Abstract
Trypanosoma brucei, the protozoan parasite responsible for sleeping sickness, evades the immune response of mammalian hosts and digestion in the gut of the insect vector by means of its coat proteins tethered to the cell surface via glycosylphosphatidylinositol (GPI) anchors. To evaluate the importance of GPI for parasite survival, we cloned and disrupted a trypanosomal gene, TbGPI10, involved in biosynthesis of GPI. TbGPI10 encodes a protein of 558 amino acids having 25% and 23% sequence identity to human PIG-B and Saccharomyces cerevisiae Gpi10p, respectively. TbGPI10 restored biosynthesis of GPI in a mouse mutant cell line defective in mouse Pig-b gene. TbGPI10 also rescued the inviability of GPI10-disrupted S. cerevisiae, indicating that TbGPI10 is the orthologue of PIG-B/GPI10 that is involved in the transfer of the third mannose to GPI. The bloodstream form of T. brucei could not lose TbGPI10; therefore, GPI synthesis is essential for growth of mammalian stage parasites. Procyclic form cells (insect stage parasites) lacking the surface coat proteins because of disruption of TbGPI10 are viable and grow slower than normal, provided that they are cultured in nonadherent flasks. In regular flasks, they adhered to the plastic surface and died. Infectivity to tsetse flies is partially impaired, particularly in the early stage. Therefore, parasitespecific inhibition of GPI biosynthesis should be an effective chemotherapy target against African trypanosomiasis.
Trypanosoma brucei is a protozoan parasite invading humans and other mammals by transmission via tsetse flies. It causes sleeping sickness in humans and nagana disease in domestic animals living in the “tsetse belt” in central Africa. These are serious medical and agricultural problems for which safe and effective therapeutic and protective measures are highly desirable (1, 2).
T. brucei has two distinct proliferative stages, a bloodstream stage living free in mammalian blood and an insect stage (or procyclic form) living in the midguts of tsetse flies. The cell surface of both stages of this unicellular parasite is covered by a large amount of glycosylphosphatidylinositol (GPI)-anchored proteins (3, 4): 107 variant surface glycoproteins per cell for the bloodstream form of the parasite and 3 × 106 to 6 × 106 procyclins (or procyclic acidic repetitive proteins) per cell of the procyclic form of the parasite (4–6), corresponding to 10% and 1–3%, respectively, of total proteins in these parasite stages (7, 8). T. brucei evades the host's immune response by expressing structurally different forms of variant surface glycoproteins (4). Procyclins are thought to protect procyclic cells from digestion by the digestive enzymes in the fly (4, 6). In addition, T. brucei expresses a number of other GPI-anchored proteins, such as transferrin receptors in the bloodstream form (3, 4). Thus, the importance of GPI anchors for the survival and infection of T. brucei has been suggested, leading to the notion that the GPI biosynthesis pathway may be a good target for chemotherapy against African trypanosomiasis (3, 9, 10). The killing of the bloodstream form of T. brucei by a myristic acid analogue that modulates fatty acid remodeling of GPI required for GPI-anchoring of variant surface glycoproteins strongly supports this notion (11). GPI has been shown to be essential in two fungi, Saccharomyces cerevisiae and Schizosaccharomyces pombe. However, it is not essential for growth of mammalian cells, although it is necessary for proper embryogenesis (12, 13). To test whether GPI is essential for T. brucei, we cloned a trypanosome gene that is required for biosynthesis of GPI and disrupted it in both the mammalian and insect stages.
Materials and Methods
Manipulation of Mammalian and Yeast Cells.
Transfection, radiolabeling, and analyses by flow cytometer and TLC were carried out as reported (14). Analysis with yeast cells was performed as reported (15).
Disruption of TbGPI10.
The bloodstream form of T. brucei brucei strain 427 clone 221a and the procyclic form derived from strain 427 (16) were transfected with an SphI–EcoRI fragment containing hygromycin (HYG) or neomycin (NEO) resistance gene (illustrated in Fig. 2A) for homologous recombination. After transfection, hygromycin-resistant (H-R) and geneticin-resistant (N-R) clones were established. One N-R clone of bloodstream form was transfected further with the HYG fragment with or without episomal TbGPI10 plasmids bearing bleomycin (BLE) resistance gene. Southern blot analysis of established geneticin- and hygromycin-resistant clones (NH-R) and geneticin-, bleomycin-, and hygromycin-resistant clones (NBH-R) was carried out by using 4 μg of SalI–XhoI cut DNA with the Sal–Sph probe (indicated in Fig. 2A). One H-R clone of the procyclic form was transfected further with the NEO fragment. To confirm the homologous recombination, Southern blot analysis of established hygromycin- and geneticin-resistant (HN-R) clones was performed.
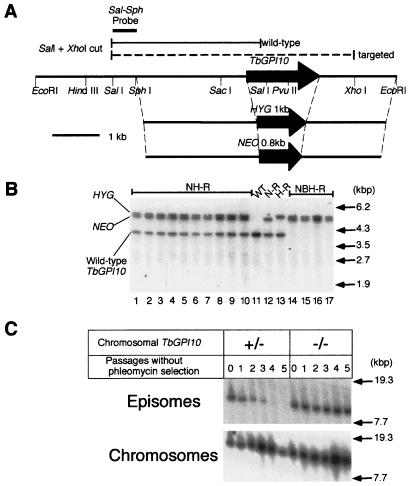
Figure 2.
Essentiality of TbGPI10 for bloodstream form of T. brucei. (A) Knockout strategy. A restriction map of TbGPI10 and its flanking regions and two targeting constructs in which TbGPI10 was replaced with HYG or NEO are shown. A probe for Southern blotting (Sal–Sph probe) and predicted fragments detected with this probe are indicated above the restriction map. (B) Southern blot analysis of drug-resistant clones. Expected positions of wild-type and homologous recombinant fragments are indicated on the left, and size markers are on the right. WT, wild-type; kbp, kilobase pair. (C) Stability of episomal plasmids in the presence or absence of chromosomal TbGPI10. TbGPI10 double knockout cells bearing episomal TbGPI10 plasmids with BLE were cultured in the absence of drug selection. As a positive control for a loss of episomal plasmids without drug selection, a neomycin-resistant, heterozygous TbGPI10 knockout procyclic clone was transfected with an episomal plasmid bearing BLE. At passages, DNA was prepared, digested with BamHI, and analyzed by Southern blot hybridization with BLE probe to detect episomal DNA and Sal–Sph probe to detect chromosomal TbGPI10. Size markers are on the right. Bands shown in each panel had the same mobility, although they are not well aligned in the figure because of uneven running.
As an episomal plasmid vector, we used pT11bs-ble that was prepared from pT11-bs (17) by exchanging NEO with BLE (18). For the bloodstream form, TbGPI10, which was flanked by splice acceptor signal and polyadenylation signal from the aldolase gene (19), was inserted in a SmaI site. For the procyclic form, TbGPI10, which was flanked by actin splice acceptor signal and polyadenylation signal (20), was inserted in a SmaI site.
Stability of Episomal Plasmids.
TbGPI10 double and single knockout cells bearing episomal TbGPI10 plasmids with BLE were cultured in the absence of drug selection. Cultures were initiated with 104 parasites per ml in HMI9 with 20% (vol/vol) heat-inactivated FCS (21). When culture density reached 0.8 × 106 to 1.3 × 106 parasites per ml, the culture was diluted 100-fold, and a new culture passage was started. The rest of the culture was used for DNA extraction.
Biosynthesis of GPI.
Membranes of procyclics were labeled with GDP-[3H]mannose (22, 23) except for the incubation condition (27°C for 1 h). Aliquots of the radiolabeled samples were subjected to TLC analysis directly or after digestion with 1.7 units/ml Jack bean α-mannosidase (Sigma) in 150 μl of buffer (24) or 20% human serum as a source of GPI-specific phospholipase D (GPI-PLD) in 150 μl of buffer (25). Samples were also digested with phospholipase A2 or phosphatidylinositol-specific phospholipase C. Identities of mannolipids were determined based on their susceptibilities to the enzymes and on known TLC profiles reported by others (22–25).
Myristic Acid Labeling of Procyclics.
[3H]myristic acid [50μCi (1 Ci = 37 GBq)] was added to 0.5 ml of SDM79 culture medium (26) containing 5 mg/ml defatted BSA. The suspension was added together subsequently with 5 ml of SDM79 with 10% (vol/vol) FCS to 5 ml of trypanosome culture (1 × 107 cells per ml) and incubated for 16 h at 27°C. After that, procyclins were concentrated (27) and analyzed by SDS/12% PAGE and autoradiography.
Analysis of Procyclins with a Pulse-Chase Experiment.
Procyclics washed in PBS were resuspended at 108 cells per ml in SDM-79 with low (133 μM) proline and 10% (vol/vol) dialyzed FCS. [14C]proline was added (10 μCi/ml, Amersham Pharmacia) and incubated for 30 min at 27°C. Chase was initiated by 10-fold dilution with prewarmed complete SDM-79 medium containing 10% (vol/vol) FCS and continued for 20 h. At each time point, 1.0 ml of culture was centrifuged, and the supernatant and the cell pellet were separated. Supernatants were filtered to eliminate parasites completely. Cell pellets were solubilized in 1.0 ml of buffer (50 mM Tris⋅HCl, pH 7.5, containing 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 1 μg/ml leupeptin, and 0.1 mM 7-amino-1-chloro-3-tosylamido-2-heptanone; ref. 28). These samples were precleared by mixing with 20 μl of 50% suspension of protein G-Sepharose and gently rotating for 1 h at 4°C. After centrifugation at 20,000 × g for 10 min, EP procyclins were immunoprecipitated from the supernatants by incubating with 10 μg of anti-EP procyclins monoclonal antibody (Cedarlane Laboratories) or an isotype-matched control monoclonal antibody for 1 h at 4°C, followed by mixing with 20 μl of 50% suspension of protein G-Sepharose and gently rotating for 1 h at 4°C. EP procyclins bound to the Sepharose were analyzed by 10–20% gradient SDS/PAGE and autoradiography.
Infection of Tsetse Flies.
Wild-type and doubly TbGPI10-disrupted procyclics were mixed with washed horse red blood cells in SDM-79 medium with 10% (vol/vol) FCS. The final concentration of trypanosomes was 107 per ml. Approximately 120 flies were infected with each procyclic clone through an artificial membrane (29). At 14 and 24 days after infection, about 40% and 60% of the flies, respectively, were dissected.
Results and Discussion
Cloning of TbGPI10, a Gene Involved in GPI Synthesis.
We found in the GenBank database a T. brucei expressed-sequence tag (accession no. N99300) that is homologous to human PIG-B, a gene involved in transferring the third mannose to a GPI anchor precursor (14). Based on this sequence, we cloned a full-length cDNA from a T. brucei cDNA library and termed the gene TbGPI10 (GenBank accession no. AB033824; GPI10 is the PIG-B orthologue of S. cerevisiae; ref. 15). TbGPI10 encodes a protein of 558 amino acids, having 25% and 23% sequence identity to human PIG-B and S. cerevisiae Gpi10p, respectively. To test whether TbGPI10 acts in transferring the third mannose like PIG-B/Gpi10p, we transfected TbGPI10 expression plasmids into mouse T lymphoma cell line S1A-b, which is defective in mouse PIG-B (14). S1A-b cells did not express the GPI-anchored protein Thy-1 because of a defect in GPI biosynthesis (Fig. 1A left). After transient transfection of human PIG-B cDNA, 13% of cells recovered the surface expression of Thy-1 as expected (Fig. 1 center). Similarly, TbGPI10 restored the surface Thy-1 expression on 7% of the S1A-b cells (Fig. 1 right), indicating restoration of GPI biosynthesis.
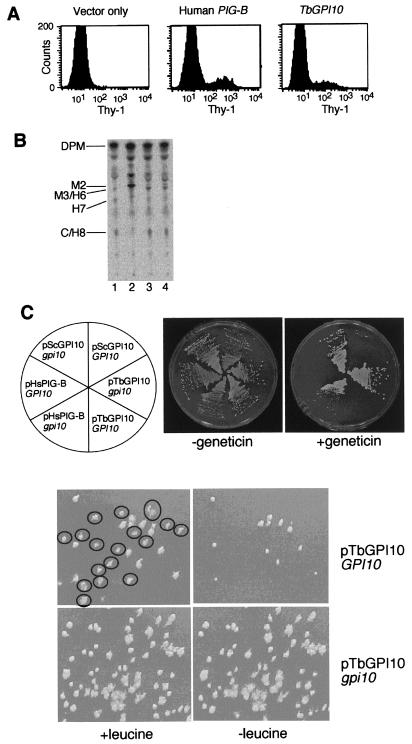
Figure 1.
TbGPI10 is a functional homologue of PIG-B/GPI10. (A) Restoration of the surface expression of GPI-anchored protein Thy-1 on mouse PIG-B-deficient S1A-b cells with TbGPI10. S1A-b cells transiently transfected with an empty vector; human PIG-B and TbGPI10 plasmids were stained for Thy-1 and analyzed in a flow cytometer. (B) Restoration of biosynthesis of the mature GPI-anchor precursors with TbGPI10 in S1A-b cells. Wild-type S1A cells (lane 1), PIG-B-deficient S1A-b cells (lane 2), and stable transfectants of S1A-b with TbGPI10 (lane 3) or human PIG-B (lane 4) were labeled with d-[3H]mannose in the presence of tunicamycin. Radiolabeled lipids were analyzed by TLC. DPM, dolichol phosphate mannose; M2 and M3/H6, intermediates with two and three mannoses, respectively; H7 and C/H8, complete GPI anchors. (C) Rescue of lethality of GPI10 knockout S. cerevisiae with TbGPI10. (Upper) Yeast multicopy vector (p425) carrying S. cerevisiae GPI10 (pScGPI10), TbGPI10 (pTbGPI10), and human PIG-B (pHsPIG-B) was introduced into wild-type (GPI10) and GPI10 knockout (gpi10) S. cerevisiae. The transformants were inoculated on plates as indicated. In the presence of geneticin, the rescued gpi10 strains, but not the GPI10 strains, can grow, because disruption of GPI10 was by replacement with kanamycin resistance gene. (Lower) Single colonies of GPI10 and gpi10 S. cerevisiae bearing pTbGPI10 were inoculated in 50 ml of SD medium in the presence of leucine (the selection marker for pTbGPI10), cultured to stationary phase, and then plated onto SD plates in the presence of leucine. The colonies were then replica plated onto SD plates with or without leucine. More than 50% of wild-type cells lost pTbGPI10 (no growth without leucine), whereas none of GPI10 knockout cells were able to lose the plasmid, confirming that GPI10 is essential for growth of S. cerevisiae. Colonies that grew on the nonselective plates but not on the selective plates were circled.
S1A-b cells accumulated a GPI intermediate M2 containing two mannoses (30) (Fig. 1B, compare lanes 1 and 2). TbGPI10, as well as human PIG-B, suppressed the accumulation of M2 and generated M3 (bearing three mannoses) and mature GPI, H7, and C/H8 (Fig. 1B, lanes 3 and 4).
GPI10 Is Essential for Growth of S. cerevisiae (15).
The inviability of GPI10.
Disrupted S. cerevisiae was rescued by TbGPI10 [Fig. 1C, middle right section in geneticin (−) plate] as well as by yeast GPI10 (upper left section) and human PIG-B (lower left section). These results indicate that TbGPI10 is a functional homologue of human PIG-B and yeast GPI10, involved in transfer of the third mannose to the GPI.
TbGPI10 Is Essential for Growth of Bloodstream Form of T. brucei.
A profile of Southern blot hybridization was consistent with TbGPI10 being a single copy gene, and hence a suitable target for a gene disruption experiment. To prepare knockout constructs, we cloned an 8-kilobase EcoRI fragment containing TbGPI10 from T. brucei genomic DNA (Fig. 2A). Because T. brucei is a diploid organism and TbGPI10 was an intronless gene, the entire TbGPI10 coding region was replaced with a HYG or a NEO resistance gene for two targetings by homologous recombination. We transfected bloodstream form parasites with an SphI–EcoRI fragment of one of the TbGPI10 knockout constructs (Fig. 2A), established drug-resistant clones, and assessed homologous recombination events by Southern blot hybridization (Fig. 2B). One of the two TbGPI10 alleles was disrupted by homologous recombination with a knockout construct bearing either NEO or HYG (Fig. 2B, lanes 12 and 13), leaving one intact allele. When the second knockout event was attempted, we obtained clones resistant to both hygromycin and geneticin; however, all of them still retained an intact allele in addition to two disrupted alleles (Fig. 2B, lanes 1–10). This result suggested that chromosomal TbGPI10 was amplified in some cells and that only those cells could form clones because of the essentiality of TbGPI10.
To confirm that TbGPI10 is essential for growth of the bloodstream form of trypanosomes, we introduced episomal plasmids carrying TbGPI10 and the BLE resistance gene into single knockout clones and then disrupted the second allele. By Southern blotting analysis of clones resistant to the three drugs, we confirmed that the two chromosomal TbGPI10 alleles were replaced with drug-resistance genes successfully (Fig. 2B, lanes 14–17). Disruption of TbGPI10 from both alleles in NBH-R clones was confirmed by PCR by using primers for TbGPI10 flanking region (data not shown). When we used other single knockout clones generated by using either the neomycin or hygromycin resistance gene, the results were essentially similar. This result indicates that TbGPI10 double knockout clones can be obtained only when an episomal copy of the gene is present. Next, we tested whether the episomal TbGPI10 plasmids can be lost in the absence of chromosomal TbGPI10. We cultured single and double knockout clones bearing episomal TbGPI10 plasmids without phleomycin selection, prepared DNA samples periodically, and analyzed them by Southern blot hybridization. When chromosomal TbGPI10 was present, the episomal plasmid disappeared after four passages (Fig. 2C). In contrast, when chromosomal TbGPI10 was disrupted, the episomal plasmid was maintained stably without drug selection for at least 10 passages (data shown only for 5 passages). These results indicate that biosynthesis of GPI anchors is essential for viability of the bloodstream form of T. brucei. GPI could be essential, because it is required for the surface expression of GPI-anchored proteins. In the absence of the variant surface glycoprotein coat, the cell might be too fragile to grow. Additionally or alternatively, a lack of transferrin receptors (31) might result in a fatal iron deficiency. The possibility that a GPI intermediate or intermediates, accumulated because of a lack of transfer of the third mannose, might be toxic is not excluded.
TbGPI10 Is Not Essential but Important for Growth of Procyclic Form of T. brucei.
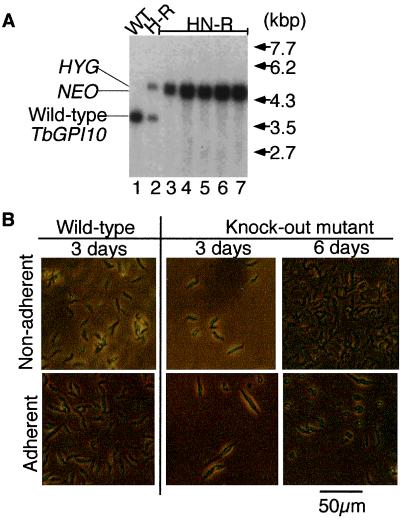
In contrast to results with the bloodstream form of the parasite, we could find conditions for the procyclic form where TbGPI10 could be deleted (Fig. 3A, lanes 3–7). At first, we were not successful in disruption of the second TbGPI10 allele. Then, we found that a double disruption was possible if the procyclic parasites were kept in flasks treated for nonadherent cultures (for example, Sumilon MS-2005R, Tokyo, Japan or Falcon 3009). As shown in Fig. 3B, TbGPI10-disrupted mutant parasites grown in a flask treated for nonadherent cultures had a shape similar to that wild-type cells, although their growth rate was only one-half that of wild-type cells (doubling time 30.1 h compared with 15.8 h; Fig. 3 B Upper). When the mutant procyclic cells were cultured in a normal culture flask, they adhered to the plastic surface, became elongated, and died within several days (Fig. 3 B Lower). It is likely that TbGPI10-disrupted procyclic cells had an abnormally adhesive surface because of a lack of procyclins (see below).
Figure 3.
Disruption of TbGPI10 in the procyclic form of T. brucei. (A) Southern blot analysis of homologous recombination. Samples of SalI and XhoI cut DNA of wild-type strain 427 (WT, lane 1), single TbGPI10 knockout clone (H-R, lane 2), and five double knockout clones derived from H-R (HN-R, lanes 3–7) were hybridized with a Sal–Sph probe. Expected positions of wild-type and homologous recombinant fragments are indicated on the left, and size markers are on the right. (B) Microscopic observation of the TbGPI10 knockout procyclics in culture. Wild-type and the knockout mutant procyclics were inoculated into flasks treated for nonadherent culture (Sumilon, Tokyo, MS-2005R; Upper; nonadherent) and regular nontreated flasks for adherent culture (Iwaki, Chiba, Japan, 3100-025; Lower; adherent) at a concentration of 105 per ml. On days 3 and 6 of culture at 27°C, procyclics were observed under a phase contrast microscope.
Disruption of TbGPI10 should result in a loss of complete GPI anchors, and this loss should result in a lack of GPI attachment to procyclins and their defective surface expression. To test this prediction, GPI biosynthesis was analyzed by labeling cell lysates with GDP-[3H]mannose and separating mannolipids by TLC (22). The mannolipids were characterized by treatments with Jack bean α-mannosidase, GPI-PLD (Fig. 4A), phospholipase A2, and phosphatidylinositol-specific phospholipase C (data not shown). Four major mannolipids, M3(acyl), PP3, PP1, and A′-like, were generated by lysates of wild-type cells as reported (Fig. 4A, lane 1; refs. 3, 32, and 33). M3(acyl) is an intermediate bearing acylated inositol and three mannoses, but its third mannose is not yet linked to ethanolaminephosphate. A′-like and PP3 are intermediates close to the complete GPI anchor, PP1, which is added to proteins posttranslationally. The latter three mannolipids have ethanolaminephosphate linked to the third mannose. Consistent with these structures, PP3, PP1, and A′-like, but not M3(acyl), were resistant to α-mannosidase (Fig. 4A, lane 2), whereas all of them were sensitive to GPI-PLD (Fig. 4A, lane 3). When TbGPI10 was disrupted, the profile of mannolipids was very different, showing accumulation of several other mannolipids (Fig. 4A, lane 4) all of which were sensitive to α-mannosidase (Fig. 4A, lane 5) and GPI-PLD (Fig. 4A, lane 6). Their identities were confirmed by treatments with phospholipase A2 and phosphatidylinositol-specific phospholipase C (data not shown). Thus, intermediates with a terminal ethanolaminephosphate were not observed, consistent with blocking the transfer of the third mannose. Transfection of TbGPI10 returned GPI biosynthesis to normal (Fig. 4A, lanes 7–9).
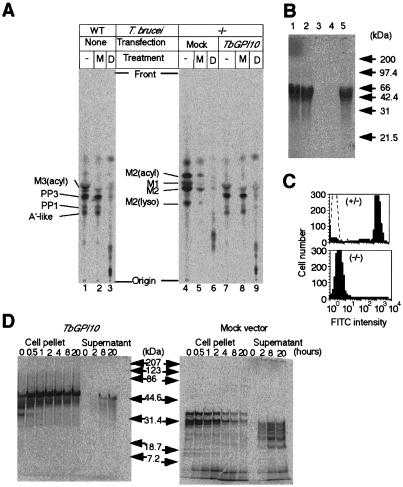
Figure 4.
Defective GPI biosynthesis, GPI anchoring, and surface expression of procyclins in TbGPI10 knockout procyclics. (A) GPI biosynthesis. Wild-type (WT) and doubly disrupted mutant (−/−), which were transformed with an empty vector (Mock) or TbGPI10 plasmid (TbGPI10), were used. The membranes were incubated with GDP-[3H]mannose to label GPI, and aliquots were subjected to TLC directly (−) or after digestion with α-mannosidase (M) or GPI-PLD (D). Identities of mannolipids are shown on the left of chromatograms. Designations of mannolipids from TbGPI10-disruptant are tentative. M1 and M2, intermediates containing one and two mannoses; M2(acyl) and M2(lyso), M2 species with acylation on inositol and with a lack of sn-2 fatty acid; M3(acyl), an intermediate bearing three mannoses with acylation on inositol; A′-like, an intermediate bearing three mannoses with ethanolamine phosphate on the third mannose; PP3, A′-like intermediate with acylation on inositol; PP1, complete GPI precursor (a lyso form of PP3). The spots that appeared after GPI-PLD-treatments (lanes 3, 6, and 9) are inositol-acylated GPI glycans. (B) Incorporation of myristic acid into procyclins. Lane 1, wild-type; lane 2, single TbGPI10-disruptant; lane 3, double TbGPI10-disruptant; lane 4, double TbGPI10-disruptant bearing an empty plasmid; lane 5, double TbGPI10-disruptant bearing TbGPI10 plasmid. Size markers are on the right. (C) Surface expression of EP procyclins. Single and double TbGPI10-disruptant clones were stained with anti-EP procyclins (shaded lines) or control (dotted line) monoclonal antibodies and analyzed in a FACScan. (D) Pulse-chase analysis of EP procyclins. Double TbGPI10-disrupted mutant bearing TbGPI10 (Left) or empty (Right) plasmid was pulse-labeled with [14C]proline for 30 min and chased for indicated time periods. At each time point, aliquots of samples were separated into supernatants and cell pellets, solubilized by detergent, and immunoprecipitated with anti-EP procyclins antibody. Immunoprecipitates were analyzed by SDS/PAGE and autoradiography.
We then tested whether GPI anchor attachment and surface expression of procyclins were defective in mutant procyclics (Fig. 4 B and C). We labeled the cells with [3H]myristic acid, which is known to be incorporated into the GPI anchor (31, 33). Radiolabeled procyclic cells were delipidated, and procyclins were extracted from the insoluble residues with 9% (vol/vol) butan-1-ol (27). The extracts were analyzed by SDS/PAGE and autoradiography (Fig. 4B). Wild-type and single TbGPI10 disrupted cells incorporated [3H]myristic acid into a 45- to 50-kDa band of EP procyclins (refs. 5, 27, and 34; Fig. 4B, lanes 1 and 2). T. brucei procyclic cells can express two isoforms of procyclins, EP procyclins bearing tandem repeat units of glutamic acid and proline (EP) and GPEET procyclins bearing internal pentapeptide (GPEET) repeats (7, 35–37). It is known that ratios of these two kinds of procyclins are different among strains and even among clones in the same strain, and can vary with culture length (27, 34). Apparently, this clone of T. brucei procyclics expresses only EP procyclins. In the TbGPI10-disrupted clone, [3H]myristic acid was not incorporated into EP procyclins (Fig. 4B, lane 3). EP procyclins reincorporated [3H]myristic acid on transfection of TbGPI10 plasmids (Fig. 4B, lane 5) but not empty plasmids (Fig. 4B, lane 4). Therefore, the TbGPI10 knockout mutant did not attach GPI to procyclins.
To test for cell surface expression of EP procyclins, we used flow cytometric analysis (Fig. 4C). High level expression of EP procyclins was observed (Fig. 4C Upper) on the surface of a single disrupted clone, whereas no cell surface expression was observed on the double disruptant (Fig. 4C Lower). The results show that EP procyclins, and most likely all GPI-anchored proteins, are not expressed on the surface of TbGPI10-disrupted T. brucei because of a lack of GPI anchoring, suggesting that the procyclics do not have GPI-anchored receptors essential for growth.
To investigate the fate of non-GPI-anchored EP procyclins, mutant cells were transfected with TbGPI10 vectors or empty vectors and pulse-labeled with [14C]proline for 30 min and chased for various times. EP procyclins were immunoprecipitated from all extracts or medium and analyzed by SDS/PAGE (Fig. 4D). In TbGPI10-transfected (GPI-sufficient) cells, radiolabeled proline was incorporated into 35-, 40-, and 50-kDa EP procyclin polypeptides that were chased into a mature 50-kDa band that was stable for 20 h (Fig. 4D left). A small amount of the latter band was found in the medium. In GPI-deficient cells, the mature 50-kDa EP procyclins were not produced, but several smaller polypeptides were seen. On chase, their signals were reduced (Fig. 4D right). Several smaller polypeptides were found secreted into the culture medium, indicating that non-GPI-anchored EP procyclins were secreted into culture medium where they were found degraded. These low molecular mass bands are specific, because we did not detect any bands in samples immunoprecipitated with a control antibody in a similar way (data not shown). There is a report that procyclic form parasites could not lose all of the procyclins (26). This report is in contrast to the present result that procyclic cells lacking procyclins can grow if they are kept in flasks for nonadherent cultures. Most likely, regular flasks were used in the previous study (26), which did not allow depletion of all procyclins.
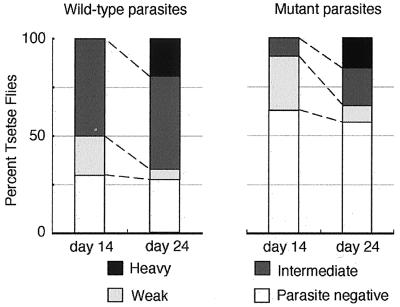
Next, we tested whether the lack of GPI-anchored proteins affects the ability of procyclics to infect tsetse flies. It is known that when procyclic form trypanosomes are mixed with red blood cells and fed to tsetse flies through an artificial membrane, they are capable of establishing an infection in the midgut with an efficiency similar to that of bloodstream forms (29). Thus, we fed tsetse flies with procyclic forms of wild-type and TbGPI10 knockout clones together with horse red blood cells. At 3 days after the feeding, we dissected three flies each and confirmed the presence of the comparable numbers of viable trypanosomes in their midguts. On days 14 and 24 after infection, we dissected about 60 flies each and examined the presence of parasites in the midguts (Fig. 5). On day 14, 50% of flies fed with wild-type procyclics had an intermediate infection (3–99 parasites per field). However, only 10% of flies fed with TbGPI10-disrupted procyclics had an intermediate infection, and the rest had none or less than three parasites per field. Clearly, TbGPI10-disrupted parasites had a lower ability to survive in the midgut during the first 2 weeks of infection. Similar results were obtained in two other experiments done in a similar way (data not shown). These results are consistent with a previous report that procyclics depleted of EP procyclins had much less ability in the infection to tsetse fly midgut (29).
Figure 5.
Effect of defective GPI anchor biosynthesis on the infection of procyclic T. brucei in tsetse fly midgut. Flies were infected with procyclic forms, thereafter fed three times per week in vitro with defibrinated horse blood, and dissected on days 14 and 24. Midguts were scored for degrees of infection as heavy (100–300 trypanosomes per field in 10 fields with the ×20 objective, black section), intermediate (between “heavy” and “weak”, dark gray section), weak (less than three trypanosomes, light gray section), and negative (no trypanosome detectable, white section).
A total fraction of infected flies (heavy, intermediate, and weak) stayed similar on days 14 and 24, in both groups (70% vs. 72% in wild-type and 37% vs. 43% in TbGPI10-disruptant). However, on day 24, extents of infection increased in both clones, suggesting that, once infection was established, procyclics can proliferate even in the absence of GPI-anchored proteins. The midgut is separated into two compartments by a peritrophic membrane (38). Trypanosomes first enter the endoperitrophic space surrounded by the peritrophic membrane where digestive enzymes are active. Then, they migrate into the ectoperitrophic space that is between the peritrophic membrane and gut epithelium. In this space, digestive enzymes are inactive, and the procyclics are thought to divide actively (38–40). The fact that TbGPI10-disrupted procyclics had a lower ability to establish infection during the initial stage suggests that they are more sensitive to digestive enzymes, consistent with the idea that GPI-anchored procyclins play a role in the resistance to digestion. The results also suggest that GPI-deficient procyclics can grow in the ectotrophic space. We did not follow the infected flies after day 24, because it is known that procyclics of strain 427 are not capable of differentiating into the metacyclic form that is infectious to mammalian hosts (29).
The experiments reported here demonstrate the importance of GPI anchors for T. brucei. In the bloodstream form, GPI is essential for growth under all conditions tested. The GPI-deficient procyclic form of T. brucei grows only under nonadherent culture conditions and at a slower rate. Furthermore, mutant procyclics were less competent for establishment of infection in the tsetse fly midgut. These results support the idea that a compound that would selectively inhibits GPI biosynthesis in T. brucei would be useful for control of African trypanosomiasis. The GPI biosynthesis pathways of T. brucei and mammalian cells are similar, but there are significant differences (41). Characterization of enzymes responsible for the different biosynthetic reactions is required for development of effective chemotherapy. A compound that inhibits GPI synthesis in mammalian cells and yeast but not in T. brucei has already been developed (42). It should, therefore, be possible to develop compounds that inhibit GPI synthesis in T. brucei but not in mammalian cells (43).
Acknowledgments
We thank P. Bütikofer, A. Acosta-Serrano, and Y. Morita for discussion, N. Inoue and J. Takeda for critically reading the manuscript, and K. Kinoshita and C. Kunz Renggli for excellent technical assistance. This work was supported by grants from the Human Frontier Science Program; the Ministry of Education, Sports, Science, and Culture; the Science and Technology Agency of Japan; and the University of Basel.
Abbreviations
- GPI
glycosylphosphatidylinositol
- HYG
hygromycin
- NEO
neomycin
- BLE
bleomycin
- H-R
hygromycin-resistant
- N-R
geneticin-resistant
- NH-R
geneticin- and hygromycin-resistant
- NBH-R
geneticin-, bleomycin, and hygromycin-resistant
- GPI-PLD
GPI-specific phospholipase D
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB033824).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180230697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180230697
References
- 1.Kuzoe F A. Acta Trop. 1993;54:153–162. doi: 10.1016/0001-706x(93)90089-t. [DOI] [PubMed] [Google Scholar]
- 2.Hursey B S, Slingenbergh J. World Anim Rev. 1995;3:67–73. [Google Scholar]
- 3.McConville M J, Ferguson M A J. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pays E, Nolan D P. Mol Biochem Parasitol. 1998;91:3–36. doi: 10.1016/s0166-6851(97)00183-7. [DOI] [PubMed] [Google Scholar]
- 5.Clayton C E, Mowatt M R. J Biol Chem. 1989;264:15088–15093. [PubMed] [Google Scholar]
- 6.Ferguson M A J, Murray P, Rutherford H, McConville M J. Biochem J. 1993;291:51–55. doi: 10.1042/bj2910051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mowatt M R, Clayton C E. Mol Cell Biol. 1987;7:2838–2844. doi: 10.1128/mcb.7.8.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross G A M. Annu Rev Immunol. 1990;8:83–110. doi: 10.1146/annurev.iy.08.040190.000503. [DOI] [PubMed] [Google Scholar]
- 9.Englund P T. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita T, Takeda J. Parasitol Today. 1994;10:139–143. doi: 10.1016/0169-4758(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 11.Doering T L, Raper J, Buxbaum L U, Adams S P, Gordon J I, Hart G W, Englund P T. Science. 1991;252:1851–1854. doi: 10.1126/science.1829548. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita T, Ohishi K, Takeda J. J Biochem (Tokyo) 1997;122:251–257. doi: 10.1093/oxfordjournals.jbchem.a021746. [DOI] [PubMed] [Google Scholar]
- 13.Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Lab Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- 14.Takahashi M, Inoue N, Ohishi K, Maeda Y, Nakamura N, Endo Y, Fujita T, Takeda J, Kinoshita T. EMBO J. 1996;15:4254–4261. [PMC free article] [PubMed] [Google Scholar]
- 15.Sütterlin C, Escribano M V, Gerold P, Maeda Y, Mazon M J, Kinoshita T, Schwarz R T, Riezman H. Biochem J. 1998;332:153–159. doi: 10.1042/bj3320153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nozaki T, Haynes P A, Cross G A M. Mol Biochem Parasitol. 1996;82:245–255. doi: 10.1016/0166-6851(96)02741-7. [DOI] [PubMed] [Google Scholar]
- 17.Patnaik P K, Kulkarni S K, Cross G A M. EMBO J. 1993;12:2529–2538. doi: 10.1002/j.1460-2075.1993.tb05908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferies D, Tebai P, Ray D L, Pays E. Nucleic Acids Res. 1994;21:191–195. doi: 10.1093/nar/21.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hug M, Carruthers V B, Hartmann C, Sherman D S, Cross G A M, Clayton C. Mol Biochem Parasitol. 1993;61:87–96. doi: 10.1016/0166-6851(93)90161-p. [DOI] [PubMed] [Google Scholar]
- 20.Patnaik P K, Fang X, Cross G A M. Nucleic Acids Res. 1994;22:4111–4118. doi: 10.1093/nar/22.20.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirumi H, Hirumi K. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 22.Masterson W J, Doering T L, Hart G W, Englund P T. Cell. 1989;56:793–800. doi: 10.1016/0092-8674(89)90684-3. [DOI] [PubMed] [Google Scholar]
- 23.Doering T L, Laurence J R, Buxbarum U, Hart G W, Englund P T. Methods Companion Methods Enzymol. 1990;1:288–296. [Google Scholar]
- 24.Güther M S, Masterson W J, Ferguson M A J. J Biol Chem. 1994;269:18694–18701. [PubMed] [Google Scholar]
- 25.Hirose S, Ravi L, Hazra S V, Medof M E. Proc Natl Acad Sci USA. 1991;88:3762–3766. doi: 10.1073/pnas.88.9.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brun R, Schönenberger M. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 27.Bütikofer P, Ruepp S, Boschung M, Roditi I. Biochem J. 1997;326:415–423. doi: 10.1042/bj3260415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bangs J D, Brouch E M, Ransom D M, Roggy J L. J Biol Chem. 1996;271:18387–18393. doi: 10.1074/jbc.271.31.18387. [DOI] [PubMed] [Google Scholar]
- 29.Ruepp S, Furger A, Kurath U, Renggli C K, Hemphill A, Brun R, Roditi I. J Cell Biol. 1997;137:1369–1379. doi: 10.1083/jcb.137.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puoti A, Desponds C, Fankhauser C, Conzelmann A. J Biol Chem. 1991;266:21051–21059. [PubMed] [Google Scholar]
- 31.Bitter W, Gerrits H, Kieft R, Borst P. Nature (London) 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 32.Field M C, Menon A K, Cross G A M. J Biol Chem. 1991;266:8392–8400. [PubMed] [Google Scholar]
- 33.Field M C, Menon A K, Cross G A M. J Biol Chem. 1992;267:5324–5329. [PubMed] [Google Scholar]
- 34.Treumann A, Zitzmann N, Hülsmeier A, Prescott A R, Almond A, Sheehan J, Ferguson M A J. J Mol Biol. 1997;269:529–547. doi: 10.1006/jmbi.1997.1066. [DOI] [PubMed] [Google Scholar]
- 35.Roditi I, Carrington M, Turner M. Nature (London) 1987;325:272–274. doi: 10.1038/325272a0. [DOI] [PubMed] [Google Scholar]
- 36.Mowatt M R, Clayton C E. Mol Cell Biol. 1988;8:4055–4062. doi: 10.1128/mcb.8.10.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mowatt M R, Wisdom G S, Clayton C E. Mol Cell Biol. 1989;9:1332–1335. doi: 10.1128/mcb.9.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller N, Lehane M J. Parasitol Today. 1993;9:45–50. doi: 10.1016/0169-4758(93)90030-j. [DOI] [PubMed] [Google Scholar]
- 39.Vickerman K. Br Med Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 40.Vickerman K, Teley L, Hendry K A K, Turner C M R. Biol Cell. 1988;64:109–119. doi: 10.1016/0248-4900(88)90070-6. [DOI] [PubMed] [Google Scholar]
- 41.Ferguson M A J. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- 42.Sütterlin C, Horvath A, Gerold P, Schwarz R T, Wang Y, Dreyfuss M, Riezman H. EMBO J. 1997;16:6374–6383. doi: 10.1093/emboj/16.21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith T K, Sharma D K, Crossman A, Brimacombe J S, Ferguson M A J. EMBO J. 1999;18:5922–5930. doi: 10.1093/emboj/18.21.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]