Abstract
Hypermethylated in cancer-1 (HIC1) is a tumor suppressor frequently targeted for promoter hypermethylation in medulloblastoma, an embryonal tumor of the cerebellum. Recently, we showed that HIC1 is a direct transcriptional repressor of ATOH1, a proneural transcription factor required for normal cerebellar development, as well as for medulloblastoma cell viability. Because demethylating agents can induce reexpression of silenced tumor suppressors, restoring HIC1 function may present an attractive therapeutic avenue in medulloblastoma by exploiting an apparent addiction to ATOH1.
Introduction
During cerebellar development, neuronal progenitors known as granule cell precursors (GCP) proliferate in response to activation of the Hedgehog (Hh) pathway, driven by Sonic Hedgehog (Shh) ligand secreted by Purkinje cells (1). This GCP population is a potential substrate for the development of medulloblastoma, a primitive neuroectodermal tumor primarily associated with childhood, although it is also believed that a subset of medulloblastomas may arise from the deep-seated ventricular zone of the posterior medullary vellum (2). Although the Hh pathway is critical for the normal expansion of the GCP population, <25% of medulloblastoma cases are associated with activating mutations in the Hh pathway (3). The imperative to better understand the pathogenesis of this tumor is driven by two important issues. First, medulloblastoma and its treatment can cause devastating effects on the lives of patients and families. Second, and of broader significance, is the idea that medulloblastoma represents one of the best models for understanding how developmental processes can promote and sustain the growth of tumors, and how such pathways might be targeted for therapeutic benefit. Studying both a mouse model and human tumor cell lines, we have recently identified how epigenetic gene silencing of an important tumor suppressor can deregulate developmental pathways critical to the genesis of this tumor.
Chromosome 17p Is Frequently Deleted in Medulloblastoma
Loss of chromosomal arm 17p occurs commonly in sporadic human medulloblastoma. However, mutations in p53, the best-characterized tumor suppressor located on 17p, are rarely detected (3). Analysis of a commonly deleted region associated with poor prognosis in medulloblastoma, 17p12-13.3, suggested that novel tumor suppressor genes may be present at this locus (4, 5). Two such genes have now been described as follows: KCTD11 (also known as RENKCTD11), located at 17p13.2, and hypermethylated in cancer 1 (HIC1), located at 17p13.3. KCTD11 functions as a hedgehog pathway antagonist; and its overexpression causes a decrease in medulloblastoma cell viability, making it a putative tumor suppressor in medulloblastoma (6). HIC1 is a zinc finger transcriptional repressor hypermethylated in a variety of pediatric and human tumors. Until recently, HIC1 had only one known transcriptional target, SIRT1, a class III histone deacetylase that functions to protect cellular longevity by partially inactivating p53 by deacetylation (7). Recent work by our group has since shown Hic1 to be a specific transcriptional repressor of Atoh1, a proneural transcription factor necessary for both tumor cell viability and for the formation of GCPs (8). Atoh1 up-regulation is commonly found in mouse models of medulloblastoma in which the tumors exhibit deregulated Shh signaling pathway expression (9–12). This relationship is maintained in a subset of human medulloblastomas in which tumors that express ATOH1 also express GLI1, but it is uncertain whether this is an exclusive relationship (13). Because Atoh1 is a likely target of Hh signaling, this finding provides a developmental framework for understanding how Hic1 might function as a tumor suppressor in medulloblastoma.
Characterization of HIC1 as a Tumor Suppressor Gene
HIC1 was first identified in a region of chromosome 17p frequently targeted for allelic loss and CpG island hypermethylation in cancer (14). The structure of the mouse and human loci consists of a large single coding exon, which translates an NH2-terminal BTB/POZ domain, and 5 Kruppel-like zinc fingers. There are at least three first exons, each driven by separate promoters of variable CpG island density (15). Aberrant CpG island methylation in these promoter regions has been described in a wide variety of human tumors (16–22).
Analysis of HIC1 and the 17p13.3 locus reveals that they are commonly methylated in medulloblastoma and do not seem to be limited to any specific clinical or pathologic subgroup (5, 23–25). Hypermethylation of the HIC1 gene in medulloblastoma also correlates with a reduction or absence of gene expression. Methylation-dependent reexpression of HIC1 in medulloblastoma cell lines after treatment with the DNA methyltransferase inhibitor, 5-aza-2′-deoxycytidine, provides further evidence for a causal role of hypermethylation in regulating expression (23, 25). The frequency of HIC1 epigenetic silencing in medulloblastoma makes it an attractive tumor suppressor candidate in medulloblastoma; and its potential role in regulating differentiation through transcriptional repression of ATOH1 provide a mechanistic link to better understand how HIC1 might function in this context.
Hic1-Mediated Suppression of Atoh1 in Development and Medulloblastoma
As GCPs mature, they descend from the outer surface of the cerebellum known as the external granule cell layer (EGL), and ultimately take up residence as mature postmitotic mature neurons in the internal granule cell layer (IGL; ref. 1). This period of neuronal development provides a highly informative model of neuronal differentiation based on the topography of the developing cerebellum. Using immunohistochemical assays, we showed that as GCPs descend, they express Hic1 in a regulated fashion, corresponding precisely to the developmental phase in which Atoh1 expression is lost (9). Because Atoh1 is critically linked to the specification and proliferation of GCPs (8), we investigated the idea that Hic1 plays a key role in suppressing Atoh1 expression in this progenitor pool.
The proneural bHLH transcription factor Atoh1 is the earliest marker of GCPs (8). Atoh1 is critical for proper cerebellar development as loss of the Atoh1 murine ortholog blocks the development and proliferative ability of the EGL, as well as the production of mature granule cells in the IGL (8, 26). Our data in cultured GCPs shows that Hic1-mediated repression of Atoh1 expression is dominant to the induction of Atoh1 by Shh. In other words, Hic1 can act downstream of Shh to repress Atoh1 expression; thus, its expression may serve to render GCPs insensitive to Shh, and therefore permit terminal differentiation despite exposure to high levels of Shh during their migration (Fig. 1). This may explain a long-standing paradox in cerebellar development: although GCPs require Purkinje cell–derived Shh for their growth, postmitotic GCPs are insensitive to Shh, although they are exposed to the highest levels of Shh as they migrate past the Purkinje cell layer to take residence in the IGL (27).
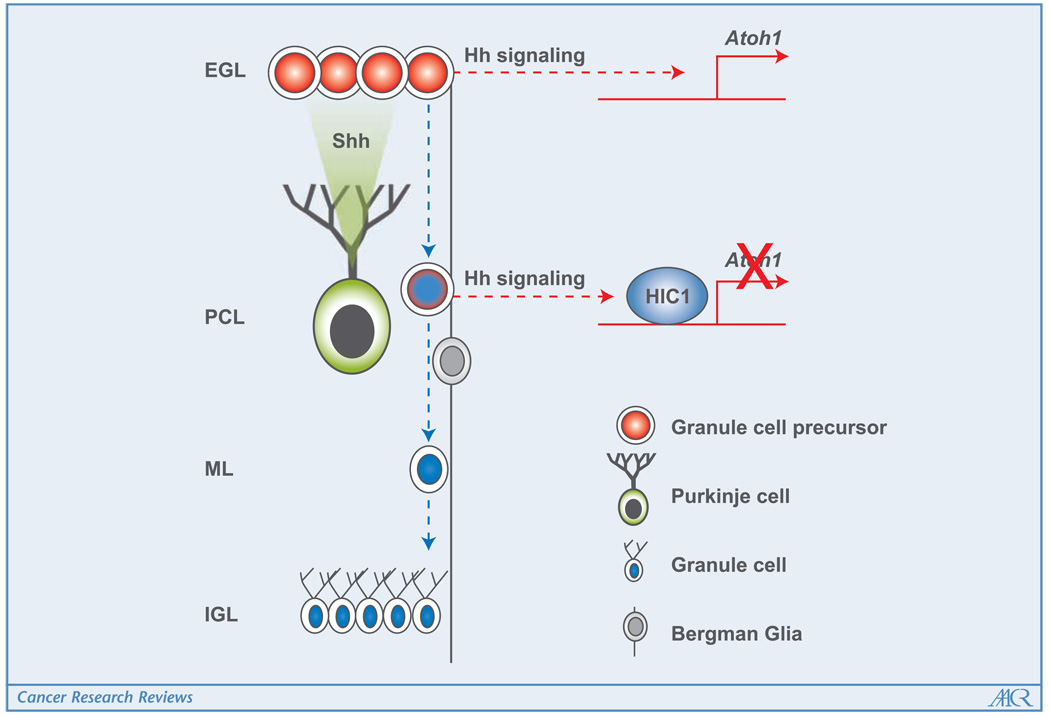
Figure 1.
Simplified representation of the developing cerebellum, showing the relationship between Atoh1 and Hic1 expression in the differentiation of granule neurons. GCPs in the EGL proliferate in response to Shh ligand produced byPurkinje cells (green) and express high levels of Atoh1 (red) driven byactivation of the Hh pathway. Hic1 (blue) is expressed bycells lining the inner EGL, and more strongly as the differentiating postmitotic granule cells migrate down the Bergmann glia through the molecular layer (ML) and Purkinje cell layer (PCL), to the IGL. Hic1 directlyrepresses transcription of Atoh1 in maturing granule cells downstream of the Hh pathway, making them unable to respond to high concentrations of Shh in the PCL.
A more rigorous test of this idea is a mouse model, in which we crossed Hic1+/− mice to animals heterozygous for a loss of function mutation in Patched homolog 1 (Ptch1; ref. 9). In keeping with the role of Ptch1 as a critical negative regulator of the Hh pathway, 10% to 15% of Ptch1+/− animals develop medulloblastoma (28). We showed that mice doubly heterozygous for mutations in Hic1 and Ptch1 have a marked increase in the incidence of medulloblastoma; and that in each case, the remaining wild-type Hic1 allele was silenced by dense promoter hypermethylation (9). As predicted by our model, Atoh1 expression is deregulated in these tumors. Additionally, we show that Atoh1 expression is required for medulloblastoma cell viability (9), results that were independently confirmed by the Dr. Martine F. Roussel team, St. Jude Children’s Research Hospital (29).
Available evidence suggests that ATOH1 may not be a classic oncogene, but that its overexpression and functional requirement in medulloblastoma is more consistent with the concept of “lineage addiction.” Here, cancer cells become addicted to the expression of transcription factors required for the development of an organ-specific progenitor (30). Typically, these genes cannot act as oncogenes but are required for the maintenance of a progenitor cell phenotype in cancer. This phenomenon is exemplified by MITF, a transcription factor expressed in, and required for, the maintenance of melanocytes and melanoma (31). ATOH1 is similar in that it directs cerebellar granule cell lineage survival during development and protects the proliferative potential and survival of medulloblastoma; thus, ATOH1 may function as a lineage-survival oncogene in medulloblastoma. Deregulation of ATOH1 expression through loss of HIC1 function may well promote medulloblastoma formation by preventing terminal differentiation and, thus, expand the pool of transformation-competent GCPs during development.
Epigenetic Therapy in Medulloblastoma?
Regardless of whether ATOH1 is a traditional oncogene or a lineage-survival oncogene, it is a potential therapeutic target due to its promotion of medulloblastoma viability. It is known that many pathways interact with Atoh1 during cerebellar development, including the hedgehog-signaling pathway and the Notch pathway (1, 32), suggesting that Atoh1 is tightly regulated both spatially and temporally. Although transcription factors are notoriously difficult targets for small molecule drug development, recent interest in epigenetic therapies that remove silencing marks such as DNA hypermethylation can be effective in restoring expression of genes such as HIC1 (23, 25). Atoh1 expression can be blocked by restoring Hic1 function, even when Atoh1 expression is stimulated by pathways including Hh signaling. Because pharmaceutical approaches to reverse DNA promoter methylation are now used clinically in the treatment of myelodysplastic syndrome (33, 34), it is possible that therapies targeting epigenetic events regulating HIC1 expression may be of use in medulloblastoma.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 2.Fan X, Eberhart CG. Medulloblastoma stem cells. J Clin Oncol. 2008;26:2821–2827. doi: 10.1200/JCO.2007.15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eberhart CG. Medulloblastoma in mice lacking p53 and PARP: all roads lead to Gli. Am J Pathol. 2003;162:7–10. doi: 10.1016/S0002-9440(10)63792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogen PH, Daneshvar L, Metzger AK, Duyk G, Edwards MS, Sheffield VC. Involvement of multiple hromosome 17p loci in medulloblastoma tumorigenesis. Am J Human Genet. 1992;50:584–589. [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbertson R, Wickramasinghe C, Hernan R, et al. Clinical and molecular stratification of disease risk in medulloblastoma. Br J Cancer. 2001;85:705–712. doi: 10.1054/bjoc.2001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Marcotullio L, Ferretti E, De Smaele E, et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci U S A. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Arie N, Bellen HJ, Armstrong DL, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 9.Briggs KJ, Corcoran-Schwartz IM, Zhang W, et al. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22:770–785. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JY, Nelson AL, Algon SA, et al. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev Biol. 2003;263:50–66. doi: 10.1016/s0012-1606(03)00434-2. [DOI] [PubMed] [Google Scholar]
- 11.Lin W, Kemper A, McCarthy KD, et al. Interferon-γ induced medulloblastoma in the developing cerebellum. J Neurosci. 2004;24:10074–10083. doi: 10.1523/JNEUROSCI.2604-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver TG, Read TA, Kessler JD, et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 13.Salsano E, Croci L, Maderna E, et al. Expression of the neurogenic basic helix-loop-helix transcription factor NEUROG1 identifies a subgroup of medulloblastomas not expressing ATOH1. Neuro-Oncol. 2007;9:298–307. doi: 10.1215/15228517-2007-014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wales MM, Biel MA, el Deiry W, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 15.Pinte S, Guerardel C, Deltour-Balerdi S, Godwin AK, Leprince D. Identification of a second G-C-rich promoter conserved in the human, murine and rat tumor suppressor genes HIC1. Oncogene. 2004;23:4023–4031. doi: 10.1038/sj.onc.1207504. [DOI] [PubMed] [Google Scholar]
- 16.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 17.Fujii H, Biel MA, Zhou W, Weitzman SA, Baylin SB, Gabrielson E. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16:2159–2164. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- 18.Issa JP, Baylin SB, Herman JG. DNA methylation changes in hematologic malignancies: biologic and clinical implications. Leukemia. 1997;11 Suppl 1:S7–S11. [PubMed] [Google Scholar]
- 19.Kanai Y, Hui AM, Sun L, et al. DNA hypermethylation at the D17S5 locus and reduced HIC-1 mRNA expression are associated with hepatocarcinogenesis. Hepatology. 1999;29:703–709. doi: 10.1002/hep.510290338. [DOI] [PubMed] [Google Scholar]
- 20.Kanai Y, Ushijima S, Ochiai A, Eguchi K, Hui A, Hirohashi S. DNA hypermethylation at the D17S5 locus is associated with gastric carcinogenesis. Cancer Lett. 1998;122:135–141. doi: 10.1016/s0304-3835(97)00380-7. [DOI] [PubMed] [Google Scholar]
- 21.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–3740. [PubMed] [Google Scholar]
- 22.Strathdee G, Appleton K, Illand M, et al. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158:1121–1127. doi: 10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsey JC, Lusher ME, Anderton JA, et al. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 24.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- 25.Waha A, Koch A, Meyer-Puttlitz B, et al. Epigenetic silencing of the HIC-1 gene in human medulloblastomas. J Neuropathol Exp Neurol. 2003;62:1192–1201. doi: 10.1093/jnen/62.11.1192. [DOI] [PubMed] [Google Scholar]
- 26.Jensen P, Smeyne R, Goldwitz D. Analysis of cerebellar development in math1 null embryos and chimeras. J Neurosci. 2004;24:2202–2211. doi: 10.1523/JNEUROSCI.3427-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahmane N, Ruiz-i-Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 28.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 31.Garraway LA, Weir BA, Zhao X, et al. "Lineage addiction" in human cancer: lessons from integrated genomics. Cold Spring Harb Symp Quant Biol. 2005;70:25–34. doi: 10.1101/sqb.2005.70.016. [DOI] [PubMed] [Google Scholar]
- 32.Gazit R, Krizhanovsky V, Ben-Arie N. Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway. Development. 2004;131:903–913. doi: 10.1242/dev.00982. [DOI] [PubMed] [Google Scholar]
- 33.Lubbert M, Wijermans P, Kunzmann R, et al. Cytogenetic responses in high risk myelodysplastic syndrome following low-dose treatment with the DNA methylation inhibitor 5-aza-2′-deoxyazacytidine. Br J Haematol. 2001;114:349–357. doi: 10.1046/j.1365-2141.2001.02933.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuendgen A, Graf T, Zohren F, et al. Induction of complete remission in a patient with acute myeloid leukemia refractory to high dose chemotherapy through treatment with 5-azacytidine. Leuk Res. 2007;31:407–409. doi: 10.1016/j.leukres.2006.06.017. [DOI] [PubMed] [Google Scholar]