker PCR [19-26,33,34]. These methods are laborious because the mutation hotspots in exons 2, 3 and 4 have to be screened separately. Furthermore, the common mutations seen at codons 12, 13, 61 and 146 are not the result of any single nucleotide change; instead, every base at these codons has 3 or 4 types of base changes. It is therefore difficult to use the above-mentioned methods to precisely detect all the base changes without using direct sequencing or hybridization. Although the probe hybridization method requires no direct sequencing, it requires several probes to accurately determine base changes. With our method, the detection of base changes in three key exons of K-RAS can be combined into one assay, which allows a sample to be screened for all K-RAS mutation hotspots simultaneously. Because the technique is a sequencing-based approach, additional sequencing is not necessary. Meanwhile, the advantage of using primer extension over hybridization with allele-specific oligonucleotide probes to distinguish sequence variants is based on the high accuracy of the nucleotide incorporation reaction catalyzed by a DNA polymerase compared with the caveats of hybridization. These caveats include differences in thermal stability between mismatched and perfectly matched hybrids formed with the allele-specific oligonucleotide probes. Current products of the thermostable enzymes used in primer extension reaction have very low error rates and have increased efficiency and specificity for ddNTPs [35]. These characteristics provide negligible primer misincorporation and excellent discrimination between wild, heterozygous and homozygous genotypes. Additionally, the same reaction conditions can be used to detect any variable nucleotide regardless of the nucleotide sequence flanking the variable site. Another advantage of the primer extension reaction is its multiplexing capability, with several mutations being detected in a single reaction tube. Although multiplex PCR-SSCP- or PCR-ARMS- based methods can also simultaneously detect several mutations, PCR-SSCP require further confirmation by direct sequencing, and using PCR-ARMS to detect all the changes at codons 12, 13, 61 and 146 require more primers than are possible for one reaction tube. Our multiplex primer extension methods can detect more than 40 types of changes at these 4 codons. Compared with direct sequencing, primer extension reaction assays are faster because automated fluorescent capillary electrophoresis of the products requires only 25 minutes in comparison with more than one hour for capillary electrophoresis require for standard sequencing. However, we should point out that our proposed method can only detect K-RAS mutations in a DNA mixture containing at least 5% mutant DNA. Moreover, our method can not detect K-RAS mutations occurred outside the codons that we targeted although direct sequencing we performed did not find any either. In the past, primer extension-based methods were used for detecting a single base change across several conditions, and they have rarely been used to detect the three base changes at a given codon [36-40]. In this study, we extend the application to detect all the base changes at a given codon of K-RAS gene simultaneously and established it as a simple and fast way to screen K-RAS gene. Our results indicated a strong association between K-RAS mutation in exon 2 and clinical outcome in terms of survival, lymph node involvement, and tumor size. Mutated exon 2 of K-RAS, thus represents a molecular lesion in the development of more aggressive disease. The effectiveness of cetuximab in colorectal cancer is strongly associated with K-RAS mutation status [12]. The method that we demonstrated in this report provides a simple, fast, and reliable way to identify K-RAS mutation for the purpose of clinical evaluation in colorectal cancer. Meanwhile, the principle of the method can also be used to detect H-RAS and N-RAS mutations, or to detect the mutation at a given codon in other genes in colorectal and other cancers
Methods
Samples
Resected primary colorectal cancers were obtained from 228 patients in Changhua Christian Hospital. All the cases were adenocarcinomas and were classified as "poorly-differentiated" to "well-differentiated". All patients were staged according to the 2002 American Joint Committee on Cancer staging system [41]. The tumor specimens were frozen immediately after surgical resection and stored in liquid nitrogen until DNA was extracted. The age of the patients ranged from 36–72 years with a mean of 49 years. This study was approved by the Institute Review Board of Changhua Christian Hospital.
DNA extraction, PCR and direct sequencing of the K-RAS gene
DNA extraction was performed as previously described [33]. Three pairs of primers were used to amplify the exon-intron junctions and coding regions of exons 2, 3, and 4 of the K-RAS gene (see Additional file 1). The PCR was performed with a denaturing step at 94°C for 5 min, then 30 sec at 94°C, 30 sec at 56°C, and 1 min at 72°C for 35 cycles, followed by a final 5 min at 72°C. The PCR products were visualized on a 2.5% agarose gel. These PCR products were then subjected to direct sequencing using the same primers, and all mutations were confirmed by sequences originating from both the upstream and downstream primers. Direct sequencing was performed on a Beckman Coulter CEQ 8000 Series Genetic Analysis System (Beckman Coulter Inc., Fullerton, CA, USA) according to manufacturer instructions.
Multiplex PCR and primer extension analysis of mutations in K-RAS codons 12, 13, 61 and 146
Multiplex PCR was used to amplify exons 2, 3 and 4 of K-RAS gene in a single tube. The primers used for the multiplex PCR were identical to those used for the direct sequencing analysis. After multiplex PCR amplification, the PCR products were purified to remove the remaining primers and unincorporated deoxynucleotide triphosphates, using the PCR-M™ Clean Up System (Viogene-biotek Co., Sunnyvale, CA, USA) or a simple treatment with exonuclease I and shrimp alkaline phosphatase. After removing the primers, the products were subjected to primer extension analysis. The primers ranged from 20–65 bases, and most probed the sense strand (see Additional file 2). The exception was when there were adjacent mutations; in this case both sense and antisense strand primers were used. Degenerate bases were used in the primers to identify adjacent mutations. Various concentrations of probe for either codon 12, 13, 61 or 146, were added to the tube containing purified PCR products, as well as 4 μl of SNPStart Master Mix (Beckman Coulter Inc., Fullerton, CA, USA) containing Taq DNA polymerase and fluorescently labelled ddNTPs. Each 10-μl mixture was subjected to 30 single-base extension cycles consisting of a denaturation step at 90°C for 10 sec, and primer annealing and extension at 55°C for 20 sec. After cycle extension, unincorporated fluorescent ddNTPs were incubated with 0.25 μl of shrimp alkaline phosphatase (SAP) (United States Biochemical Co., Cleveland, USA), 1.3 μl SAP buffer, and 1.45 μl ddH2O at 37°C for 30 min, followed by enzyme deactivation at 65°C for 15 min.
The primer extension reaction products were resolved by automated capillary electrophoresis on a capillary electrophoresis platform. Briefly, 38.75 μl of sample loading solution and 0.25 μl of Size Standard 80 (Beckman Coulter Inc., Fullerton, CA, USA) were added to 1 μl of primer extension products. The fluorescently tagged extended products in the mixture were electrophoretically separated across a 36-cm capillary containing POP-4 for 17 min and analyzed using GeneScan application software (Beckman Coulter Inc., Fullerton, CA, USA)
Statistics
Comparison between clinicopathological features and status of K-RAS mutation in colorectal cancers were analyzed by Chi-square test or Fisher's exact test. Independent prognostic factors were analyzed by the Cox proportional harzards regression model. Variables in the model included tumor size, histologic grade, and lymph node metastasis. A P value of less than 0.05 was considered statistically significant.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YSC performed the experiments and drafted the manuscript. KTY helped to design the study. TJC and NCH participated in the statistical analysis. CC helped to design the study. HCL participated in the coordination of the study. JGC design the study. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Primers used to amplify exon-intron junctions and coding regions of exons 2, 3, and 4 of the K-RAS gene. This table includes the primers used in the multiplex PCR of K-RAS.
The sequences of primers used for detection of hotspot mutations of K-RAS. This table lists the primers used in the mutation analysis of K-RAS by the method of primer extension.
Contributor Information
Ya-Sian Chang, Email: 970494@ms.kmuh.org.tw.
Kun-Tu Yeh, Email: 10159@cch.org.tw.
Tien-Jye Chang, Email: tjchang@dragon.nchu.edu.tw.
Connie Chai, Email: connie.chai@duke.edu.
Hsiu-Chin Lu, Email: lu40130960@gmail.com.
Nicholas C Hsu, Email: nc.hsu@msa.hinet.net.
Jan-Gowth Chang, Email: jgchang@ms.kmuh.org.tw.
Acknowledgements
We thank Shu-Hui Lin for technical advice. This study was supported by the National Science Council of Taiwan (NSC-95-2320-B-037-050-MY3), a grant from Changhua Christian Hospital, and a grant from Kaohsiung Medical University Hospital.
References
- Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- Singh A, Sowjanya AP, Ramakrishna G. The wild-type Ras: road ahead. FASEB J. 2005;19:161–170. doi: 10.1096/fj.04-2584hyp. [DOI] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Minamoto T, Mai M, Ronai Z. K-ras mutation: early detection in molecular diagnosis and risk assessment of colorectal, pancreas, and lung cancers – a review. Cancer Detect Prev. 2000;24:1–12. [PubMed] [Google Scholar]
- Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochim Biophys Acta. 2005;25(1756):127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Imamura N, Kuramoto A, Ishihara H, Shimizu S. Detection of high incidence of H-RAS oncogene point mutations in acute myelogenous leukemia. Am J Hematol. 1993;43:151–153. doi: 10.1002/ajh.2830430217. [DOI] [PubMed] [Google Scholar]
- Hongyo T, Buzard GS, Palli D, Weghorst CM, Amorosi A, Galli M, Caporaso NE, Fraumeni JF Jr, Rice JM. Mutations of the K-ras and p53 genes in gastric adenocarcinomas from a high-incidence region around Florence, Italy. Cancer Res. 1995;55:2665–2672. [PubMed] [Google Scholar]
- Suzuki Y, Orita M, Shiraishi M, Hayashi K, Sekiya T. Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene. 1990;5:1037–1043. [PubMed] [Google Scholar]
- Tsukuda K, Tanino M, Soga H, Shimizu N, Shimizu K. A novel activating mutation of the K-ras gene in human primary colon adenocarcinoma. Biochem Biophys Res Commun. 2000;278:653–658. doi: 10.1006/bbrc.2000.3839. [DOI] [PubMed] [Google Scholar]
- Wang JY, Wang YH, Jao SW, Lu CY, Kuo CH, Hu HM, Hsieh JS, Chong IW, Cheng TL, Lin SR. Molecular mechanisms underlying the tumorigenesis of colorectal adenomas: correlation to activated K-ras oncogene. Oncol Rep. 2006;16:1245–1252. [PubMed] [Google Scholar]
- Akagi K, Uchibori R, Yamaguchi K, Kurosawa K, Tanaka Y, Kozu T. Characterization of a novel oncogenic K-ras mutation in colon cancer. Biochem Biophys Res Commun. 2007;352:728–732. doi: 10.1016/j.bbrc.2006.11.091. [DOI] [PubMed] [Google Scholar]
- Poehlmann A, Kuester D, Meyer F, Lippert H, Roessner A, Schneider-Stock R. K-ras mutation detection in colorectal cancer using the Pyrosequencing technique. Pathol Res Pract. 2007;203:489–497. doi: 10.1016/j.prp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- van Heek NT, Clayton SJ, Sturm PD, Walker J, Gouma DJ, Noorduyn LA, Offerhaus GJ, Fox JC. Comparison of the novel quantitative ARMS assay and an enriched PCR-ASO assay for K-ras mutations with conventional cytology on endobiliary brush cytology from 312 consecutive extrahepatic biliary stenoses. J Clin Pathol. 2005;58:1315–1320. doi: 10.1136/jcp.2004.022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taback B, Bilchik AJ, Saha S, Nakayama T, Wiese DA, Turner RR, Kuo CT, Hoon DS. Peptide nucleic acid clamp PCR: a novel K-ras mutation detection assay for colorectal cancer micrometastases in lymph nodes. Int J Cancer. 2004;111:409–414. doi: 10.1002/ijc.20268. [DOI] [PubMed] [Google Scholar]
- Lleonart ME, Ramón y Cajal S, Groopman JD, Friesen MD. Sensitive and specific detection of K-ras mutations in colon tumors by short oligonucleotide mass analysis. Nucleic Acids Res. 2004;32:e53. doi: 10.1093/nar/gnh051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JD, Chan EC, Shih CL, Chen TL, Liang Y, Hwang TL, Chiou CC. Detection of rare mutant K-ras DNA in a single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe. Nucleic Acids Res. 2006;34:e12. doi: 10.1093/nar/gnj008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørheim J, Lystad S, Lindblom A, Kressner U, Westring S, Wahlberg S, Lindmark G, Gaudernack G, Ekstrøm P, Røe J. Mutation analyses of KRAS exon 1 comparing three different techniques: temporal temperature gradient electrophoresis, constant denaturant capillary electrophoresis and allele specific polymerase chain reaction. Mutat Res. 1998;403:103–112. doi: 10.1016/s0027-5107(98)00057-8. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shiesh SC, Wu SJ. Rapid detection of K-ras mutations in bile by peptide nucleic acid-mediated PCR clamping and melting curve analysis: comparison with restriction fragment length polymorphism analysis. Clin Chem. 2004;50:481–489. doi: 10.1373/clinchem.2003.024505. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Nagaoka T, Taniguchi T, Higashi H, Sugimura H, Sugano K, Yonekawa H, Satoh T, Horii T, Shirai N. Three-dimensional microarray compared with PCR-single-strand conformation polymorphism analysis/DNA sequencing for mutation analysis of K-ras codons 12 and 13. Clin Chem. 2004;50:1322–1327. doi: 10.1373/clinchem.2004.032060. [DOI] [PubMed] [Google Scholar]
- Orita S, Higashi T, Kawasaki Y, Harada A, Igarashi H, Monden T, Morimoto H, Shimano T, Mori T, Miyoshi J. A novel point mutation at codon 146 of the K-ras gene in a human colorectal cancer identified by the polymerase chain reaction. Virus Genes. 1991;5:75–79. doi: 10.1007/BF00571733. [DOI] [PubMed] [Google Scholar]
- Sloan SR, Newcomb EW, Pellicer A. Neutron radiation can activate K-ras via a point mutation in codon 146 and induces a different spectrum of ras mutations than does gamma radiation. Mol Cell Biol. 1990;10:405–408. doi: 10.1128/mcb.10.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edkins S, O'Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakman N, Borel Rinkes IH, Voest EE, Kranenburg O. Control of colorectal metastasis formation by K-Ras. Biochim Biophys Acta. 2005;1756:103–114. doi: 10.1016/j.bbcan.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Castagnola P, Giaretti W. Mutant KRAS, chromosomal instability and prognosis in colorectal cancer. Biochim Biophys Acta. 2005;1756:115–125. doi: 10.1016/j.bbcan.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Chen PH, Wang CK, Liu JD, Siauw CP, Chen YJ, Yang MJ, Liu MH, Chen TC, Chang JG. Mutation analysis of K-ras oncogenes in gastroenterologic cancers by the amplified created restriction sites method. Am J Clin Pathol. 1993;100:686–689. doi: 10.1093/ajcp/100.6.686. [DOI] [PubMed] [Google Scholar]
- McKinzie PB, Parsons BL. Detection of rare K-ras codon 12 mutations using allele-specific competitive blocker PCR. Mutat Res. 2002;517:209–220. doi: 10.1016/s1383-5718(02)00077-3. [DOI] [PubMed] [Google Scholar]
- Syvänen AC. From gels to chips: "minisequencing" primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum Mutat. 1999;13:1–10. doi: 10.1002/(SICI)1098-1004(1999)13:1<1::AID-HUMU1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Kurg A, Metspalu A, Peltonen L, Syvänen AC. Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 1997;7:606–614. doi: 10.1101/gr.7.6.606. [DOI] [PubMed] [Google Scholar]
- Wang W, Kham SK, Yeo GH, Quah TC, Chong SS. Multiplex minisequencing screen for common Southeast Asian and Indian beta-thalassemia mutations. Clin Chem. 2003;49:209–218. doi: 10.1373/49.2.209. [DOI] [PubMed] [Google Scholar]
- Makridakis NM, Reichardt JK. Multiplex automated primer extension analysis: simultaneous genotyping of several polymorphisms. Biotechniques. 2001;31:1374–1380. doi: 10.2144/01316md05. [DOI] [PubMed] [Google Scholar]
- Sanchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D. A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis. 2006;27:1713–1324. doi: 10.1002/elps.200500671. [DOI] [PubMed] [Google Scholar]
- Meagher RJ, Coyne JA, Hestekin CN, Chiesl TN, Haynes RD, Won JI, Barron AE. Multiplexed p53 mutation detection by free-solution conjugate microchannel electrophoresis with polyamide drag-tags. Anal Chem. 2007;79:1848–1854. doi: 10.1021/ac061903z. [DOI] [PubMed] [Google Scholar]
- Greene FL, Page DL, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M, Eds. American Joint Committee on Cancer Staging Manual. 6. Philadelphia: Springer; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used to amplify exon-intron junctions and coding regions of exons 2, 3, and 4 of the K-RAS gene. This table includes the primers used in the multiplex PCR of K-RAS.
The sequences of primers used for detection of hotspot mutations of K-RAS. This table lists the primers used in the mutation analysis of K-RAS by the method of primer extension.