Abstract
Background
The Nijmegen Breakage Syndrome is a chromosomal instability disorder characterized by microcephaly, growth retardation, immunodeficiency, and increased frequency of cancers. Familial studies on relatives of these patients indicated that they also appear to be at increased risk of cancer.
Methods
In a candidate gene study aiming at identifying genetic determinants of breast cancer susceptibility, we undertook the full sequencing of the NBN gene in our cohort of 97 high-risk non-BRCA1 and -BRCA2 breast cancer families, along with 74 healthy unrelated controls, also from the French Canadian population. In silico programs (ESEfinder, NNSplice, Splice Site Finder and MatInspector) were used to assess the putative impact of the variants identified. The effect of the promoter variant was further studied by luciferase gene reporter assay in MCF-7, HEK293, HeLa and LNCaP cell lines.
Results
Twenty-four variants were identified in our case series and their frequency was further evaluated in healthy controls. The potentially deleterious p.Ile171Val variant was observed in one case only. The p.Arg215Trp variant, suggested to impair NBN binding to histone γ-H2AX, was observed in one breast cancer case and one healthy control. A promoter variant c.-242-110delAGTA displayed a significant variation in frequency between both sample sets. Luciferase reporter gene assay of the promoter construct bearing this variant did not suggest a variation of expression in the MCF-7 breast cancer cell line, but indicated a reduction of luciferase expression in both the HEK293 and LNCaP cell lines.
Conclusion
Our analysis of NBN sequence variations indicated that potential NBN alterations are present, albeit at a low frequency, in our cohort of high-risk breast cancer cases. Further analyses will be needed to fully ascertain the exact impact of those variants on breast cancer susceptibility, in particular for variants located in NBN promoter region.
Background
Pathogenic mutations in BRCA1, BRCA2, TP53, ATM, CHEK2, BRIP1 and PALB2 have been associated with an increased breast cancer risk and, together, are found in less than 25% of breast cancer families showing a clear pattern of inheritance (high-risk families) [1]. It is thus clear that other susceptibility alleles remain to be identified to explain the increased risk in the remnant high-risk families. As the number and characteristics of such alleles are undetermined, a focussed candidate gene approach based on genes closely interacting with the known susceptibility genes such as BRCA1 and BRCA2, the two major susceptibility genes identified yet, constitutes a study design of choice to identify rare-moderate-penetrance susceptibility alleles.
In the cell, nibrin, encoded by the NBN gene (also known as NBS1), participates in pathways of double strand breaks (DSB)-induced DNA repair and, together with its partners MRE11A and RAD50, is required for activation of these pathways in response to DNA damages [2]. In fact, nibrin is at the crossroad of several pathways implicating genes already associated with breast cancer susceptibility and/or chromosomal instability disorders [2,3]. Individuals homozygous for hypomorphic mutations in NBN suffer from the Nijmegen Breakage Syndrome (NBS), an autosomal recessive chromosomal instability disorder characterized by microcephaly, growth retardation, immunodeficiency and hyper-radiosensitivity [4]. Cancers, in particular haematological malignancies, are common adverse events in patients with NBS, as almost 40% of them develop a malignancy before the age of 21 years, and this correlates with a marked impairment in DSB repair observed in cells from these patients [5].
Some studies have associated an heterozygous NBN status with numerous types of cancers, including breast cancer [6-10], suggesting that being a carrier of a deleterious mutation in NBN may confer an increased risk of approximately 2 to 3-fold [6]. This was also supported by the observation that relatives of NBS patients display a higher than expected rate of cancers [4,11]. However, other studies failed to find an association with an increased risk of cancer [12,13].
In support of a role of NBN in tumor formation, evidence from mouse models demonstrated that Nbn heterozygosity predisposes cells to malignancies, as they display a wide variety of tumors: liver, mammary gland, prostate, lung as well as lymphomas [14]. Indeed, cells from these mice displayed an elevated frequency of chromosomal aberrations. These observations were correlated by studies of NBN heterozygous mutation carriers demonstrating that cell lines from these individuals showed spontaneous chromosomal instability (chromatid and chromosomes breaks, and chromosomes rearrangements) [15,16] as well as increased sensitivity to radiation-induced chromosomal aberrations [17]. Thus, it has been hypothesized that in cells of carriers of deleterious mutations in DNA repair genes such as NBN, a decrease in DNA repair capabilities resulting from a gene dosage effect (i.e. lower gene expression) may be sufficient to create a permissive environment for tumor development [18,19]. It has also been suggested that these DNA repair genes may show differences in tissue-specific protein-dosage thresholds, below which they may fail to operate normally [20].
Thus, based on the close relation of NBN and known breast cancer susceptibility genes in the cell DNA repair pathways, and the studies suggesting a possible involvement of NBN alterations in cancer susceptibility, we undertook the analysis of the entire coding sequence, intron/exon junctions, as well as the proximal promoter region of the NBN gene. We therefore performed a thorough re-sequencing of a series of 97 breast cancer cases selected from high-risk families from the French Canadian population, and 74 unrelated healthy controls from the same origin for sequence variations that could possibly modulate breast cancer risk.
Methods
Ascertainment of families
All 97 non-BRCA1/2 individuals from high-risk French Canadian breast and ovarian cancer families participating in this study were part of a larger interdisciplinary program termed INHERIT BRCAs [21]. All participants were at least 18 years of age and had to sign an informed consent form. Ethics committees reviewed the research project at the 7 participating institutions from which the patients were referred. The details regarding selection criteria of the breast cancer cases as well as the experimental and clinical procedures have been described previously [21,22].
PCR amplification and direct sequencing
PCR amplification of NBN (NM_002485.4) coding sequence, as well as flanking intronic regions, was performed on breast cancer cases and controls using primer pairs as described in Table 1. Sequencing reactions and sequence analysis were performed as described previously [22]. Alternative splice screening was also performed on cDNA on a subset of breast cancer cases using primers as described in Table 1.
Table 1.
Oligonucleotides used for NBN PCR amplification and sequencing.
| F/R1 | Oligonucleotide sequence (5'→3') | Annealing temp. (°C) | PCR lenght (bp) | MgCl2 final conc. (mM) | Comments | |
|---|---|---|---|---|---|---|
| Genomic sequence | ||||||
| Exon 1 | F | GACGTTAAGACAAGTTGATTTGAACTTAGA* | 60 | 965/ | 0.5 | 2% DMSO added to the PCR reaction |
| R | TCCGCCCATGCTAACTTCCT | 1113 | ||||
| F2 | TTTAGTAGTGCGCAGGATGTAGAC | |||||
| Exon 2 | F | CCTTTGATAGCCTTCAGTGAGGC | 64 | 743 | 1.0 | Sequencing primer: |
| R | AGCCAGAGTCATGAAGGTCTGTTC | CCACTGGTACCACTGCCACA | ||||
| Exon 3 | F | GTCAGGAGAATCCCCACTGACTT* | 60 | 548 | 1.5 | |
| R | GGCACAGAGTCCAATACTGTGCT | |||||
| Exon 4 | F | TGGGAAGTTACATTTCTTCGATTCC* | 58 | 728 | 1.5 | |
| R | GCACTGTCATAACCTTCTCGGTG | |||||
| Exon 5 | F | GCAGTGACCAAAGACCGACTTCTA* | 58 | 561 | 1.5 | |
| R | TGAGGTTACCTCAGTGCCATTTACT* | |||||
| Exon 6 | F | AAACGCGATTAGATGCTTTTTGTC* | 60 | 630 | 1.5 | |
| R | ACCCCACTTTGGTACACAGAAC | |||||
| Exon 7 | F | CCACAGAGAGTGTAACAGTTCCAGG* | 63 | 1100 | 1.0 | 2% DMSO added to the PCR reaction |
| R | TTAATTCTTGTATCGGCCGGG | |||||
| Exon 8 | F | TAACAGTGCCCCAGCGAGTAAG* | 58 | 785 | 1.5 | |
| R | TCCTCTTACACTGTCGACCCTTAGA | |||||
| Exon 9 | F | TTAGATAAGCCCGTCATAGATGCC* | 58 | 516 | 1.5 | |
| R | TAACTACTCGCCGCTCCTTTACA | |||||
| Exon 10 | F | GTTTGTCAGTCGTCTATAGTGGAGCA* | 58 | 763 | 1.5 | |
| R | AATTGCGGCAAGTAAAATAACACG | |||||
| Exon 11 | F | CCCTGCCCACAACCTTACTACG* | 60 | 1500 | 1.5 | |
| R | GACCACAGCCATGAATGAGTGG * | |||||
| Exon 12 | F | TCCTACCATCTACAGACAACCATGG | 58 | 619 | 1.5 | |
| R | CCATGATCAATCCATTTCAAGGC * | |||||
| Exon 13 | F | GCAAACAGTGCTGAGATTTTGTGTC | 58 | 850 | 1.5 | |
| R | CCTGAGCTAAAGAACCTCCTCAAGTAG * | |||||
| Exon 14 | F | CAACATCTCCTGCTTGGACTCTG | 60 | 638 | 1.0 | Sequencing primer: |
| R | GAAGAATTTGCTTGAAGGCCACC | GATGGGTTTAGAACAGAGTTACTGCT | ||||
| Exon 15 | F | GAACTCAGATGTGGTGACCTCCAG | 58 | 441 | 1.5 | |
| R | CAATTTCACACAATTCGGGAACC * | |||||
| Exon 16 | F | GAGGAATGGGGATCTTTGAAGC | 58 | 457 | 1.5 | Sequencing primer: |
| R | GTAACTTAAATCGCTTCTATACAC | AAGCAACATCAAAGGGATACATGA | ||||
| cDNA | ||||||
| PCR 1 | F | GTTACGCGGTTGCACGTCG | 64 | 998 | 1.5 | Exons 1 to 8 |
| R | GTCATGAAAATCACCGCCAATC | |||||
| PCR 2 | F | TCTTTTTGGCTCCGGGAACG | 62 | 567 | 1.5 | Exons 7 to 10 |
| R | GCTGCTGCTGAGAAGCCCTATC | Overlap of 169 pb with PCR1 | ||||
| PCR 3 | F | CCCACTGTAAAGGAGTCCTGCA | 62 | 634 | 1.5 | Exons 10 to 12 |
| R | TACTTTCTGGTACTGCTTCATCACT | Overlap of 151 pb with PCR2 | ||||
| PCR 4 | F | CCATAGAAGATGAAGTATTGGA | 62 | 700 | 1.5 | Exons 11 to 16 |
| R | GTAACTTAAATCGCTTCTATACAC | Overlap of 165 pb with PCR3 | ||||
| Promoter cloning | ||||||
| F | GACGACGCTAGCGACGTTAAGACAAGTTGATTTGAACTTAGA | 62 | 515 | 1.0 | 2% DMSO added to the PCR reaction Added restriction sites are underlined |
|
| R | GACGACAAGCTTATCGGTCCGGCTCCTCAGGGCTG | |||||
1 Primer direction: forward (F) or reverse (R) 2 Alternative primer Oligonucleotides used as sequencing primers are marked with an (*)
Variants characterization and haplotype estimations
Deviation from Hardy-Weinberg equilibrium (HWE) and allelic difference between both series was evaluated using a two-sided Chi Square test with 1 degree of freedom. The possible effect of a given variant on exonic splicing enhancers was assessed using the ESEfinder 3.0 program [23] and on splice consensus sites using NNSPLICE [24] and Splice Site Finder [25] web based programs. The potential impact of the promoter variant and pairwise linkage disequilibrium were evaluated as described elsewhere [26]. Haplotype analysis was performed using the WHAP program [27] implementing a regression-based association test allowing for evaluation of haplotype-specific association. Haplotype analyses were performed on all variants identified as well as on variants showing a minor allele frequency (MAF) greater than 5%.
Luciferase promoter assays
A portion of 401 bp of the NBN promoter region, and the entire 5'UTR sequence (110 bp), was amplified by PCR from a breast cancer individual carrier of the c.-242-110delAGTA deletion using primers introducing a NheI or a HindIII restriction site (Table 1). PCR products were then digested and introduced in the pGL3 basic vector (Promega Corporation, Madison, WI, USA). Direct sequencing of clones was performed to confirm the presence of the reference sequence allele as well as the four nucleotides deletion. Transient transfections in MCF-7, LNCaP, HeLa and HEK293 cells and dual luciferase reporter assays were performed as described previously [26], with cells harvested 24–48 h after transfection. ATBF1 expression levels were measured in these four cell lines by quantitative real-time PCR (QRT-PCR) as described previously [28], using RNA extracted by the TriReagent method (Molecular Research Center Inc, Cincinnati, OH, USA) and specific primers (sens: 5'-TGCAACTAAACCGCCCACATATA-3'; antisens 5'-CCCCAAGTGAGATAAAGCTAAACAAA-3'). Levels of expression were normalized using the housekeeping gene HPRT1, and are indicated relative to the MCF-7 cell line (sens primer: 5'-AGTTCTGTGGCCATCTGCTTAGTAG-3'; antisens primer: 5'-AAACAACAATCCGCCCAAAGG-3').
Results
Sequence variations in the NBN gene
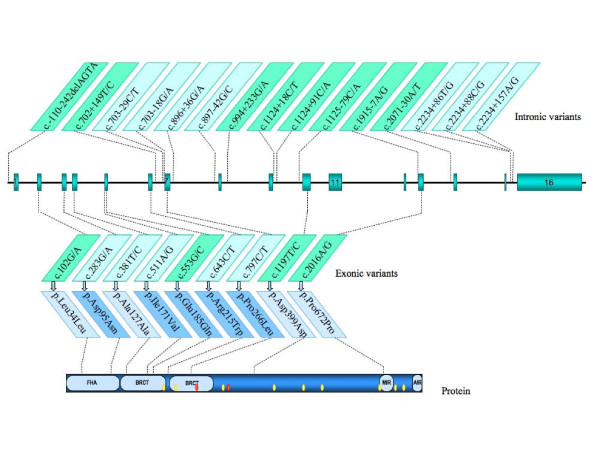
Direct sequencing of NBN entire coding region, adjacent intronic sequences as well as the proximal promoter region was performed on 97 affected individuals from French Canadian breast and ovarian cancer families (one individual per family). Twenty-four variants were identified (Tables 2, 3 and Figure 1), including a new rare synonymous change at codon 127 (c.381T/C) and a new variant located in intron 15 (c.2234+86T/G), both not reported in the literature and databases, and found exclusively among breast cancer cases. Variants identified in the case dataset were also genotyped in 74 healthy individuals from the same population. All SNPs genotyped were in HWE. Nine out of the 24 variations were located in the coding region (Table 2), while the remaining 15 were located in untranslated regions (Table 3). Among all variants identified, half (12) were considered as common (MAF ≥ 5%) while the remaining 12 were rare variations. Two rare non-synonymous exonic variants (c.511A/G and c.797C/T) were observed only once in the case series, of which p.Ile171Val is located in the first BRCA1 C-terminal (BRCT) domain (Figure 1). Of these two variants, only c.511A/G could be genotyped in an additional family member, i.e. an unaffected male cousin, who was found non-carrier (data not shown). Two other rare variants, c.283G/A and c.643C/T, were both observed in one breast cancer case and one control. For the c.643C/T variant, a DNA sample from another available family member affected with ovarian cancer was analyzed and turned out to be wild type (data not shown). Among the variants located in untranslated regions (Table 3), the c.2234+86T/G variant was present exclusively in two breast cancer cases. When genotype frequencies of all variants were compared between both series, only the c.-242-110delAGTA variant in the promoter showed a statistically significant association with breast cancer (OR 3.4, 95% CI: 1.1–10.5; p = 0.029).
Table 2.
Exonic sequence variations of the NBN gene in French Canadian breast cancer cases and healthy controls.
| SNP | SNP ID1 (dbSNP ID) | Protein change | Location | Series (No.) | MAF2 | Common homozygote No. (expected)3 | Heterozygote No. (expected)3 | Rare homozygote No. (expected)3 | HWE p-value | OR (95% CI)4 | Reported MAF in Caucasian |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.102G/A | p.Leu34Leu | Exon 2 | Cases (97) | 0.284 | 52 (49.80) | 35 (39.41) | 10 (7.80) | 0.27 | 0.7 | 0.283–0.4505 |
| (rs1063045) | Controls (71) | 0.310 | 33 (33.82) | 32 (30.37) | 6 (6.82) | 0.65 | (0.4–1.3) | ||||
| 2 | c.283G/A | p.Asp95Asn | Exon 3 | Cases (97) | 0.005 | 96 (96.00) | 1 (0.99) | 0 (0) | 0.96 | 0.7 | |
| (NA)6 | Controls (71) | 0.007 | 70 (70.00) | 1 (0.99) | 0 (0) | 0.95 | (0.0–11.9) | ||||
| 3 | c.381T/C | p.Ala127Ala | Exon 4 | Cases (97) | 0.005 | 96 (96.00) | 1 (0.99) | 0 (0) | 0.96 | 2.3 | |
| (NA) | Controls (72) | 0.000 | 72 (72.00) | 0 (0) | 0 (0) | 1.00 | (0.1–56.1) | ||||
| 4 | c.511A/G | p.Ile171Val | Exon 5 | Cases (97) | 0.005 | 96 (96.00) | 1 (0.99) | 0 (0) | 0.96 | 2.3 | |
| (NA)6 | Controls (73) | 0.000 | 73 (73.00) | 0 (0) | 0 (0) | 1.00 | (0.1–57.0) | ||||
| 5 | c.553G/C | p.Glu185Gln | Exon 5 | Cases (97) | 0.268 | 54 (51.97) | 34 (38.06) | 9 (6.97) | 0.29 | 0.6 | |
| (rs1805794) | Controls (73) | 0.308 | 34 (34.93) | 33 (31.13) | 6 (6.93) | 0.61 | (0.3–1.2) | ||||
| 6 | c.643C/T | p.Arg215Trp | Exon 6 | Cases (97) | 0.005 | 96 (96.00) | 1 (0.99) | 0 (0) | 0.96 | 0.8 | |
| (rs34767364) | Controls (73) | 0.007 | 72 (72.00) | 1 (0.99) | 0 (0) | 0.95 | (0.0–12.2) | ||||
| 7 | c.797C/T | p.Pro266Leu | Exon 7 | Cases (97) | 0.005 | 96 (96.00) | 1 (0.99) | 0 (0) | 0.96 | 2.3 | 0.0005 |
| (rs769420) | Controls (73) | 0.000 | 73 (73.00) | 0 (0) | 0 (0) | 1.00 | (0.1–57.0) | ||||
| 8 | c.1197T/C | p.Asp399Asp | Exon 10 | Cases (97) | 0.320 | 47 (44.91) | 38 (42.19) | 12 (9.91) | 0.33 | 0.9 | 0.28–0.455 |
| (rs709816) | Controls (72) | 0.313 | 34 (34.03) | 31 (30.94) | 7 (7.03) | 0.99 | (0.5–1.7) | ||||
| 9 | c.2016A/G | p.Pro672Pro | Exon 13 | Cases (97) | 0.284 | 52 (49.80) | 35 (39.41) | 10 (7.80) | 0.27 | 0.8 | 0.2835 |
| (rs1061302) | Controls (70) | 0.300 | 34 (34.30) | 30 (29.40) | 6 (6.30) | 0.86 | (0.4–2.2) | ||||
1 According to the nomenclature guidelines of the Human Genome Variation Society (reference sequence NM_002485.4), nucleotide number one being the A from the ATG codon.
2 Minor allele frequency
3 As expected under Hardy-Weinberg equilibrium
4 Odds ratios for comparison of heterozygotes versus common homozygotes
5 From NCBI dbSNP data.
6 No entry in NCBI dbSNP database, although reported in the literature.
Table 3.
Promoter and intronic sequence variations of the NBN gene in French Canadian breast cancer cases and healthy controls.
| SNP | SNP ID1 (dbSNP ID) |
Location | Series (No.) | MAF2 | Common homozygote No. (expected)3 | Heterozygote No. (expected)3 | Rare homozygote No. (expected)3 | HWE p-value | OR (95% CI)4 | Reported MAF in Caucasian |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | c.-110-242delAGTA | Promoter | Cases (97) | 0.082 | 81 (81.66) | 16 (14.68) | 0 (0.66) | 0.38 | 3.4 | |
| (rs36226237) | Controls (72) | 0.028 | 68 (68.06) | 4 (3.89) | 0 (0.06) | 0.81 | (1.1–10.5) | |||
| 11 | c.702+149T/C | Intron 6 | Cases (97) | 0.082 | 83 (81.66) | 12 (14.68) | 2 (0.66) | 0.07 | 0.7 | 0.108–0.1175 |
| (rs3026271) | Controls (73) | 0.089 | 60 (60.58) | 13 (11.84) | 0 (0.58) | 0.40 | (0.3–1.6) | |||
| 12 | c.703-29C/T | Intron 6 | Cases (97) | 0.015 | 94 (94.02) | 3 (2.95) | 0 (0.02) | 0.88 | 1.1 | 0.0215 |
| (NA)6 | Controls (73) | 0.014 | 71 (71.01) | 2 (1.97) | 0 (0.01) | 0.91 | (0.2–7.0) | |||
| 13 | c.703-18G/A | Intron 6 | Cases (97) | 0.026 | 92 (92.06) | 5 (4.87) | 0 (0.06) | 0.79 | 3.9 | |
| (rs769418) | Controls (73) | 0.007 | 72 (72.00) | 1 (0.99) | 0 (0) | 0.95 | (0.4–34.2) | |||
| 14 | c.896+36G/A | Intron 7 | Cases (97) | 0.026 | 92 (92.06) | 5 (4.87) | 0 (0.06) | 0.79 | 3.9 | 0.021–0.055 |
| (rs1805826) | Controls (73) | 0.007 | 72 (72.00) | 1 (0.99) | 0 (0) | 0.95 | (0.4–34.2) | |||
| 15 | c.897-42G/C | Intron 7 | Cases (97) | 0.015 | 94 (94.02) | 3 (2.95) | 0 (0.02) | 0.76 | 0.4 | |
| (NA)6 | Controls (73) | 0.034 | 68 (68.09) | 5 (4.83) | 0 (0.09) | 0.76 | (0.1–1.9) | |||
| 16 | c.994+233G/A | Intron 8 | Cases (97) | 0.294 | 50 (48.37) | 37 (40.25) | 10 (8.37) | 0.43 | 0.7 | 0.285 |
| (rs6990969) | Controls (74) | 0.311 | 34 (35.15) | 34 (31.70) | 6 (7.15) | 0.53 | (0.4–1.4) | |||
| 17 | c.1124+18C/T | Intron 9 | Cases (97) | 0.278 | 52 (50.52) | 36 (38.97) | 9 (7.52) | 0.45 | 0.7 | 0.09–0.3065 |
| (rs2234744) | Controls (74) | 0.304 | 35 (35.84) | 33 (31.32) | 6 (6.84) | 0.64 | (0.4–1.4) | |||
| 18 | c.1124+91C/A | Intron 9 | Cases (97) | 0.273 | 53 (51.24) | 35 (38.52) | 9 (7.24) | 0.37 | 0.7 | 0.283–0.3065 |
| (rs1805818) | Controls (74) | 0.304 | 35 (35.84) | 33 (31.32) | 6 (6.84) | 0.64 | (0.4–1.3) | |||
| 19 | c.1125-79C/A | Intron 9 | Cases (97) | 0.284 | 52 (49.80) | 35 (39.41) | 10 (7.80) | 0.27 | 0.8 | 0.3235 |
| (rs1805786) | Controls (72) | 0.292 | 36 (36.13) | 30 (29.75) | 6 (6.13) | 0.94 | (0.4–1.5) | |||
| 20 | c.1915-7A/G | Intron 12 | Cases (97) | 0.284 | 52 (49.80) | 35 (39.41) | 10 (7.80) | 0.27 | 0.8 | 0.20–0.3125 |
| (rs2308962) | Controls (70) | 0.300 | 34 (34.30) | 30 (29.40) | 6 (6.30) | 0.86 | (0.4–1.5) | |||
| 21 | c.2071-30A/T | Intron 13 | Cases (97) | 0.284 | 52 (49.80) | 35 (39.41) | 10 (7.80) | 0.27 | 0.7 | |
| (rs3736639) | Controls (71) | 0.310 | 33 (33.82) | 32 (30.37) | 6 (6.82) | 0.65 | (0.4–1.3) | |||
| 22 | c.2234+86T/G | Intron 15 | Cases (97) | 0.010 | 95 (95.01) | 2 (1.98) | 0 (0.01) | 0.92 | 3.8 | |
| (NA) | Controls (72) | 0.000 | 72 (72.00) | 0 (0) | 0 (0) | 1.00 | (0.2–80.3) | |||
| 23 | c.2234+88C/G | Intron 15 | Cases (97) | 0.015 | 94 (94.02) | 3 (2.95) | 0 (0.02) | 0.88 | 0.4 | 0.017–0.0425 |
| (rs13312970) | Controls (71) | 0.035 | 66 (66.09) | 5 (4.82) | 0 (0.09) | 0.76 | (0.1–1.8) | |||
| 24 | c.2234+157A/G | Intron 15 | Cases (97) | 0.046 | 88 (88.21) | 9 (8.58) | 0 (0.21) | 0.63 | 2.3 | 0.000–0.0255 |
| (rs13312971) | Controls (71) | 0.021 | 68 (68.03) | 3 (2.94) | 0 (0.03) | 0.86 | (0.6–8.9) | |||
1 According to the nomenclature guidelines of the Human Genome Variation Society (reference sequence NM_002485.4), nucleotide number one being the A from the ATG codon. 2 Minor allele frequency 3 As expected under Hardy-Weinberg equilibrium 4 Odds ratios for comparison of heterozygotes versus common homozygotes 5 From NCBI dbSNP data. 6 No entry in NCBI dbSNP database, although reported in the literature.
Figure 1.
Schematic representation of the NBN gene on chromosome 8q21. The upper part shows variants identified in the NBN gene. Light green boxes refer to SNPs with MAF ≥ 5% in the cases set, and variants name are indicated according to the reference sequence NM_002485.4. Corresponding coding variations are indicated relative to the protein structure in the bottom part of the figure. Light blue boxes refer to synonymous variations. FHA: Forkhead-associated domain. BRCT: BRCA1 C-terminal domain. MIR: MRE11 interacting region. AIR: ATM interacting region. Yellow dots: acetylated lysine residues (K208, K233, K334, K441, K504, K544, K665, K690, K698 and K715). Red dots: phosphorylated serine residues (S278 and S343).
Identification of NBN alternative splice forms
Analysis of NBN cDNA was also performed on a subset of these breast cancer cases and highlighted the presence of two distinct alternative splice events. The first splice variant involved the insertion of 50 bp of the intron between exons 2 and 3 and is expected to result in a premature stop codon [29]. The other alternative splice form identified involves the skipping of exons 12 to 14 which is expected to produce an in-frame deletion of 113 amino acids in the region of the NBN protein involved in the interaction with its partner MRE11A (Figure 1). However, QRT-PCR of this specific form was performed on cDNA samples from a subset of 10 breast cancer cases from our cohort and showed a very low expression relative to the main isoform (data not shown), which is consistent with previous work [30].
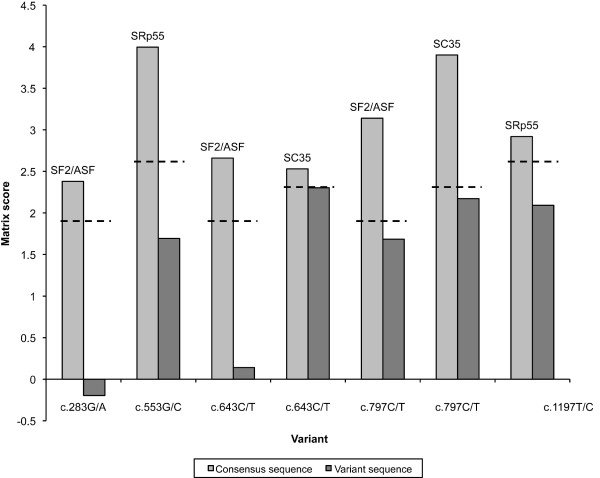
In silico analysis of the putative impact of NBN exonic and intronic sequence variants on these splice events was also performed. Analysis of the nine exonic sequence variants using ESEfinder indicated that, while five variants (c.283G/A, c.553G/C, c.643C/T, c.797C/T, and c.1197T>C) might have an impact on putative score motifs of four SR proteins, a closer examination of these results shows that these scores, while below the program thresholds, remain relatively high and thus are unlikely to affect NBN constitutive splicing (Figure 2). According to the Splice Site prediction program, the intronic variant c.896+36G/A might slightly increase a putative acceptor site (score from 0.66 to 0.75). However, this was not confirmed by the Splice Site Finder program, which predicted the abolition of a putative donor site with a score of 71.2. As for the c.1124+91C/A intronic variant, only the Splice Site Prediction program predicted the abolition of a weak acceptor site (score of 0.45) while for the c.2234+157A/G intronic variant, the Splice Site Finder program estimated the creation of a putative acceptor site with a score of 77.2. Hence, none of the variants identified are expected to be responsible for the alternative splice events observed.
Figure 2.
In silico analysis of the effect of coding variants on putative exonic splicing enhancer (ESE) motifs by the ESEfinder program. Only scores for predicted motifs potentially abolished by exonic variants are shown. Light gray: motif matrix scores for consensus sequences. Dark gray: motif matrix scores for variant sequences. The name of the SR protein potentially involved is indicated for each score pair. Doted line indicates the default threshold used by ESEfinder for the corresponding motif.
Effect of the c.-242-110delAGTA variant on luciferase reporter gene expression
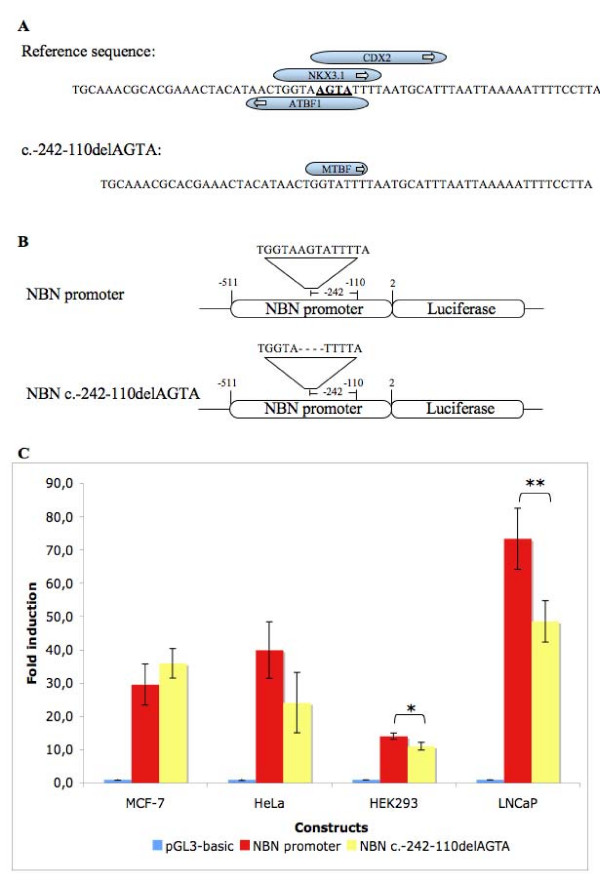
The putative effect of the c.-242-110delAGTA variant located in the promoter region was also assessed by the MatInspector program, which predicts that this variant may abolish recognition motifs for the ZFHX3/ATBF1 (Zinc finger homeobox 3, or AT motif-binding factor 1), NKX3-1 (NK3 homeobox 1) and CDX2 (Caudal-type homeobox 2) transcription factors, and create a potential binding site for the MTBF (Muscle-specific MT binding factor) transcription factor (Figure 3A). The effect of the c.-242-110delAGTA variant on NBN expression was therefore further assessed using a dual reporter gene system. MCF-7, LNCaP, HeLa and HEK293 cells were then transiently transfected with a construct containing the consensus NBN sequence or with the variant sequence, together with the Renilla reporter plasmid as an internal transfection control (Figure 3B). While no significant difference in expression was observed between both constructs in MCF-7 and HeLa cells, a slight diminution of luciferase expression was observed for the variant construct relative to the reference sequence construct in both the HEK293 and LNCaP cell lines (Figure 3C). Interestingly, QRT-PCR measures indicated that ATBF1 mRNA is 2.4 times more expressed in HeLa cells, and more than 4.3 times more expressed in LNCaP cells, than in MCF-7 cells (data not shown).
Figure 3.
Effect of the c.-242-110delAGTA deletion on NBNpromoter activity. Panel A shows the results of the in silico predictions of transcription factors binding sites affected by the c.-242-110delAGTA variant obtained from the MatInspector program. Panel B shows the schematic representation of the two reporter gene constructs containing the NBN promoter used in this study. The positions relative to the nucleotide A of the NBN reference sequence ATG codon are indicated. Panel C shows the results of the luciferase reporter assay. MCF-7, LNCaP, HeLa and HEK293 cells were transiently co-transfected with the Renilla reporter plasmid (pRL) as a transfection control. Each data represents mean ± standard deviation of triplicates. Data are shown as fold of induction compared to the activity of cells transfected with the empty pGL3-basic luciferase reporter vector. *p = 0.0245 **p = 0.0181
LD analysis across NBN genomic sequence and haplotypes determination
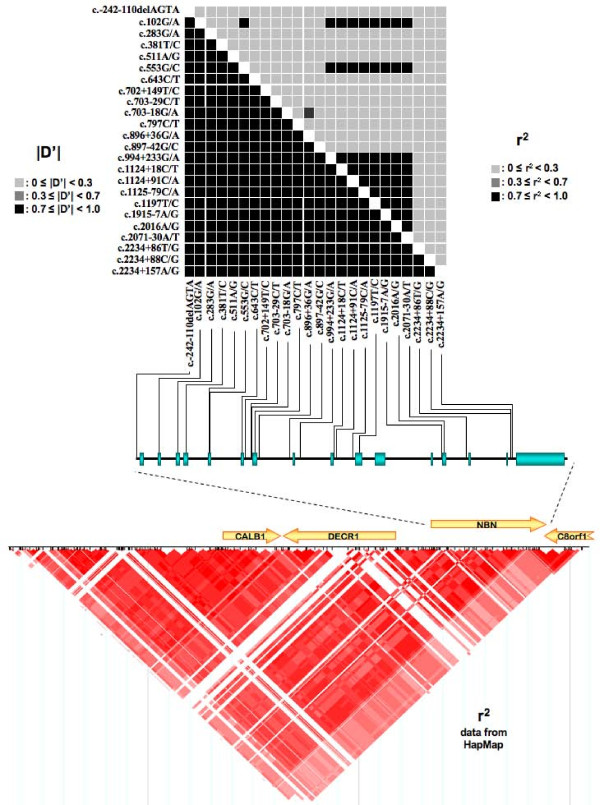
Pairwise LD measures of all variants using the control dataset are presented in Figure 4. |D'| values show a high degree of LD for all SNP pairs while the more stringent r2 measure, dependent upon allele frequency, shows a limited block of LD involving mainly SNPs located in the second half of the NBN gene. The bottom part of Figure 4 shows the r2 HapMap CEU data in the vicinity of NBN on chromosome 8, and suggests that NBN may overlap two blocks of LD. The major part of NBN seems to be in a strong block of LD extending 5' of the gene, while its 3' extremity spans over another smaller block.
Figure 4.
Linkage disequilibrium across the NBN gene and neighbouring chromosomal region. Upper part shows linkage disequilibrium (LD) results (r2 and |D'|) obtained from the control genotypes data used in our study and their relative position along NBN sequence. In the bottom part of the figure are indicated r2 data from the International HapMap project (release 21a) for the genomic region encompassing NBN. The intensity of the box color is proportional to the strength of LD. r2 = 0 in white; 0 < r2 < 1 in shades of pink/red; r2 = 1 in bright red.
Haplotype phasing of NBN using the WHAP program with all 24 variants identified in our datasets indicated that 14 haplotypes are present with an estimated frequency greater than 0.5%. Although WHAP did not estimate a significant difference between both groups, the p-value observed was borderline significant (p = 0.0588). The majority of these estimated haplotypes (84.5%) have a frequency greater than 2%. Two haplotypes (#4 and #7 in Table 4) showed a weak significant association with breast cancer, being more frequent among cases (p = 0.0205 and p = 0.0403, respectively). The same analysis, using only common variations (MAF >5%), suggested that one haplotype is more frequent among cases than controls (WH4, p = 0.0227), with another haplotype showing a non-significant trend of over-representation in cases (WH5, p = 0.0883) (Table 5). Global estimation of haplotypes was further confirmed using the PHASE 2.1.1 program, with concordant results (data not shown).
Table 4.
Estimated haplotypes using the WHAP program and all variants identified, for cases and controls combined.
| Estimated Haplotype Frequencies | |||||
|---|---|---|---|---|---|
| Haplotype | SNPs1 10-1-2-3-4-5-6-11-12-13-7-14-15-16-17-18-19-8-20-9-21-22-23-24 |
Cases | Controls | Combined | p-value2 |
| 1 | TGGTAGCTCGCGGGCCCTAAATCA | 0.481 | 0.486 | 0.483 | 0.92 |
| 2 | TAGTACCTCGCGGATAACGGTTCA | 0.232 | 0.255 | 0.244 | 0.636 |
| 3 | TGGTAGCCCGCGGGCCCTAAATCA | 0.065 | 0.097 | 0.078 | 0.29 |
| 4 | AGGTAGCTCGCGGGCCCTAAATCA | 0.089 | 0.030 | 0.052 | 0.0205 |
| 5 | TAGTAGCTCGCGGATAACGGTTCA | 0.036 | 0.007 | 0.022 | 0.147 |
| 6 | TGGTAGCTCGCGCGCCCTAAATCA | 0.012 | 0.036 | 0.021 | 0.146 |
| 7 | TAGTACCTCGCGGATAACGGTTGA | 0.006 | 0.037 | 0.019 | 0.0403 |
| 8 | TGGTAGCTCGCGGGCCCTAAATCG | 0.019 | 0.022 | 0.015 | 0.846 |
| 9 | TGGTAGCTCACAGGCCCCAAATCA | 0.013 | 0.000 | 0.011 | 0.11 |
| 10 | TGGTACCTCGCGGGCCCTAAATCA | 0.012 | 0.008 | 0.010 | 0.749 |
| 11 | TGGTAGCTTGCGGGCCCTAAATCA | 0.006 | 0.014 | 0.008 | 0.47 |
| 12 | TGGTAGCTCGCGGACCCTAAATCA | 0.006 | 0.007 | 0.006 | 0.897 |
| 13 | TGGTAGCTTGCGGGCCCTAAATCG | 0.011 | 0.000 | 0.006 | 0.126 |
| 14 | TGGTAGCTCACAGGCCCCAAATCG | 0.011 | 0.008 | 0.006 | 0.128 |
1SNPs identification as in Table 2 and 3. 2 p-value for haplotype-specific test, each haplotype versus all other haplotypes.
Table 5.
Estimated haplotypes using the WHAP program and SNPs with a frequency greater than 5% for cases and controls combined.
| Estimated Haplotype Frequencies | ||||||
|---|---|---|---|---|---|---|
| Haplotype | SNPs1 10-1-5-11-16-17-18-19-8-20-9-21 |
Cases | Controls | Combined | p-value2 | OR (95%CI) |
| WH1 | TGGTGCCCTAAA | 0.511 | 0.558 | 0.538 | 0.361 | REF |
| WH2 | TACTATAACGGT | 0.250 | 0.293 | 0.264 | 0.399 | 0.9 (0.6–1.6) |
| WH3 | TGGCGCCCTAAA | 0.065 | 0.096 | 0.080 | 0.277 | 0.7 (0.3–1.7) |
| WH4 | AGGTGCCCTAAA | 0.087 | 0.030 | 0.059 | 0.0227 | 3.2 (1.1–9.2) |
| WH5 | TGGTGCCCCAAA | 0.038 | 0.008 | 0.027 | 0.0883 | 5.1 (0.9–30.1) |
| WH6 | TAGTATAACGGT | 0.033 | 0.007 | 0.021 | 0.176 | 7.8 (0.8–30.5) |
| WH7 | TGCTGCCCTAAA | 0.017 | 0.008 | 0.011 | 0.542 | 2.4 (0.3–16.1) |
1SNPs identification as in Table 2 and 3.2p-value for haplotype-specific test, each haplotype versus all other haplotypes.
A closer examination of these two haplotypes (WH4 and WH5) showed that the only variation present on WH4 is the c.-242-110delAGTA variant, which on its own shows a significant difference in frequency between both groups. Thus, this variant is likely to be responsible of the association observed as any further breakdown of this haplotype using a sliding window analysis shows that all combinations involving this variant display a significant association (data not shown). As for the WH5 haplotype carrying the c.1197T/C variant in exon 10, it does not show any significant difference in frequency between the both groups and, despite its strong linkage disequilibrium with the adjacent variants, it shows only a tendency towards significativity when present in combination with the major alleles at all other positions (Table 5).
Discussion
Although the relevance of mutations in several DNA repair genes (BRCA1, BRCA2, TP53, PTEN, PALB2, BRIP1) in breast cancer susceptibility have been well established, the association of variants in other genes, potentially accounting for the remaining familial clustering, is not well defined. For example, heterozygous carriers of deleterious mutations in the NBN gene, in particular the Slavic founder mutation 657del5, has been associated with a 2- to 3-fold increased risk of cancer [6]. However the impact of other NBN variants on cancer risk is unclear. Nonetheless, studies have demonstrated an increased spontaneous chromosomal instability in cells from heterozygous carriers of NBN mutations [7,16]. In addition, the presence of a specific pattern of gene expression involving pathways of DNA repair and damage bypass, mitotic checkpoint and apoptosis was demonstrated, suggesting that cells from these carriers may display a much less efficient DNA repair system [31]. In this regard, a recent study by Someya et al. [32] showed a correlation between persistent radiation-induced NBN foci and both chromosomal instability and sporadic breast cancer risk. To address the possible implication of NBN sequence variants on breast cancer risk, we took advantage of our resource of high-risk breast cancer families drawn from the French Canadian population. In order to increase the statistical power of our investigation, one affected individual from each family was selected for analysis [33], each thoroughly screened for BRCA1 and BRCA2 mutations or large genomic rearrangements in these genes [21].
The majority of the coding variants identified in our cohort of breast cancer cases are situated in the forkhead-associated (FHA) domain and the two BRCT domains, and these domains have been demonstrated to be essential to NBN binding to the histone γ-H2AX [34,35]. One of the rare variants identified in this study, p.Ile171Val, was first described in acute lymphoblastic leukemia (ALL) patients [36]. This variant has been previously reported to be over-represented in some cancer cohorts [9,10,37,38], and has also been previously described at the homozygous state in a Japanese NBS patient affected with aplastic anemia and genomic instability [39]. Interestingly, analysis of the patient's and her father's lymphoblastoid cell lines demonstrated a higher frequency of spontaneous chromosomal aberrations compared with healthy controls (6- and 4-fold increase, respectively). These results suggest that the p.Ile171Val variation may be deleterious, also supported by the fact that this variant alters an amino acid which is conserved in species such as the chicken and the fruitfly [39]. However, the impact of the p.Ile171Val substitution remains to be clarified as a large study, including cases from Germany and the Republic of Belarus, did not find any association with breast cancer in those populations [40].
Another rare variant, p.Arg215Trp, has been considered pathogenic in previous studies based on its highly conserved position and the change introduced. Moreover, this variant has been identified in monozygotic twins compound heterozygotes for 657del5/p.Arg215Trp and affected with a severe form of NBS [41]. NibrinTrp215 appears to affect the correct nibrin function, and cells carrying this variant shows delayed DNA DSB rejoining [41]. Modelisation of the tandem BRCT domains suggests that the Arg215 residue is required for correct orientation of the BRCT domains and recognition of γ-H2AX [34]. In their experiment, nibrinTrp215 seems to partially interfere with nibrinArg215 activity and may act in a co-dominant fashion. It is also interesting to note that although this variant is relatively frequent in several studies, to our knowledge no NBS cases homozygous for this variant have been reported.
Of the other rare variants present in our cohort of breast cancer cases, the p.Pro266Leu variant was observed in one breast cancer case, and the p.Asp95Asn variant was found in one breast cancer case and one control. Although these variants involve conserved residues, their putative effect on protein function remains unclear. In a previous study on non-Hodgkin lymphoma, both variants were detected only in healthy controls [42]. Regarding the p.Asp95Asn variant first identified in an ALL patient [36], no association was found in a study of larynx cancer [38] or ALL [37]. This variant was found in one out of 613 and 121 unselected and familial prostate cancer cases, respectively, but not in controls [43]. However, p.Asp95Asn is not predicted to be highly damaging as its expression in NBS cells seems to have a similar activity to the wild type protein [42]. The only other non-synonymous change identified in our cohort is the p.Glu185Gln common variant (MAF >25%), which has been genotyped in several association studies. Although the majority of the studies analyzing this variant found no association with cancer [12,38,42-44], some studies reported an association with breast cancer [8], basal cell carcinoma in men [45] and a recent meta-analysis of this variant in bladder cancer revealed a significant association after controlling for potential bias [46]. In addition, Musak et al. [47] reported that this variant may modulate the frequency of chromatid-type aberrations among tire plant workers. However, our data do not allow us to confirm any positive association.
Among the non-coding variants, a possible association of the c.-242-110delAGTA variant with an increased risk of breast cancer could be observed (OR 3.4 95% CI: 1.1–10.5). This association was further confirmed by the estimation of the haplotype diversity in our dataset, which highlighted the presence of the haplotype bearing the c.-242-110delAGTA variant at a higher frequency among the breast cancer case subset. In silico prediction of putative transcription factor binding sites indicated that of the four transcription factor binding sites potentially affected by the deletion, some are predicted to involve proteins not expressed in the breast. MTBF acts in the muscle regulation of the myostatin gene [48], while CDX2 is mainly expressed in the gut although its ectopic expression was also reported in cases of non-gastrointestinal carcinomas [49]. As for the transcription factor NKX3-1, it is mainly expressed in the prostate although it is also found at low levels in the mammary gland and breast tumors, and has been suggested to act as a haplo-insufficient tumor suppressor in prostate cancer [50]. The fourth transcription factor potentially affected is ATBF1 (also known as ZFHX3). ATBF1 was shown to suppress the expression of the alpha-fetoprotein [51] and the MYB oncogene [52]. ATBF1 also cooperates with p53 to activate the CDKN1A promoter and trigger cell cycle arrest [53]. ATBF1 is expressed in the mammary gland and breast tumors (NCBI's UNIGENE data), and the expression of ATBF1 mRNA was correlated with a better prognosis in 153 patients with invasive carcinomas of the breast [54]. This might suggest that deregulation of genes controlled through ATBF1 could have an important impact on breast cancer formation and progression.
Luciferase reporter gene assay using the promoter sequence proximal to the ATG codon indicated a similar level of expression of the c.-242-110delAGTA variant allele as compared to the reference sequence construct in the breast cancer cell line MCF-7 and in HeLa cells. However, a slight decrease in luciferase expression was observed in both HEK293 and LNCaP cells, which might indicate that this variant could have an impact on NBN expression in a cell type manner. The analysis of ATBF1 gene expression indicated that this transcription factor is weakly expressed, in particular in the MCF-7 cells. This is comparable to a recent study performed on a panel of 32 breast cancer cell lines, which indicated that in 75% of these cell lines, ATBF1 is expressed at 50% or less relative to the normal breast [55]. This could in turn explain the lack of effect observed for the luciferase assay in the MCF-7 cell line. On the other hand, one could hypothesize that NBN (or transcription factors affecting its expression) may be regulated following irradiation or other genotoxic stress. Additional research will however be needed to address this possibility, as well as binding experiments to confirm ATFB1 binding to this region of the promoter. However, it is also important to keep in mind that the promoter sequence cloned here, although supporting NBN expression, is unlikely to constitute the entire NBN promoter. It is indeed plausible that transcription factors acting on the proximal sequence might be influenced by other transcription factors acting further upstream. As such, despite the lack of variation in expression induction found in MCF-7 by luciferase assay, it is possible that the c.-242-110delAGTA variant could be in LD with another functional variant located elsewhere in the promoter, as a high degree of LD is present across the whole NBN gene. Unfortunately, the promoter variant could not be associated by LD with another variant in the coding region of the gene, which would have allowed to directly measure NBN allelic expression.
Conclusion
Our analysis suggests that the variant c.-242-110delAGTA identified in our cohort of breast cancer individuals and located in the promoter region of the NBN gene may be associated with an increased risk of breast cancer. However, the functional analysis by luciferase promoter assay does not support a causative effect of this variant in the breast cancer cell line MCF-7, although a reduction of luciferase expression driven by the NBN promoter can be observed in two other cell lines tested. As the NBN genomic region displays a high degree of LD, we cannot rule out the effect of other variants located upstream of the region analyzed here. Alternatively, the effect of this variant may be dependent upon induction by a genotoxic stress such as irradiation. Although NBS is a rare syndrome, carriers of deleterious alterations in this gene are more common and may display subtle manifestations in cellular pathways predisposing NBN-mutated carriers to malignancy. Further analyses will therefore be needed to ascertain the impact of rare variants and promoter variants of NBN on breast cancer susceptibility in other populations.
Abbreviations
AIR: ATM interacting region; ALL: Acute lymphoblastic leukemia; ATBF1: AT motif-binding factor 1; BRCT: BRCA1 C-terminal; CDX2: Caudal-type homeobox 2; CI: Confidence interval; DSB: Double strand breaks; ESE: Exonic splicing enhancer; FHA: Forkhead-associated; HWE: Hardy-Weinberg equilibrium; LD: Linkage disequilibrium; MAF: Minor allele frequency; MIR: MRE11A interacting region; MTBF1: Muscle-specific MT binding factor; NBS: Nijmegen breakage syndrome; NKX3-1: NK3 homeobox 1; OR: Odds ratio; QRT-PCR: Quantitative real-time PCR; ZFHX3: Zinc finger homeobox 3.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SD participated in the molecular analysis study, performed the reporter gene assays experiments and bioinformatics analyses, and drafted the manuscript. CJB participated in the molecular analysis study. YL provided technical assistance and helped draft the manuscript. GO performed DNA and RNA extractions, cell maintenance and participated in the luciferase assays experiments. FD conceived the study, its design and coordination, and authored the final version of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Sylvie Desjardins, Email: sylvie.desjardins@crchul.ulaval.ca.
Joly Charles Beauparlant, Email: charles.joly-beauparlant@crchul.ulaval.ca.
Yvan Labrie, Email: yvan.labrie@crchul.ulaval.ca.
Geneviève Ouellette, Email: genevieve.ouellette@crchul.ulaval.ca.
INHERIT BRCAs, Email: jacques.simard@crchul.ulaval.ca.
Francine Durocher, Email: francine.durocher@crchul.ulaval.ca.
Acknowledgements
The authors would like to thank all individuals and families who participated in this study. We thank C. Brousseau, Dr. S. Délos, M.-A. Lajoie, P. Léger, J. Rhéaume, C. Samson, M. Tranchant, Dr. M. Dumont, G. Leblanc, H. Malouin, N. Bolduc, A. MacMillan and T. Babineau of the Cancer Genomics Laboratory for genetic counselling, sample management and mutation screening. We also thank F. Guénard for advices regarding luciferase assays, M. Ouellet and A.-M. Bureau-Blouin for skillful technical help and Dr. D. Labuda and C. Moreau at the Centre de Cancérologie Charles Bruneau of Ste-Justine Hospital for help with control DNA samples.
Other members of INHERIT BRCAs involved in this study are as follow: Paul Bessette: Service de gynécologie, Centre Hospitalier Universitaire de Sherbrooke, Fleurimont, QC, Canada; Jocelyne Chiquette: Clinique des maladies du sein Deschênes-Fabia, Hôpital du Saint-Sacrement, Québec, QC, Canada; Rachel Laframboise: Service de médecine génétique, CHUQ, Pavillon CHUL, Québec, QC, Canada; Jean Lépine: Centre Hospitalier régional de Rimouski, Rimouski, QC, Canada; Bernard Lespérance, Roxane Pichette: Service d'hémato-oncologie, Hôpital du Sacré-Cœur, Montréal, QC, Canada; Marie Plante: Service de gynécologie, CHUQ, L'Hôtel-Dieu de Québec, Québec, Canada; Jacques Simard: Director and instigator of the integrated research program INHERIT BRCAs, and chairholder of the Canada Research Chair in Oncogenetics. Cancer Genomics Laboratory, Oncology and Molecular Endocrinology Research Centre, Centre Hospitalier Universitaire de Québec and Laval University, Québec, Canada.
This work was supported by the Canadian Institutes of Health Research (CIHR) and Institute of Cancer and Institute of Gender and Health for the INHERIT BRCAs research program, the Fond de la Recherche en Santé du Québec (FRSQ)/Réseau de Médecine Génétique Appliquée (RMGA), the Canadian Breast Cancer Research Alliance (CBCRA) and the CURE Foundation. S.D. holds studentships from Fondation René Bussières and Fondation Desjardins, C.J.B. holds a Frederick Banting and Charles Best Canada Graduate Scholarships – Master's Award studentship from CIHR, and F.D. is a recipient of a chercheur-boursier from the Fonds de la Recherche en Santé du Québec (FRSQ) and Research Career Award in the Health Sciences from CIHR/R&D Health Research Foundation.
References
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85(4):509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Durocher F, Guénard F, Desjardins S, Ouellette G, Labrie Y. In: Molecular genetics of cancer. Sinnett D, editor. Kerala: Research Signpost; 2005. Inherited susceptibility to breast cancer: accomplishments and challenges; pp. 9–93. [Google Scholar]
- Seemanova E. An increased risk for malignant neoplasms in heterozygotes for a syndrome of microcephaly, normal intelligence, growth retardation, remarkable facies, immunodeficiency and chromosomal instability. Mutat Res. 1990;238(3):321–324. doi: 10.1016/0165-1110(90)90024-6. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Scuric Z, D'Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair (Amst) 2006;5(2):251–257. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Feshchenko S, Schurmann P, Waltes R, Wieland B, Hillemanns P, Rogov YI, Dammann O, Bremer M, Karstens JH. Nijmegen Breakage Syndrome mutations and risk of breast cancer. Int J Cancer. 2008;122(4):802–806. doi: 10.1002/ijc.23168. [DOI] [PubMed] [Google Scholar]
- Heikkinen K, Rapakko K, Karppinen SM, Erkko H, Knuutila S, Lundan T, Mannermaa A, Borresen-Dale AL, Borg A, Barkardottir RB. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27(8):1593–1599. doi: 10.1093/carcin/bgi360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HM, Wang HC, Chen ST, Hsu GC, Shen CY, Yu JC. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Biomarkers Prev. 2007;16(10):2024–2032. doi: 10.1158/1055-9965.EPI-07-0116. [DOI] [PubMed] [Google Scholar]
- Nowak J, Mosor M, Ziolkowska I, Wierzbicka M, Pernak-Schwarz M, Przyborska M, Roznowski K, Plawski A, Slomski R, Januszkiewicz D. Heterozygous carriers of the I171V mutation of the NBS1 gene have a significantly increased risk of solid malignant tumours. Eur J Cancer. 2008;44(4):627–630. doi: 10.1016/j.ejca.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Roznowski K, Januszkiewicz-Lewandowska D, Mosor M, Pernak M, Litwiniuk M, Nowak J. I171V germline mutation in the NBS1 gene significantly increases risk of breast cancer. Breast Cancer Res Treat. 2008;110(2):343–348. doi: 10.1007/s10549-007-9734-1. [DOI] [PubMed] [Google Scholar]
- Seemanova E, Jarolim P, Seeman P, Varon R, Digweed M, Swift M, Sperling K. Cancer risk of heterozygotes with the NBN founder mutation. J Natl Cancer Inst. 2007;99(24):1875–1880. doi: 10.1093/jnci/djm251. [DOI] [PubMed] [Google Scholar]
- Forsti A, Angelini S, Festa F, Sanyal S, Zhang Z, Grzybowska E, Pamula J, Pekala W, Zientek H, Hemminki K. Single nucleotide polymorphisms in breast cancer. Oncol Rep. 2004;11(4):917–922. [PubMed] [Google Scholar]
- Millikan RC, Player JS, Decotret AR, Tse CK, Keku T. Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14(10):2326–2334. doi: 10.1158/1055-9965.EPI-05-0186. [DOI] [PubMed] [Google Scholar]
- Dumon-Jones V, Frappart PO, Tong WM, Sajithlal G, Hulla W, Schmid G, Herceg Z, Digweed M, Wang ZQ. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63(21):7263–7269. [PubMed] [Google Scholar]
- Stumm M, Neubauer S, Keindorff S, Wegner RD, Wieacker P, Sauer R. High frequency of spontaneous translocations revealed by FISH in cells from patients with the cancer-prone syndromes ataxia telangiectasia and Nijmegen breakage syndrome. Cytogenet Cell Genet. 2001;92(3–4):186–191. doi: 10.1159/000056900. [DOI] [PubMed] [Google Scholar]
- Tanzanella C, Antoccia A, Spadoni E, di Masi A, Pecile V, Demori E, Varon R, Marseglia GL, Tiepolo L, Maraschio P. Chromosome instability and nibrin protein variants in NBS heterozygotes. Eur J Hum Genet. 2003;11(4):297–303. doi: 10.1038/sj.ejhg.5200962. [DOI] [PubMed] [Google Scholar]
- Neubauer S, Arutyunyan R, Stumm M, Dork T, Bendix R, Bremer M, Varon R, Sauer R, Gebhart E. Radiosensitivity of ataxia telangiectasia and Nijmegen breakage syndrome homozygotes and heterozygotes as determined by three-color FISH chromosome painting. Radiat Res. 2002;157(3):312–321. doi: 10.1667/0033-7587(2002)157[0312:ROATAN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Alt FW. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001;20(40):5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11(11):S52–59. doi: 10.1016/S0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- Fodde R, Smits R. Cancer biology. A matter of dosage. Science. 2002;298(5594):761–763. doi: 10.1126/science.1077707. [DOI] [PubMed] [Google Scholar]
- Simard J, Dumont M, Moisan AM, Gaborieau V, Malouin H, Durocher F, Chiquette J, Plante M, Avard D, Bessette P. Evaluation of BRCA1 and BRCA2 mutation prevalence, risk prediction models and a multistep testing approach in French-Canadian families with high risk of breast and ovarian cancer. J Med Genet. 2007;44(2):107–121. doi: 10.1136/jmg.2006.044388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher F, Labrie Y, Soucy P, Sinilnikova O, Labuda D, Bessette P, Chiquette J, Laframboise R, Lepine J, Lesperance B. Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families. BMC Cancer. 2006;6:230. doi: 10.1186/1471-2407-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4(3):311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenard F, Labrie Y, Ouellette G, Joly Beauparlant C, Simard J, Durocher F. Mutational analysis of the breast cancer susceptibility gene BRIP1/BACH1/FANCJ in high-risk non-BRCA1/BRCA2 breast cancer families. J Hum Genet. 2008;53(7):579–591. doi: 10.1007/s10038-008-0285-z. [DOI] [PubMed] [Google Scholar]
- Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23(2):255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- Ivanga M, Labrie Y, Calvo E, Belleau P, Martel C, Pelletier G, Morissette J, Labrie F, Durocher F. Fine temporal analysis of DHT transcriptional modulation of the ATM/Gadd45g signaling pathways in the mouse uterus. Mol Reprod Dev. 2009;76(3):278–288. doi: 10.1002/mrd.20949. [DOI] [PubMed] [Google Scholar]
- Takai K, Sakamoto S, Sakai T, Yasunaga J, Komatsu K, Matsuoka M. A potential link between alternative splicing of the NBS1 gene and DNA damage/environmental stress. Radiat Res. 2008;170(1):33–40. doi: 10.1667/RR1191.1. [DOI] [PubMed] [Google Scholar]
- Varon R, Dutrannoy V, Weikert G, Tanzarella C, Antoccia A, Stockl L, Spadoni E, Kruger LA, di Masi A, Sperling K. Mild Nijmegen breakage syndrome phenotype due to alternative splicing. Hum Mol Genet. 2006;15(5):679–689. doi: 10.1093/hmg/ddi482. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Ewens WJ. Heterozygous carriers of Nijmegen Breakage Syndrome have a distinct gene expression phenotype. Genome Res. 2006;16(8):973–979. doi: 10.1101/gr.5320706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya M, Sakata K, Tauchi H, Matsumoto Y, Nakamura A, Komatsu K, Hareyama M. Association of ionizing radiation-induced foci of NBS1 with chromosomal instability and breast cancer susceptibility. Radiat Res. 2006;166(4):575–582. doi: 10.1667/RR0638.1. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: Implications for design of association studies. Genet Epidemiol. 2003;25(3):190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- di Masi A, Viganotti M, Polticelli F, Ascenzi P, Tanzarella C, Antoccia A. The R215W mutation in NBS1 impairs gamma-H2AX binding and affects DNA repair: molecular bases for the severe phenotype of 657del5/R215W Nijmegen breakage syndrome patients. Biochem Biophys Res Commun. 2008;369(3):835–840. doi: 10.1016/j.bbrc.2008.02.129. [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12(21):1846–1851. doi: 10.1016/S0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Varon R, Reis A, Henze G, von Einsiedel HG, Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL) Cancer Res. 2001;61(9):3570–3572. [PubMed] [Google Scholar]
- Mosor M, Ziolkowska I, Pernak-Schwarz M, Januszkiewicz-Lewandowska D, Nowak J. Association of the heterozygous germline I171V mutation of the NBS1 gene with childhood acute lymphoblastic leukemia. Leukemia. 2006;20(8):1454–1456. doi: 10.1038/sj.leu.2404285. [DOI] [PubMed] [Google Scholar]
- Ziolkowska I, Mosor M, Wierzbicka M, Rydzanicz M, Pernak-Schwarz M, Nowak J. Increased risk of larynx cancer in heterozygous carriers of the I171V mutation of the NBS1 gene. Cancer Sci. 2007;98(11):1701–1705. doi: 10.1111/j.1349-7006.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Shimizu K, Mimaki S, Sakiyama T, Mori T, Shimasaki N, Yokota J, Nakachi K, Ohta T, Ohki M. First case of aplastic anemia in a Japanese child with a homozygous missense mutation in the NBS1 gene (I171V) associated with genomic instability. Hum Genet. 2004;115(5):372–376. doi: 10.1007/s00439-004-1155-1. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Schurmann P, Waltes R, Feshchenko S, Zalutsky IV, Bremer M, Dork T. NBS1 variant I171V and breast cancer risk. Breast Cancer Res Treat. 2008;112(1):75–79. doi: 10.1007/s10549-007-9820-4. [DOI] [PubMed] [Google Scholar]
- Seemanova E, Sperling K, Neitzel H, Varon R, Hadac J, Butova O, Schrock E, Seeman P, Digweed M. Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability. J Med Genet. 2006;43(3):218–224. doi: 10.1136/jmg.2005.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerosaletti KM, Morrison VA, Sabath DE, Willerford DM, Concannon P. Mutations and molecular variants of the NBS1 gene in non-Hodgkin lymphoma. Genes Chromosomes Cancer. 2002;35(3):282–286. doi: 10.1002/gcc.10114. [DOI] [PubMed] [Google Scholar]
- Hebbring SJ, Fredriksson H, White KA, Maier C, Ewing C, McDonnell SK, Jacobsen SJ, Cerhan J, Schaid DJ, Ikonen T. Role of the Nijmegen breakage syndrome 1 gene in familial and sporadic prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(5):935–938. doi: 10.1158/1055-9965.EPI-05-0910. [DOI] [PubMed] [Google Scholar]
- Figueroa JD, Malats N, Rothman N, Real FX, Silverman D, Kogevinas M, Chanock S, Yeager M, Welch R, Dosemeci M. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28(8):1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- Thirumaran RK, Bermejo JL, Rudnai P, Gurzau E, Koppova K, Goessler W, Vahter M, Leonardi GS, Clemens F, Fletcher T. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006;27(8):1676–1681. doi: 10.1093/carcin/bgi381. [DOI] [PubMed] [Google Scholar]
- Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, Rosa F, Di Gregorio A, Polidoro S, Saletta F, Ioannidis JP. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101(1):24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- Musak L, Soucek P, Vodickova L, Naccarati A, Halasova E, Polakova V, Slyskova J, Susova S, Buchancova J, Smerhovsky Z. Chromosomal aberrations in tire plant workers and interaction with polymorphisms of biotransformation and DNA repair genes. Mutat Res. 2008;641(1–2):36–42. doi: 10.1016/j.mrfmmm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Du R, An X, Chen Y, Qin J. Functional analysis of the Myostatin gene promoter in sheep. Sci China C Life Sci. 2007;50(5):648–654. doi: 10.1007/s11427-007-0085-2. [DOI] [PubMed] [Google Scholar]
- Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3(7):593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- Mogal AP, Meer R van der, Crooke PS, Abdulkadir SA. Haploinsufficient prostate tumor suppression by Nkx3.1: a role for chromatin accessibility in dosage-sensitive gene regulation. J Biol Chem. 2007;282(35):25790–25800. doi: 10.1074/jbc.M702438200. [DOI] [PubMed] [Google Scholar]
- Morinaga T, Yasuda H, Hashimoto T, Higashio K, Tamaoki T. A human alpha-fetoprotein enhancer-binding protein, ATBF1, contains four homeodomains and seventeen zinc fingers. Mol Cell Biol. 1991;11(12):6041–6049. doi: 10.1128/mcb.11.12.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar P, Dvorakova M, Kralova J, Pajer P, Kozmik Z, Dvorak M. Myb-interacting protein, ATBF1, represses transcriptional activity of Myb oncoprotein. J Biol Chem. 1999;274(20):14422–14428. doi: 10.1074/jbc.274.20.14422. [DOI] [PubMed] [Google Scholar]
- Miura Y, Kataoka H, Joh T, Tada T, Asai K, Nakanishi M, Okada N, Okada H. Susceptibility to killer T cells of gastric cancer cells enhanced by Mitomycin-C involves induction of ATBF1 and activation of p21 (Waf1/Cip1) promoter. Microbiol Immunol. 2004;48(2):137–145. doi: 10.1111/j.1348-0421.2004.tb03491.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Kawaguchi M, Miura Y, Iwase H. ATBF1-a messenger RNA expression is correlated with better prognosis in breast cancer. Clin Cancer Res. 2005;11(1):193–198. [PubMed] [Google Scholar]
- Sun X, Zhou Y, Otto KB, Wang M, Chen C, Zhou W, Subramanian K, Vertino PM, Dong JT. Infrequent mutation of ATBF1 in human breast cancer. J Cancer Res Clin Oncol. 2007;133(2):103–105. doi: 10.1007/s00432-006-0148-y. [DOI] [PubMed] [Google Scholar]