Abstract
Here, we describe the identification and characterization of a nuclear body (matrix-associated deacetylase body) whose formation and integrity depend on deacetylase activity. Typically, there are 20–40 0.5-μM bodies per nucleus, although the size and number can vary substantially. The structure appears to contain both class I and the recently described class II histone deacetylases (HDAC)5 and 7 along with the nuclear receptor corepressors SMRT (silencing mediator for retinoid and thyroid receptor) and N-CoR (nuclear receptor corepressor). Addition of the deacetylase inhibitors trichostatin A and sodium butyrate completely disrupt these nuclear bodies, providing a demonstration that the integrity of a nuclear body is enzyme dependent. We demonstrate that HDAC5 and 7 can associate with at least 12 distinct proteins, including several members of the NuRD and Sin3A repression complexes, and appear to define a new but related complex.
Nuclear hormone receptors are evolutionarily conserved ligand-dependent transcription factors that influence the biological processes of cell proliferation and differentiation and animal physiology (1). Their ability to target and bind to sequence-specific DNA elements and to act as either potent transcriptional activators in the presence of their cognate ligand or transcriptional repressors in the absence of a ligand allows this gene family to be used as a model system to study and characterize the initiation and process of gene transcription. In the absence of ligand, the nuclear corepressors SMRT (silencing mediator for retinoid and thyroid receptors) and N-CoR (nuclear receptor corepressor) bind to nuclear hormone receptors and act as “platform proteins” recruiting a large protein complex that includes class I and II histone deacetylases (HDAC) and Sin3A (2). It is hypothesized that the ability of this complex to deacetylate histones results in an altered chromatin state that is inhibitory to transcription (3). Because of the isolation of SMRT and N-CoR through their interaction with nuclear hormone receptors, extensive studies have now determined that these corepressors are also recruited by an increasing number of diverse transcription factors, including CBF1/RBPJK, PLZF, BCL6, MYOD Bach2, and Pbx1 (3).
Class II HDACs localize to distinct nuclear bodies within the cell nucleus and have only recently been shown to interact with SMRT/N-CoR. Their potential role in transcriptional repression by SMRT/N-CoR is yet to be fully characterized. In this study, we have investigated the role of class II HDACs in the recruitment of additional core deacetylase factors to the SMRT/N-CoR complex. The findings described here suggest that protein members of both the NuRD and Sin3A repression complexes can interact with the corepressors SMRT/N-CoR, indicating the existence of a related repression complex. In addition, we have investigated and characterized the class II nuclear bodies and have subsequently identified a unique nuclear body, which we have termed matrix-associated deacetylase (MAD) body, that depends on deactylase activity for its structural integrity.
Methods
Constructs.
The plasmids pCMX, pCMX-GAL4 DBD, pCMX-SMRT, and pMH100-TK-luc have been described elsewhere (2). Standard PCR amplifications of the appropriate cDNA and subcloning techniques were used to generate pCMX carboxyl-terminal hemagglutinin (HA) or Flag-epitope tagged and GAL4 or yellow fluorescence protein (YFP) fusion constructs. All constructs were verified by double-stranded sequencing to confirm identity and reading frame. Detailed information regarding each construct is available on request. Mutations were generated by using the QuickChange kit (Stratagene) according to the manufacturer's directions. Mutations were verified by sequencing.
Transfections.
Monkey CV-1 cells were grown in DMEM supplemented with 10% FBS, 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate at 37°C in 7% CO2. CV-1 cells (60–70% confluence, 48-well plate) were cotransfected with 16.6 ng of pCMXGAL4 and pCMXGAL4-HDAC constructs, 100 ng of pMH100-TK-Luc, and 100 ng of pCMX-LacZ in 200 μl of DMEM containing 10% FBS by a N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate-mediated procedure. After 24 h, the medium was replaced, and cells were harvested and assayed for luciferase activity 36–48 h after transfection. The luciferase activity was normalized by the level of β-galactosidase activity. Each transfection was performed in triplicate and repeated at least three times.
Coimmunoprecipitation.
For immunoprecipitations, 293 cells were transfected with 15 μg of the appropriate plasmid with Targefect F1 (Targeting Systems, San Diego, CA). Cells were harvested 48 h later by lysing in 50 mM Tris (pH 8.0)/150 mM NaCl/10% glycerol/0.5% Triton X-100/1 mM PMSF, and protease inhibitors. Cells were lysed for 15 min at 4°C, scraped, and centrifuged for 15 min at 12,000 × g. Supernatant was kept as whole cell extract. After preclearing by incubation with A/G agarose (Santa Cruz Biotechnology), immunoprecipitations were carried out with either HA-agarose (Santa Cruz) or M2-agarose (Sigma) and proceeded for 2 h at 4°C. Beads were washed three to four times in lysis buffer without Triton for HDAC assays and PBS with 0.1% Nonidet P-40 for coimmunoprecipitations. For coimmunoprecipitations, samples were separated on SDS-polyacrylamide gels, transferred to nitrocellulose membrane, and probed with the appropriate antibodies.
HDAC Assays.
HDAC assays were performed according to standard protocols (3). Briefly, 60,000 cpm of 3H-labeled histones was incubated with immunoprecipitates for 2 h at 37°C. Reactions were stopped by addition of acetic acid/HCl to a final concentration of 0.12/0.72 M and extracted with two volumes of ethyl acetate. Samples were centrifuged, and the supernatant was counted in a scintillation counter. Each reaction represents approximately one-third of a transfected 10-cm plate of cells.
Green Fluorescence and Immunofluorescence Microscopy.
CV-1 cells were plated into two-well chamber slides and transfected with Targefect F1 (Targeting Systems). After 48 h, cells were washed in PBS, fixed in 3.7% paraformaldehyde, and permeabilized with 1% Triton X-100. For immunostaining, fixed cells were incubated with antibodies against SMRT (Affinity Bioreagents, Neshanic Station, NJ), CREB-binding protein (CBP) (Upstate Biotechnology, Lake Placid, NY), or SC-35 (American Type Culture Collection) for 1 h, washed, and incubated with secondary antibody (Cy3 or Cy5) for 1 h. Cells were washed and mounted with Permount. 4′,6-diamidino-2-phenylindole was included in the final wash to visualize nuclei. Images were visualized with an Olympus 1 × 70 inverted system microscope equipped with charge-coupled device. The resulting images were deconvolved with deltavision2 software (Applied Precision, Issaquah, WA). CV-1 nuclear matrix preparations were done by using established procedures at high salt extraction (final wash 2 M NaCl).
Results and Discussion
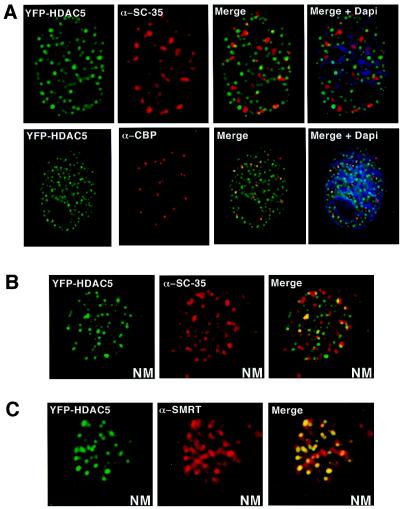
We have recently isolated two members of the class II HDAC family, mHDAC5 and 7, through their direct interaction with the nuclear hormone receptor corepressors, SMRT and N-CoR (3, 4). The subcellular distribution revealed that SMRT localizes to a nuclear structure in a class II HDAC- (but not class I)-dependent manner. The size and morphology of these nuclear structures are reminiscent of promyelocytic leukemia (PML) oncogenic domains (PODs) (5) and RNA splicing bodies (SBs) (6), which have been shown to harbor distinct interacting proteins. This raised the possibility that the structure itself, if unique, could be used to identify HDAC-associated proteins. To examine the potential overlap of the HDAC domains with established nuclear substructures, CV-1 cells transiently expressing a YFP-tagged HDAC5 were subjected to immunohistochemistry with antibodies against SC-35, PML, and CBP to visualize PODs and SBs. The SC-35 antibody revealed specific structures that do not colocalize with the HDAC5 fluorescence, indicating that the distribution of HDAC5 bodies is distinct from that of interchromatin granule clusters (IGCs) (SBs) (Fig. 1A). The staining pattern of both PML (not shown) and CBP also failed to overlap with that of HDAC5, providing further evidence that HDAC structure represents a unique nuclear domain (Fig. 1A). Previous studies have demonstrated that HDAC activity can be found in the nuclear matrix (7), although the basis for this activity remains unknown. To test whether the class II HDAC5 and 7 are associated with the nuclear matrix, nuclei from cells expressing YFP-HDAC were prepared and subjected to high-salt extraction (8, 9) to isolate nuclear matrix proteins. We find that HDAC5 and 7 are tightly associated with the nuclear matrix and retain a subcellular dot-like structure that is distinct from both IGCs (Fig. 1B) and PODs (not shown). These data suggest the existence of a nuclear body that is tightly associated with the nuclear matrix. We refer to these structures as matrix-associated deacetylase or MAD bodies.
Figure 1.
HDAC5 colocalizes with SMRT in the nuclear matrix. (A) YFP-HDAC5 subnuclear structures do not colocalize with SC-35-postive IGCs or PODs identified by the CBP antibody. CV-1 cells were plated into two-well chamber slides and transfected with YFP-HDAC5. For immunostaining, fixed cells were incubated with antibodies against SC-35 (ATCC) for 1 h, washed, and incubated with secondary antibody (Cy3 or Cy5) for 1 h. Cells were washed and mounted with Permount. 4′,6-diamidino-2-phenylindole was included in the final wash. Images were visualized with an Olympus 1 × 70 inverted system microscope equipped with charge-coupled device. The resulting images were deconvolved. (B) YFP-HDAC5 is retained in the nuclear matrix of transfected CV-1 cells by using established nuclear matrix preparation procedures. SC-35 was also present in the nuclear matrix but does not colocalize with YFP-HDAC5. NM, nuclear matrix. (C) SMRT colocalizes with YFP-HDAC5 in the nuclear matrix. Antibody to SMRT (Affinity Bioreagents) colocalizes with YFP-HDAC5 in the nuclear matrix preparations.
Analysis of PODs has indicated that all proteins found to interact with PML in vitro also localize to PODs in vivo. On the basis of the analogy to PODs, we asked whether the corepressor SMRT associates with HDAC5 and 7 in the nuclear matrix. Indeed, as shown in Fig. 1C, endogenous SMRT is found to be retained in the nuclear matrix via its association with HDAC5.
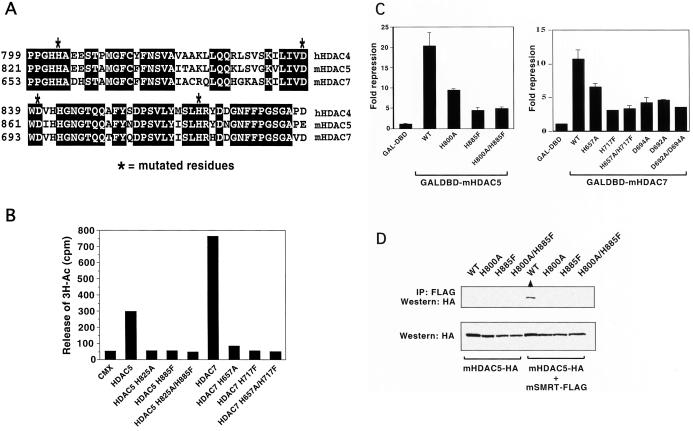
Because the deacetylation domain is the most conserved region between class II HDACs, we asked whether mutations that block deacetylation activity affect SMRT recruitment. There is some precedence for this in that mutations within the catalytic site of HDAC1 block association with corepressors Sin3A and RbAp48 (10–12). On the basis of sequence conservation between the class I and II HDACs, we generated putative inactivating mutations in the HDAC5 and 7 deacetylation domains (Fig. 2A, mutation denoted by ∗) and examined their effect on deacetylation activity, transcriptional repression, SMRT association, and subcellular localization. To measure HDAC activity, epitope-tagged (HA) wild-type and HDAC mutants were purified from transfected 293 cells. The enzymatic activity of the immunoprecipitated protein was measured in vitro by the release of 3H-acetate from labeled histones (10, 12). Although wild-type HDAC5 and 7 displayed robust histone deacetylation activity, only background levels were found in each of the mutants (Fig. 2B). The ability of the mutant proteins to repress basal transcription of a heterologous promoter was next assessed through cotransfection experiments in CV-1 cells (2, 13). Strong repression was seen with both wild-type HDAC5 (20-fold) and HDAC7 (9-fold). In contrast, the ability of the mutants to repress transcription was significantly impaired, although not completely lost (Fig. 2C). These results support the link between HDAC activity and transcriptional repression; the residual repression activity observed most likely reflects the continued ability of the mutant proteins to recruit endogenous wild-type deacetylase.
Figure 2.
Mutants of HDAC5 and 7 affect their HDAC activity, transcriptional repression ability, and interaction with SMRT. (A) An amino acid sequence alignment of a conserved region within the deacetylase domain of human HDAC4 and mouse HDAC5 and HDAC7. * indicates where residues were mutated. (B) Mutations within the deacetylase domain of HDAC5 and 7 inhibit their HDAC activity. Whole-cell extracts from 293 cells prepared from cells expressing vector alone, wild type, and mutants of mHDAC5 and mHDAC7 were immunoprecipitated with anti-HA antibodies conjugated agarose beads (Santa Cruz Biotechnology). Immunoprecipitates were then assayed for release of 3H-Ac by using established procedures. (C) Mutations within the deacetylation domains of HDAC5 or HDAC7 inhibit their ability to repress basal transcriptional repression. Reporter activity (pMH100-TK-Luc) in CV-1 cells transfected with pCMXGAL4 or pCMXGAL4-HDAC constructs. Fold repression was determined relative to the basal transcription activity of the reporter in the presence of GAL4 DBD. Each transfection was preformed in triplicate and repeated at least three times. (D) Mutations within the deacetylase domain of HDAC5 and 7 abolish protein interaction with SMRT. Whole-cell extracts prepared from 293 cells transfected with wild-type or mutant HDAC5-HA with or without mSMRTa-Flag expression vector were incubated with anti-Flag antibodies conjugated with agarose beads. Immunoprecipitates were subjected to Western blot analysis and probed with anti-HA antibody.
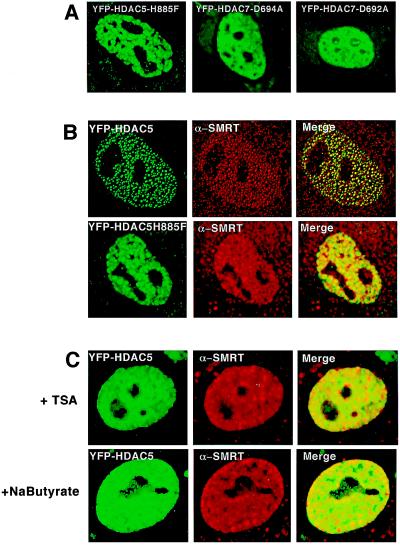
The ability of HDAC5 mutants to interact with SMRT was tested by using both coimmunoprecipitation and immunofluorescence colocalization techniques. A FLAG epitope-tagged SMRT (2) was introduced into 293 cells in the presence of wild-type or mutant HDAC5 expressing a HA tag. Only wild-type HDAC5 was precipitated in the presence of SMRT-Flag, indicating that the mutations made within the deacetylase domain of HDAC5 abolished direct interaction with SMRT (Fig. 2D). Experiments were also conducted with HDAC7, and similar results were obtained (data not shown). We next addressed the key issue of whether the deacetylase mutation affects the localization of HDAC5 and SMRT to MAD bodies. The two results seem clear; first, HDAC5 loses its ability to concentrate in highly focal MAD bodies (Fig. 3A). Rather, it shows a much expanded cotton-ball appearance with only residual structure left. Second, endogenous SMRT also seems to display almost complete dissociation from the body (Fig. 3B). Because the results in Fig. 2D suggest that the SMRT no longer interacts with HDAC5 mutants, we speculate that the diffuse pattern seen with the mutants might be an indirect effect of the mutant on other components of the MAD body. Additional mutations in HDAC7, D692A, and D694A, showed similar results (e.g., Fig. 3A 2 and 3). These data indicate that a functional deacetylase domain is required not only for interaction with SMRT but also for proper localization of the class II deacetylases to the subnuclear structures.
Figure 3.
Mutants of HDAC5 or 7 and the deacetylase inhibitor TSA or sodium butyrate disrupt the subcellular nuclear localization of HDAC5 and 7. (A) Direct fluorescence detection of mutant YFP-HDAC5 and YFP-HDAC7 in CV-1 cells shows the disruption of the characteristic subnuclear structures to a diffuse nuclear pattern. (B) Comparison of endogenous SMRT pattern when either wild-type YFP-HDAC5 or mutated YFP-HDAC5 is present CV-1 cells. The results are obtained with the use of direct and indirect immunofluorescence by using a SMRT antibody. Cells transfected with mutatedYFP-HDAC5H885F in contrast to wild-type-YFP-HDAC5 show a diffuse nuclear pattern that does not colocalize with endogenous SMRT in the nucleus. (C) Addition of the deacetylase inhibitors TSA and sodium butyrate disrupt the subcellular dot-like nuclear localization of HDAC5 and 7. Experiments were carried out by using direct fluorescence detection of wild-type YFP-HDAC5 in CV-1 cells after addition of TSA (100 nM; Upstate Biotechnology) or sodium butyrate (10 mM; Sigma). Images were viewed on an Olympus 1 × 70 inverted system microscope before deconvolation with deltavision2 software.
The finding that mutations in the deacetylase domain abolished the integrity of the MAD bodies led to speculation that the enzymatic activity itself may be crucial in their formation. We tested this notion by treating the cells with the deacetylase inhibitors sodium butyrate and trichostatin A (TSA) (14) and by examining the effects on YFP-HDAC5 localization. Both TSA and sodium butyrate led to rapid and virtually complete disintegration of the body, resulting in soluble HDAC5 distributed through the nucleus (Fig. 3D). In contrast, TSA and sodium butyrate had no effect on the formation or integrity of CBP-associated PODs (data not shown), indicating that the effects were linked to the structure and were not because of a nonspecific loss of nuclear organization. Similarly, the polymerase II inhibitor's actinomycin D and α-amanitin also had no effect on MAD bodies, indicating they do not depend on active transcription (data not shown). In aggregate, these experiments clearly identify the MAD body as a unique nuclear compartment in which deacetylase activity is a requirement for formation.
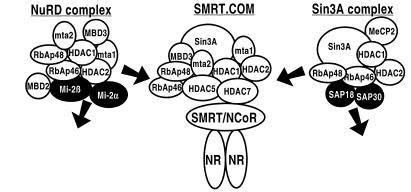
Having established that class II HDACs are able to recruit SMRT into MAD bodies, we speculated that they might comprise a unique repressor complex. In particular, we wished to understand their relationship to the recently characterized Sin3/HDAC (15) and the NuRD/Mi2 complexes (16–19). The NuRD/Mi2 complex includes MTA1/2 and MBD3 and the shared proteins HDAC1/2 and RbAp46/48 (15, 20) but not Sin3A, SMRT, or N-CoR. In contrast, the Sin3/HDAC complex includes SMRT and N-CoR, but not MTA1/2 or MBD3 (16, 17, 19). Of particular relevance, class II deacetylases have not yet been assigned to either complex (21–23). Thus, we conducted a series of immunoprecipitations to identify potential class II-associated proteins.
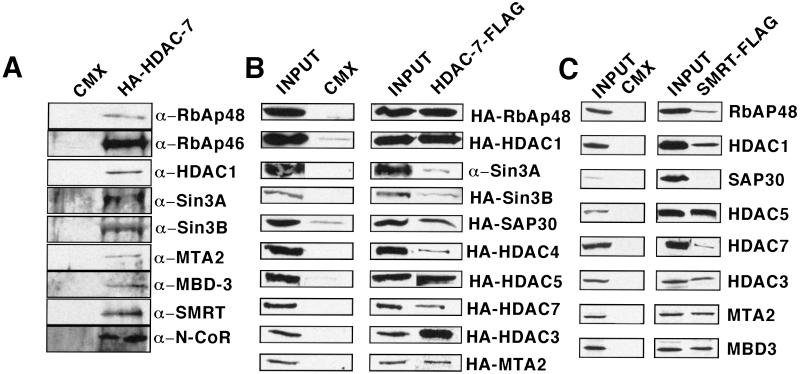
As expected, HA-tagged HDAC7 coimmunoprecipitated endogenous HDAC1, SMRT/N-CoR, Sin3A/B, and RbAp46/48 from 293 cell extracts. Surprisingly, MTA1/2, and MBD3 were also coimmunoprecipitated, providing the first in vivo association of these proteins with Sin3A/B, SMRT, and N-CoR. None of these proteins were identified in the vector-only immunoprecipitate, consistent with a specific association with HDAC7 (Fig. 4A). Similar results were observed with HDAC5 extending to the above observations (results not shown). Interestingly, despite many repeated attempts, we were unable to detect any endogenous Mi2 in any of the HDAC7 or 5 immunoprecipitates. Immunoprecipitation of the endogenous proteins was followed up by a series of paired pulldowns by using a Flag-tagged HDAC7 expression construct and HA-tagged cDNA expression constructs for SAP18, SAP30, RbAp48, Sin3A/B, HDAC1, 2, 3, 4, 5 and 7, and MTA2. The Flag antibody immunoprecipitated all of the proteins tested except SAP-18 (Fig. 4B; data for SAP18 not shown), whereas none of the proteins were immunoprecipitated with the Flag antibody in the vector-only transfected 293 cells (Fig. 4B). These results indicate that class II HDACs can specifically and selectively interact with components of both the NuRD/Mi2 and Sin3/HDAC complexes and thus appear to define a molecular identity referred to as SMRT.com (24).
Figure 4.
Members of the Sin3/HDAC and NuRD/Mi2 deacetylase protein complexes can interact with Class II HDAC7 and SMRTa. (A) HDAC7 complexes in 293 cells with proteins that have been characterized in the Sin3/HDAC and NuRD/Mi2 complexes. Whole-cell extracts prepared from 293 cells with either vector alone or mHDAC7-HA expression vector were incubated with anti-HA antibodies conjugated with agarose beads. Immunoprecipitates were subjected to Western blot analysis and probed with antibodies to RbAp48, RbAp46 (Upstate Biotechnology), HDAC1 (Affinity BioReagents), mSin3A/B (Santa Cruz), MTA2 (D. Reinberg, Howard Hughes Medical Institute, Piscataway, NJ), MBD3 (D. Reinberg), SMRT (Affinity BioReagents), and N-CoR (Upstate Biotechnology). (B) Results of coimmunoprecipitation experiments demonstrating interaction of HDAC7 with members of the Sin3/HDAC and NuRD/Mi2 deacetylase protein complexes. 293 cells were transfected with empty vector or Flag-tagged HDAC7 with the vector containing the DNA of interest HA-tagged. Whole-cell extracts were immunoprecipitated with FLAG antibody subjected to Western blot analysis and probed with anti-HA antibody. Inputs to the corresponding lanes (Left). (C) Results of coimmunoprecipitation experiments by using Flag-tagged SMRTa and HA-tagged members of the Sin3/HDAC and NuRD/Mi2 deacetylase protein complexes in 293 cells. Coimmunoprecipitation experiments were done as previously described
It is reasonable to speculate as to the role of SMRT in the class II HDAC complex and whether a SMRT directed pulldown would also identify the same proteins. Accordingly, a series of coimmunoprecipitation experiments were conducted by using Flag-SMRT vectors and HA-tagged cDNA constructs for RbAp48, HDAC1, HDAC3, SAP30, MTA2, MBD3, and HDAC7. The Flag antibody immunoprecipitated all of the proteins tested except SAP-30, whereas none of the proteins were immunoprecipitated with the Flag antibody in the vector-only transfected 293 cells (Fig. 4C). Although we did not detect an interaction of SAP-30 with SMRT by coimmunoprecipitation, work done by Laherty et al. (25) has demonstrated an involvement of SAP-30 in transcription repression by N-CoR.
We have provided strong evidence that mutations within the deacetylase domain of class II HDACs disrupt the association with SMRT and the integrity of the MAD bodies. To establish a role for the importance of the domain in forming the complex, we examined the coimmunoprecipitation of RbAp48, HDAC2, 3, 7, and MTA2, with wild-type-HDAC7, HDAC7-H657A, HDAC7-H717F, and HDAC7-A694D. Cell extracts were immunoprecipitated with Flag antibody and examined with HA antibody for the presence of wild-type-HDAC7 or mutated HDAC7 (Fig. 5). Surprisingly, only two proteins, MTA2 and RbAp48, were sensitive to the deacetylase mutations. In the case of the H717F, only RBAp48 was sensitive to the mutation. In contrast, all of the other proteins examined retained full association, indicating that the deacetylase domain and deacetylase activity play a highly selective role in protein association with both HDAC and non-HDAC proteins. Interestingly, MTA2, SMRT, and RbAp48 have been demonstrated to be essential for class II deacetylase activity but not HDAC1 or 3. Furthermore, these results suggest that there must be multiple and independent-binding sites on HDAC5 and 7 for the associated cofactors. Thus, it is possible that binding of selective proteins to the deacetylase domain may help to create or stabilize the catalytic site.
Figure 5.
Mutants of HDAC7 interact with HDACs but not with the cofactors RbAp48 and MTA2. 293 cells were transfected with empty vector or Flag-tagged vector containing the DNA of interest (MTA2, RBAp48, HDAC2, HDAC3, HDAC7) and wtHDAC7-HA or mutated HDAC7-HA. Whole-cell extracts were immunoprecipitated with FLAG antibody subjected to Western blot analysis and probed with anti-HA antibody. Inputs to the corresponding panels are in the panel above.
In summary, these results suggest that the MAD bodies may be comprised of a multisubunit protein complex that contains members of both the NuRD/Mi2 and Sin3/HDAC complexes. Class II HDAC-associated proteins include HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC7, SAP-30, RbAp48, Sin3A/B, SMRT/N-CoR, MBD3, and MTA2. Furthermore, these proteins also appear to associate with SMRT. Our results describe a nuclear substructure (MADs) that depends on HDAC activity and may thus represent a site of action that is physiologically relevant. One might argue that the mutations generated within the deacetylase domain of the class II HDACs might be inducing a structural change within the protein that disrupts the MADs, not disruption of deacetylase activity. However, the finding that TSA and sodium butyrate mimic the effects of the mutants leads us to believe that it is the deacetylation activity of the protein that is an absolute requirement for formation of MADs. The most compelling evidence in support of this notion comes from the recent crystal structure of the hyperthermophilic bacterium Aquifex aeolicus HDAC complexed with the small molecule inhibitor TSA (14). In this structure, the TSA is bound to the active-site pocket of the protein that normally accommodates the acetylated lysines but does not cause any major conformational changes to the protein indicating that the integrity of the MADs body indeed depends on enzymatic activity. To date, no class II HDACs have been found in the Sin3/HDAC or NuRD complexes, suggesting that the anticipated deacetylase complex containing class II HDACs and components of both Sin3/HDAC and NuRD/Mi2 is either not soluble (i.e., attached to the nuclear matrix) or is simply unstable. Our results suggest that unique classes of corepressors such as SMRT and N-CoR may serve as a template to draw together a unique and definable complex that contains components from two previously characterized entities (Fig. 6). Given that class II HDACs seem to be differentially expressed (23) in a number of cell lines, this complex is also likely to be dynamic in nature. In conclusion, our findings describe the composition and subnuclear organization of class II HDAC complexes, which are recruited by the corepressor SMRT and provide evidence of a nuclear matrix-associated structure in which deacetylation activity is an absolute requirement for its formation.
Figure 6.
Nuclear receptor mediated repression: a complex picture. Model depicting the nuclear receptor corepressor SMRT recruiting factors from the Sin3/HDAC and NuRD/Mi2 complexes to form a unique repressing deacetylase complex termed SMRT.com.
Acknowledgments
We thank Drs. D. Reinberg, E. Y. Lee, and S. L. Schreiber for cDNAs and antibodies, Dr. D. Egan for the pCMX-YFP plasmid and help with deltavision2 software and fluorescence experiments, and Dr. K. K. Walker for critical reading of the manuscript M.D. is a C. J. Martin Research fellow from the National Health and Medical Research Foundation of Australia. P.O. is supported by the George E. Hewitt Foundation for Medical Research. H.-Y.K. is a fellow of the Leukemia Society of America. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work is supported by grants from the National Institutes of Health.
Abbreviations
- HDAC
histone deacetylase
- SMRT
silencing mediator for retinoid and thyroid receptors
- N-CoR
nuclear receptor corepressor
- MAD
matrix-associated deacetylase
- HA
hemagglutinin
- YFP
yellow fluorescence protein
- PML
promyelocytic leukemia
- POD
PML oncogenic domain
- IGC
interchromatin granule cluster
- TSA
trichostatin A
References
- 1.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 2.Ordentlich P, Downes M, Xie W, Genin A, Spinner N B, Evans R M. Proc Natl Acad Sci USA. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao H-Y, Downes M, Ordentlich P, Evans R M. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 4.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 5.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 6.Spector D L, Fu X-D, Maniatis T. EMBO J. 1991;11:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davie J R, Hendzel M J. J Cell Biochem. 1994;55:98–105. doi: 10.1002/jcb.240550112. [DOI] [PubMed] [Google Scholar]
- 8.Capco D G, Wan K M, Penman S. Cell. 1982;29:847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- 9.Fey E G, Ornelles D A, Penman S. J Cell Sci Suppl. 1986;5:99–119. doi: 10.1242/jcs.1986.supplement_5.6. [DOI] [PubMed] [Google Scholar]
- 10.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadosh D, Struhl K. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finnin M S, Donigian J R, Cohen A, Richon V M, Rifkind R A, Marks P A, Breslow R, Pavletich N P. Nature (London) 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 15.Knoepfler P S, Eisenman R N. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 18.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 19.Ng H H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 20.Qian Y W, Wang Y C, Hollingsworth R E, Jr, Jones D, Ling N, Lee E Y. Nature (London) 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 21.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng J, Han J, Yang X J. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 24.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 25.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, et al. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]