Abstract
The assembly of individual mammalian proteasome subunits into catalytically active 20S proteasome is not well understood. Herein, we report the identification and characterization of human and mouse homologues of the yeast proteasome maturating factor Ump1p. We delineate the region of hUMP1 implicated in the specific interaction with proteasome precursors and show that hUMP1 protein is absent from the mature form of the 20S proteasome. We also show that the transcript level of mammalian UMP1 is increased after IFN-γ treatment and that mammalian UMP1 is functionally related to but not interchangeable with its yeast homologue.
The proteasome is one of the major nonlysosomal proteases present in the cytosol and nucleus of cells. Through its catabolic functions, it is implicated in many cellular processes including the progression of cell cycle, the removal of misfolded proteins, apoptosis, and the production of peptides for presentation by MHC class I molecules (1–3). The proteasome is composed of a 20S barrel-shaped core particle, capped on each side by a 19S protein complex. Whereas the 20S proteasome can degrade short peptides, its association with the 19S caps forms the 26S proteasome, which is able to degrade multiubiquitylated protein substrates (4).
The basic three-dimensional architecture of the 20S proteasome has been maintained from archaebacteria to humans and is characterized by four heptameric rings (5). The two outer rings are identical and are composed of α-subunits. The two inner rings, within which the catalytic centers are located, are also identical and are composed of β-subunits. Whereas the α- and β-rings of the archaebacterial 20S proteasome are formed of 7 identical α-subunits and 7 identical β-subunits, the more complex eukaryotic proteasomes contain at least 14 subunits, 7 distinct α-subunits, and 7 distinct β-subunits (6). In mammals, the existence of three additional β-subunits, which are expressed and incorporated into the 20S proteasome after IFN-γ treatment, further increases the complexity of the proteasome composition. Contrary to the archaebacterial proteasome, in which all seven β-subunits are catalytically active, only three of the seven different β-subunits (β1, β2, and β5) of the eukaryotic standard proteasome possess catalytic properties. In cells exposed to IFN-γ, these three catalytic subunits are replaced by three distinct ones (β1i/LMP2, β2i/MECL-1, and β5i/LMP7) resulting in the formation of a 20S proteasome termed immunoproteasome. The different cleavage specificity of the immunoproteasome compared with the standard proteasome has been shown to modulate the production of certain antigenic peptides presented by MHC class I molecules (2, 7).
A host of sequential reactions is required to reach the complex quaternary structure of eukaryotic proteasomes. The active β-subunits carry N-terminal propeptides, which have been shown to play an important role, not only in the assembly and maturation of the 20S proteasome (8), but also to protect the N-terminal catalytic Thr of the mature form from inactivation by Nα-acetylation (9, 10). In humans, the smallest precursor isolated is the 13S form, which contains three unprocessed subunits of the β-ring associated with a complete single α-ring (11, 12). Incorporation of the remaining four β-subunits leads to the formation of a 15S–16S hemiproteasome intermediate, composed of one α-ring and one β-ring. This form is proteolytically inactive. Formation of a proteolytically competent 20S proteasome requires the assembly of two hemiproteasomes and the autocatalytic removal of the propeptides from the catalytic β-subunits (13). A protein chaperoning the assembly of two hemiproteasomes has been identified recently in the yeast Saccharomyces cerevisiae (14). This protein, termed Ump1p (ubiquitin-mediated proteolysis), is associated with the 15S–16S intermediate and coordinates the maturation of a functional 20S complex. On completion of the assembly process, Ump1p remains trapped within the proteasome particle and is degraded. In humans, the cytosolic chaperone Hsc73 has been shown to interact specifically with purified 15S–16S intermediates but does not by itself promote the formation of a 20S particle (11).
In the present work, we have identified human and mouse homologues of the yeast Ump1p protein, termed hUMP1 and mUMP1, respectively. We show that hUMP1 specifically interacts with proteasome precursor complexes and is absent from active 20S proteasome. We also identified the region of hUMP1 that mediates its interaction with proteasome precursors, and found that the treatment of mammalian cells with IFN-γ increases the level of the UMP1 transcript.
Materials and Methods
Isolation and Cloning of hUMP1.
The sequence of hUMP1 was identified by performing a blast search of the SwissProt database with the yeast protein sequence as a query. Further blast searches identified several mouse expressed sequence tags. Based on this sequence information, hUMP1 was PCR amplified from a human melanoma cDNA derived from cell line MZ2-MEL 3.0, introducing a HindIII restriction site at the 5′ end and, at the 3′ end, an XbaI site and an influenza hemagglutinin epitope (ha) recognized by a monoclonal antibody (Babco, Emeryville, CA). Versions of hUMP1 carrying internal deletions were obtained with additional internal primers by using assembly PCR (details are available on request). The HindIII–XbaI fragments encoding hUMP1 or its derivatives were inserted into the pRc/CMV vector (Invitrogen) for expression in cell culture or into pBluescript II SK (+) (Stratagene) for in vitro transcription and translation.
Cell Culture and Transfection.
293T cells, human embryonic kidney cells transformed with simian virus 40 large T antigen, were cultured in DMEM supplemented with 10% (vol/vol) heat-inactivated FCS, 1% Hepes, and 1% streptomycin-penicillin. The plasmid pRc/hUMP1-ha was transiently transfected into 2.5 × 106 293T cells by using Lipofectamine (GIBCO) and the manufacturer-supplied protocol.
Metabolic Labeling, Immunoprecipitation, and Electrophoresis.
After transfection, cells were labeled and lysed as described (15). The volumes of supernatants were adjusted to contain equal amounts of 10% (vol/vol) trichloroacetic acid-insoluble 35S, followed by immunoprecipitation with a mixture of saturating amounts of antibodies, with either a monoclonal antibody against the ha epitope or a polyclonal rabbit anti-proteasome antibody and 20 μl of protein G Sepharose (Pierce) for 30 min at 4°C. The polyclonal anti-proteasome antibody was obtained by immunizing rabbit with purified human proteasome. The immunoprecipitated material was then washed three times in lysis buffer with 0.1% SDS and resuspended in SDS sample buffer (15). Samples were boiled and loaded on SDS/12% PAGE.
In Vitro Transcription and Translation.
[35S]Methionine-labeled hUMP1-ha was obtained by coupled T7 transcription/translation in reticulocyte lysates (TnT kit, Promega) according to the manufacturer's protocol. In short, 0.5 μg of DNA was used per 25 μl of transcription/translation reaction, which was incubated at 30°C between 2 h and 8 h. Reactions were immunoprecipitated as mentioned above by using a monoclonal anti-proteasome antibody MCP21 (European Collection of Animal Cell Culture, Salisbury, U.K.) or a monoclonal antibody against the ha epitope.
Sucrose Gradient Analysis and Peptidase Assays.
A 200-μl reaction was incubated at 30°C for in vitro transcription and translation of hUMP1 for 8 h to allow formation of proteasome and then layered on top of a 10–40% (vol/vol) linear sucrose gradient containing 20 mM Tris⋅HCl (pH 7.6) and centrifuged at 30,000 × g for 20 h at 4°C in a swing-out rotor. Fractions (350 μl) were collected and used for immunoprecipitations or peptidase assays. To determine the proteolytic activity of individual fractions, 45 μl of each fraction was incubated for 30 min at 37°C with 100 μM Bz-LLE-AMC (Bz, benzyloxycarbonyl; AMC, 7-amido-4-methyl-coumarin; Calbiochem) in a total volume of 100 μl. The reaction was then quenched with 100 μl of cold 100% (vol/vol) ethanol, and fluorescence was measured at 380/460 nm. The rest of each fraction was used for immunoprecipitation by using saturating amounts of the anti-proteasome antibody MCP21 and protein G Sepharose.
Northern Blot Analysis.
Total RNA was prepared from primary mouse (C57BL/6J) embryonic fibroblasts or human HeLa cells with or without prior treatment with 1,000 units/ml mouse or human IFN-γ for 24 h, separated in 1% agarose-formaldehyde gels, and blotted onto Hybond N nylon membranes (Amersham Pharmacia) as described (16). The blots were prehybridized for 3 h at 42°C, followed by an overnight incubation at 42°C in hybridization solution to which 32P-labeled probe generated by the random-priming method was added (17). The membrane was washed sequentially with 2× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA; SSPE) at room temperature, with 2× SSPE/2% (vol/vol) SDS at 65°C, and with 0.1× SSPE at room temperature. Hybridized labeled DNA was detected by autoradiography and quantified with the Fuji BAS-1500 Imaging System. Before reprobing, the blot was stripped by incubation (twice) for 15 min in 0.5% SDS heated to 100°C. Efficiency of stripping was verified by exposing the membrane to x-ray film for several days.
Expression in Yeast.
DNA fragments containing full-length or parts of mouse UMP1 were generated by PCR by using mouse cDNA as a template (16). The human UMP1 gene was amplified by PCR by using a human cDNA library (18), a gift from A. Varshavsky (California Institute of Technology, Pasadena). BglII and KpnI sites were introduced 5′ and 3′ of the ORF with the primers used in the PCR amplification. These sites were used for cloning the fragments into a high-copy 2-μ/LEU2-based yeast expression vector (pJDCEX2) between the copper-inducible PCUP1 promoter and a sequence encoding two copies of the ha epitope followed by the TCYC1 terminator sequences (14). Chimeric genes consisting of parts of yeast and mouse UMP1 were generated by assembly PCR (splicing by overlap extension; ref. 19). Four different chimeras were generated in which the heterologous parts were linked via conserved sequence stretches. Constructs I–III bear N-terminal parts of mouse UMP1 and C-terminal yeast Ump1p sequence. Construct I coded for a fusion protein with the sequence of mouse UMP1 extending to the conserved HPLE element (positions 46 to 49 in the mouse protein). Construct II contained a longer N-terminal sequence from mouse UMP1 extending to position 81, and construct III had mouse UMP1 sequence up to position 103. Construct IV was the reverse of construct I. Yeast strain JD59 (carrying a chromosomal deletion of UMP1), transformed with plasmids expressing human, mouse, or yeast UMP1 genes or with the chimeras of the latter two, were assayed for growth properties on SD media lacking leucin and containing 100 μM CuSO4 for promoter induction where indicated (14). All constructs were verified by sequencing, and their expression was monitored by anti-ha immunoblot analysis. For the yeast and mouse UMP1 genes, it was verified that otherwise identical constructs lacking the ha tag behaved like the ones containing it (data not shown). Coimmunoprecipitation of chimeric UMP1 proteins with proteasomal complexes from yeast extracts was carried out with polyclonal rabbit antiserum raised against yeast 20S proteasomes (a gift from K. Tanaka, Tokyo Metropolitan Institute of Medical Science, Tokyo) as described (14).
Results
Identification of hUMP1, a Human Homologue of the Yeast UMP1.
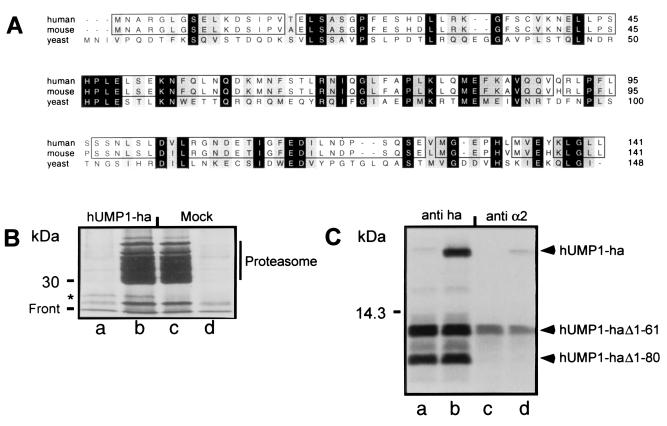
Using the available protein sequence of yeast Ump1p, we performed a blast search of the SwissProt database and identified a human protein sequence derived from a complete human cDNA (EMBL/GenBank/DDBJ accession no. AF125097). clustalw alignment of the yeast Ump1p and the newly identified protein of 141 amino acids revealed that the two sequences share a 22% identity and a 61% homology, making the human protein a likely homologue of the yeast Ump1p. We therefore named the hitherto unidentified protein hUMP1 (Fig. 1A). Further blast searches identified several mouse expressed sequence tags encoding a protein with 95% identity to hUMP1 (Fig. 1A; EMBL/GenBank/DDBJ accession no. C89498).
Figure 1.
Identification of human and mouse homologue of the yeast Ump1p protein and its interaction with mammalian proteasome. (A) CLUSTALW sequences of human and mouse protein are aligned with yeast Ump1p. Identical amino acids are labeled in black, and similar amino acids are labeled in gray. Identical amino acids between human and mouse are boxed. (B) 293T cells were transiently transfected with recombinant plasmid directing the synthesis of hUMP1-ha (lanes a and b) or mock transfected (lanes c and d) and labeled after 24 h with [35S]methionine/cysteine. Cells were then solubilized, and cleared lysate was immunoprecipitated either with a monoclonal antibody against the ha epitope (lanes a and d) or with a polyclonal antibody against the proteasome (lanes b and c). The position of hUMP1-ha is indicated by an asterisk. (C) Fragments generated from internal initiation sites were used to identify the region of hUMP1-ha important for interaction with the proteasome. The cDNA encoding the wild-type hUMP1-ha (lanes b and d) or a mutant lacking the first initiation codon (lanes a and c) was transcribed and translated in vitro. Samples were immunoprecipitated either with the anti-ha antibody (lanes a and b) or with MCP21 (lanes c and d). The positions of full-length and truncated hUMP1-ha are indicated.
To investigate whether the putative hUMP1 identified above interacts with the human proteasome, we isolated the cDNA from a human melanoma cell line and cloned it into a mammalian expression vector (see Materials and Methods for details). In the absence of available antibody, we tagged hUMP1 at the C terminus with an epitope derived from the influenza hemagglutinin, which is recognized by a specific antibody. The same strategy had yielded a functional yeast Ump1p in a previous study (14). 293T cells were either transfected with the plasmid encoding the modified hUMP1 (hUMP1-ha; Fig. 1B, lanes a and b) or mock transfected (Fig. 1B, lanes c and d). The cells were then metabolically labeled for 30 min, lysed, and processed for immunoprecipitation and SDS/PAGE analysis with either the anti-ha monoclonal antibody (lanes a and d) or the antiproteasomal subunit α2 monoclonal antibody MCP21 (Fig. 1B, lanes b and c). After immunoprecipitation with MCP21, we observed a band most likely corresponding to the endogenous 16.8-kDa hUMP1. In addition, a slightly slower migrating band corresponding to the recombinant hUMP1-ha was detectable in transfected cells, indicating that hUMP1-ha was interacting with human proteasome (Fig. 1B, lane b). Monoclonal antibody directed against the ha tag specifically recognized the recombinant hUMP1-ha in the same precipitates (Fig. 1B, lane a). As previously observed with the ha-tagged yeast Ump1p, immunoprecipitation with the anti-ha antibody did not coimmunoprecipitate proteasome subunits (14). The anti-ha antibody also recognized an unrelated protein comigrating, in mock-transfected cells, with the endogenous hUMP1 on SDS/12% PAGE (Fig. 1B, lane d). Because of our difficulties in overexpressing hUMP1 in stable transfectants (L.B. and F.L., unpublished result), we investigated the possibility of using an in vitro transcription/translation system to study the interaction between hUMP1-ha and proteasome precursors. We selected rabbit reticulocyte lysate as an experimental system, prompted by our findings that there is sufficient rabbit proteasome precursor in reticulocyte lysate and that several anti-human proteasome antibodies crossreact with rabbit proteasome.
A cDNA encoding hUMP1-ha was transcribed and translated in vitro in the presence of reticulocyte lysate and [35S]methionine. The interaction between newly translated 35S-labeled hUMP1-ha and the endogenous rabbit proteasome was detected by immunoprecipitation with the anti-proteasome antibody MCP21 (Fig. 1C). The presence of the C-terminal ha tag did not influence the interaction between hUMP1-ha and the proteasomal complexes, because untagged hUMP1 behaved identically (data not shown). As shown in Fig. 1C, a side reaction of the in vitro transcription/translation was the production of N-terminally truncated fragments that still contained an intact C terminus, because they could be immunoprecipitated efficiently with the anti-ha antibody (Fig. 1C, lanes a and b). To discriminate between the possibilities that these fragments may arise from internal initiation or proteolytic fragmentation of the full-length protein, we mutated the first initiation codon. After translating this construct, the band corresponding to the full-length protein was not detectable anymore, but we could still detect the two bands of lower apparent size, indicating that these fragments were generated by initiation of translation at Met-62 and Met-81 (Fig. 1C, lane a). Whereas full-length hUMP1-ha and the truncated form lacking the first 61 amino acids (hUMP1 Δ1–61) coimmunoprecipitated with rabbit proteasomal complexes (Fig. 1C, lanes c and d), the shorter version lacking the first 80 amino acids (hUMP1 Δ1–80) failed to do so. This result provided the first indications that the region between amino acids 62 and 80 was important for the specific interaction of hUMP1-ha with the proteasome particle.
Mammalian and Yeast UMP1 Homologues Are Functionally Related but Not Interchangeable.
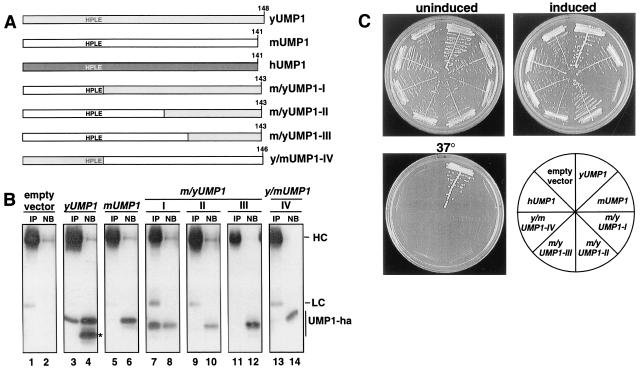
Mammalian UMP1 proteins share only 22% amino acid identity with yeast Ump1p (Fig. 1A). We therefore asked whether human and mouse UMP1 genes could functionally replace their homologue in S. cerevisiae. A yeast strain with a chromosomal deletion of UMP1 was transformed with plasmids expressing mammalian UMP1 genes. Transformants obtained with these plasmids were indistinguishable from those obtained with an empty control vector. Specifically, these transformants displayed the typical slow and temperature-sensitive growth phenotype of the ump1-Δ mutant independent of whether the expression was low (uninduced) or high (induced; Fig. 2C). We conclude that the deficiency of the yeast ump1-Δ strain could not be complemented, even at high expression levels, by the mammalian UMP1 genes. Because no growth inhibition was observed with high expression of these genes in S. cerevisiae, these results also indicated that mammalian UMP1 proteins neither interfere with the assembly of yeast proteasomes nor interact with high affinity with the heterologous proteasomal subunits. We went on to ask whether replacing parts of mouse UMP1 with the corresponding sequence from yeast UMP1 would result in functional chimeras (Fig. 2A). None of these constructs resulted in a complementation of the ump1-Δ growth defects. A chimeric UMP1 protein containing the N-terminal third of the mouse protein and the C-terminal two-thirds of yeast origin (m/yUMP1-I), however, resulted in a strong growth inhibition of yeast transformants with high, copper-induced expression (Fig. 2C). The two heterologous sequences were linked via a conserved HPLE motif, amino acids 46–49 of mUMP1 (Fig. 1A). The reverse construct (y/mUMP1-IV) as well as constructs m/yUMP1-II (linked between amino acids 81 of mUMP1 and 87 of yUMP1) and m/yUMP1-III (linked between amino acids 103 of mUMP1 and 109 of yUMP1) showed no such effect. These results indicated that, in a chimeric UMP1 protein, the presence of the C-terminal two-thirds of S. cerevisiae Ump1p, starting at the HPLE motif, is sufficient for interaction with yeast proteasome subunits. Association between the various UMP1 constructs and yeast proteasomal complexes was verified by coimmunoprecipitation with polyclonal antibodies raised against S. cerevisiae 20S proteasomes (Fig. 2B). In these experiments, yUmp1p and m/yUMP1-I coprecipitated with proteasomal complexes. In contrast, very little or no coprecipitation was observed for mUMP1 and the other chimeras analyzed. These findings are consistent with the growth phenotype of the various yeast transformants (Fig. 2C) as well as with the result that a truncated version of hUMP1 (Δ1–61) is capable of interacting with rabbit proteasomal complexes in vitro (Fig. 1C) and suggest that, in each of these species, the C-terminal domain is required for association of the respective UMP1 with precursor complexes. It is noteworthy that the faster migrating band detected in cells expressing yeast Ump1p could not be coimmunoprecipitated with the proteasome (Fig. 2B, lanes 3 and 4). This band most likely represents a product of Ump1p lacking the domain required for the interaction with proteasomes, similar to the observation made with N-terminally truncated hUMP1 obtained by in vitro translation (see above, Fig. 1C).
Figure 2.
Mammalian and yeast UMP1 proteins are not functionally interchangeable. (A) Schematic representation of constructs. (B) Coimmunoprecipitation assay to detect association of various Ump1-ha variants with proteasomal complexes. Proteins were precipitated from extracts of the same strains shown in C (except for those expressing hUMP1) with antibodies against yeast 20S proteasomes. The precipitates (IP) as well as 25% of the nonbinding material (NB) were analyzed by Western blotting with anti-ha antibody. The positions of IgG heavy chains (HC), light chains (LC), and various Ump1-ha proteins are indicated. The position of a product of yUMP1, missing the domain necessary for interaction with the proteasome, is marked with an asterisk. (C) Growth assays of yeast strain JD59 (ump1-Δ) transformed with high-copy plasmids expressing, from the copper-inducible PCUP1 promoter, human (h), mouse (m), or yeast (y) UMP1 genes or various chimeric genes of the latter two (m/yUMP1-I to m/yUMP1-III or y/mUMP1 depicted in A; see Materials and Methods and main text for details). Transformants were streaked onto selective minimal medium, with (induced) or without (uninduced) 100 μM CuSO4, and incubated at 30°C for 3 days. To assay for complementation of ump1-Δ temperature-sensitive growth phenotype, transformants were streaked onto complete medium and incubated at 37°C for 2 days.
Identification of Amino Acids Involved in hUMP1–Proteasome Interactions.
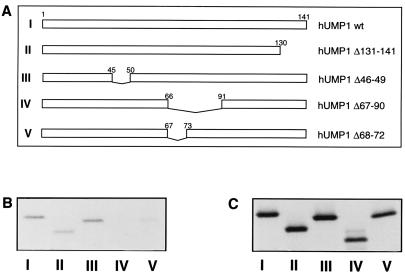
To determine further the region crucial for the physical interaction of hUMP1-ha with the proteasome, we constructed different deleted forms of hUMP1-ha (Fig. 3A) and tested them in the in vitro transcription/translation assay for association with endogenous proteasome (Fig. 3B). All constructs were expressed efficiently as observed by their detection after immunoprecipitation with the anti-ha antibody (Fig. 3C). Mutants of hUMP1-ha, lacking either the C-terminal 11 amino acids (construct II) or the highly conserved HPLE amino acid motif 46–49 (construct III) could still interact with the proteasome as efficiently as full-length hUMP1-ha (construct I). However, removal of a 24-amino acid-long stretch from position 67 to position 90 (construct IV) abolished its interaction with the proteasome, and the removal of only 5 amino acids from positions 68 to 72 (construct V) dramatically reduced its interaction. Thus, these results indicate that amino acids 68–72 play a role in the high-affinity association of hUMP1-ha to the proteasome, confirming the previous results obtained with the truncated forms of in vitro translated hUMP1-ha (Fig. 1C) and the yeast in vivo assays (Fig. 2).
Figure 3.
Delineation of the amino acids important for the interaction between hUMP1 and the proteasome. (A) Schematic description of the constructs used. Construct I represents the full-length hUMP1. Construct II depicts hUMP1 lacking the C-terminal 11 amino acids. Constructs III–V represent versions of hUMP1 lacking amino acids 46–49 (construct III), amino acids 67–90 (construct IV), or amino acids 68–72 (construct V). Constructs I–V were transcribed and translated in vitro and subjected to immunoprecipitation either by the monoclonal antibody against proteasome subunit α2 (B) or by the anti-ha antibody (C). Immunoprecipitated material was separated on SDS/15% PAGE. Constructs used are indicated at the bottom of each lane.
hUMP1 Is Absent from Mature 20S Proteasomes.
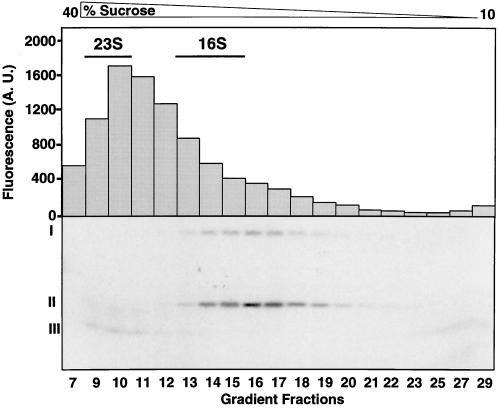
To identify the nature of the hUMP1-containing proteasome complexes, we performed sucrose gradient centrifugation experiments. Reticulocyte lysate containing in vitro translated 35S-labeled hUMP1-ha was layered onto a 10–40% (vol/vol) sucrose density gradient and centrifuged for 20 h. Fractions were then collected, and a peptidase assay and immunoprecipitation with antibody MCP21 were performed on each fraction. In parallel, ribosomal 23S and 16S RNAs were separated on an identical sucrose density gradient and served as sedimentation markers.
As observed in Fig. 4, one major peak of peptidase activity was observed in fractions 9–13, corresponding to the 20S–26S forms of the proteasome. This activity results from the peptidylglutamyl peptide hydrolyzing activity of the proteasome. Identical results were obtained by using substrates for other proteasomal activities (data not shown). Based on the sedimentation pattern of the ribosomal RNA, fractions 14–16 seem to contain catalytically inactive half proteasomes sedimenting at 15S–16S (20).
Figure 4.
hUMP1-ha is a component of proteasome precursor complexes and is absent from active 20S proteasome. In vitro translated 35S-labeled hUMP1-ha was layered onto a 10–40% (vol/vol) sucrose gradient and centrifuged at 30,000 × g for 20 h. Fractions (350 μl) were collected and assayed for proteasomal activity by incubating them with Bz-LLE-AMC and monitoring the increased fluorescence in arbitrary units (A.U.) emitted by the released fluorogenic group AMC. Each fraction was simultaneously immunoprecipitated with the polyclonal anti-proteasome antibody, followed by SDS/15% PAGE analysis and fluorography. The fractions corresponding to enriched 23S and 16S ribosomal RNA are indicated. Radioactive bands corresponding to the full-length hUMP1-ha (I) and hUMP1-ha Δ1–61 (II) identified in an earlier experiment (Fig. 1C) are indicated. The position of a degradation product of hUMP1-ha is labeled (III). See text for details.
Immunoprecipitation of the proteasomal complexes with a monoclonal anti-α2 antibody followed by separation by SDS/PAGE showed that hUMP1-ha was present in precursor complexes but absent from the 20S proteasomes. However, a faint band of low molecular mass could be reproducibly detected in fractions 9–12, enriched in 20S proteasome. This protein band most likely corresponds to a degradation product of hUMP1-ha, thus supporting the model proposed by Ramos et al. (14), in which the yeast Ump1p becomes trapped and degraded inside the 20S complex after assembly of two half proteasomes.
Mammalian UMP1 Transcript Levels Increase with IFN-γ Stimulation.
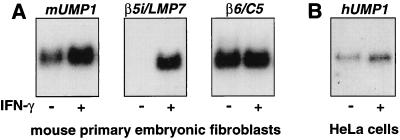
Having established that the mammalian UMP1 gene encodes a component of proteasome precursor complexes, and thus, in analogy to the yeast Ump1p, a proteasome maturation factor, we were interested in the regulation of these genes. In mammals, IFN-γ stimulation is known to result in the generation of a proteasome subtype termed immunoproteasome, which is distinguished from its standard counterpart by the incorporation of three IFN-γ-inducible β-type subunits (β1i/LMP2, β2i/MECL-1, and β5i/LMP7). Because the identified mammalian UMP1 proteins could be involved both in the maturation of standard and immunoproteasomes, we studied their response to IFN-γ stimulation. Quantification of the signals shown in Fig. 5 revealed that transcript levels of both mUMP1 and hUMP1 increased by a factor of two in response to IFN-γ stimulation (Fig. 5 A and B, respectively), whereas the transcript of β6, encoding a subunit of both types of proteasome, remained unchanged (Fig. 5A). In contrast, mouse LMP7 transcript level was detectable only after IFN-γ treatment. The moderate increase of the mammalian UMP1 transcript level in response to IFN-γ treatment would be consistent with a possible role of the encoded protein in the generation of both proteasome subtypes.
Figure 5.
Transcript levels of mouse and human UMP1 increase after IFN-γ stimulation. Northern blot analysis of UMP1 transcripts in total RNA from mouse embryonic stem cells (A) and human HeLa cells (B) treated with 1,000 units/ml IFN-γ for 24 h where indicated. Membranes were hybridized with radiolabeled probes, stripped, and rehybridized as described in Materials and Methods. Quantification of signals revealed a 2-fold induction of mUMP1 transcript and a 2.1-fold induction of hUMP1. Transcript levels of β5i/LMP7, an immunoproteasome-specific subunit, increased by a factor of 85, whereas those of β6, encoding a subunit present in both standard and immunoproteasome, remained unchanged after IFN-γ treatment.
Discussion
In this work, we have identified the human and mouse homologues of the yeast proteasome maturation factor UMP1 and shown that the human UMP1 protein interacts both with human and rabbit proteasome precursors. We also delineate the region of mammalian UMP1s implicated in the specific interaction with proteasome precursors and show that the mammalian UMP1s are functionally related but not interchangeable with their yeast homologue. As in S. cerevisiae, hUMP1 protein is absent from the 20S mature form of the proteasome. Finally, we show that the transcript level of mammalian UMP1 is increased after IFN-γ treatment.
Even though the function of yeast and mammalian UMP1 is likely to be very similar, expression of the latter in yeast cells did not lead to a complementation of the ump1Δ mutation, nor did the mammalian UMP1 associate with yeast proteasome precursors (Fig. 2). This result may be the consequence of the weak degree of identity (22%) between the primary structure of yeast and mammalian UMP1 (Fig. 1A). This low degree of identity is unexpected, because proteasome β-subunits display a higher degree of homology between yeast and humans. For instance, the catalytic β-subunits β1, β2, and β5 share, respectively, 46, 51, and 52% identity between yeast and humans. One explanation for this discrepancy may be the fact that, in addition to the high structural conservation of the 20S proteasome, each β-subunit interacts with at least five other subunits, whereas UMP1 is likely to have fewer interactions. Consequently, the sequence divergence will be much more constrained for the individual β-subunits than for UMP1 protein and is reflected in the relatively low degree of conservation between yeast and human UMP1. Alternatively, it is possible that UMP1 interacts with the propeptides of β-subunits (14), which are strikingly less well conserved than the mature parts of the β-subunits (21).
Based on the results of the yeast complementation assay and of the biochemical assays, we conclude that UMP1 contains at least two functionally distinct domains. The first one, which mediates the physical interaction between UMP1 and the proteasome precursor complexes, localizes to the C-terminal region encompassing residues 51 (in yeast) and 61 (in mammals) to 141. The sequence extending from amino acid residues 68–72 seems to be essential for this interaction. Removal of these five amino acids dramatically reduced the amount of hUMP1 associated with proteasome precursor complexes, suggesting that this region either directly contacts the proteasome or is necessary for the structural integrity of hUMP1. By using a prediction algorithm for secondary structure (22), this region is predicted to form an α-helix, which may constitute the structural feature responsible for the interaction with a particular proteasome β-subunit. A second domain that is required for the formation of a functional 20S proteasome particle from two hemiproteasomes is located N-terminally to the aforementioned domain. Indeed, exchange of the first 50 amino acids of yeast Ump1p by the mouse UMP1 protein still allowed binding of the hybrid UMP1 protein to proteasome precursor complexes but did not lead to the formation of functional 20S particles. This interpretation was suggested by a severe growth inhibition of yeast cells expressing this hybrid protein and was corroborated by coimmunoprecipitation experiments confirming its association with proteasomal complexes (Fig. 2). The functionally defined domains described above are likely to operate by interacting with distinct domains of the proteasome subunits. Whereas amino acids 68–72 of UMP1 might bind to the body of a proteasomal β-subunit, the N-terminal domain could be involved in an interaction with the prosequence of one or more β-subunits. In this model, the region encompassing residues 68–72 would mediate the incorporation of UMP1 into proteasome precursor complexes, whereas additional interactions between its N-terminal domain and β-subunit's propeptide(s) would be necessary for proteasome maturation. This interpretation is supported by the findings that the yeast β5 propeptide becomes dispensable in cells lacking Ump1p but is essential for viability in its presence (14) and by our recent findings that the in vitro binding of yUMP1 to certain β-subunits depends on the presence of their propeptides (J.H., M. London, and R.J.D., unpublished results).
We show that in both mouse and human cells, UMP1 is responsive to IFN-γ stimulation, resulting in a 2-fold increase in transcript levels. This result suggests that UMP1 proteins might be involved both in the maturation of standard proteasome and immunoproteasome. The level of mRNA coding for the subunit β6, present in both proteasome subtypes, remained unchanged, demonstrating that the IFN-γ treatment did not per se result in an induction of proteasomal subunit synthesis. A possible explanation for the elevated level of UMP1 transcript detected after IFN-γ treatment is that UMP1 might constitute, under normal conditions, a limiting factor in the assembly of proteasomes. Based on this assumption, we postulate that the IFN-γ treatment leads to a more rapid de novo assembly of immunoproteasome by increasing the level of UMP1 protein.
Acknowledgments
The authors thank N. Lévy and A.-L. Peitrequin for technical assistance; V. Jongeneel, N. Schnell, and Y. Strahm for help in database searches; S. Salvi for providing the melanoma cDNA; M. Schmidt for providing an LMP7 cDNA clone; K. Tanaka for providing an antiserum against yeast proteasomes; J.-C. Cerottini and G. Perrenoud for critical reading of the manuscript; and J. C. Howard, in whose laboratory U.B. and T.K. carried out their part of the study, for his support. This work was supported by funds from the Swiss Cancer League (to L.B. and F.L.), by a grant from the Cancer Research Institute (to F.L.), and by Deutsche Forschungsgemeinschaft Grant DO 649/1-1 (to R.J.D.).
Abbreviation
- ha
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190268597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190268597
References
- 1.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 2.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 3.Hilt W, Wolf D. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Baumeister W, Cejka Z, Kania M, Seemüller E. Biol Chem. 1997;378:121–130. doi: 10.1515/bchm.1997.378.3-4.121. [DOI] [PubMed] [Google Scholar]
- 6.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 7.Morel S, Lévy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin A-L, Monsarrat B, Van Velthoven R, Cerottini J-C, Boon T, et al. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 8.Chen P, Hochstrasser M. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 9.Arendt C S, Hochstrasser M. EMBO J. 1999;18:3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jager S, Groll M, Huber R, Wolf D, Heinemeyer W. J Mol Biol. 1999;291:997–1013. doi: 10.1006/jmbi.1999.2995. [DOI] [PubMed] [Google Scholar]
- 11.Schmidtke G, Schmidt M, Kloetzel P-M. J Mol Biol. 1997;268:95–106. doi: 10.1006/jmbi.1997.0947. [DOI] [PubMed] [Google Scholar]
- 12.Nandi D, Woodward E, Ginsburg D B, Monaco J J. EMBO J. 1997;16:5363–5375. doi: 10.1093/emboj/16.17.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nandi D, Jiang H, Monaco J J. J Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- 14.Ramos P C, Höckendorff J, Johnson E S, Varshavsky A, Dohmen R J. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 15.Valmori D, Gileadi U, Servis C, Dunbar P R, Cerottini J-C, Romero P, Cerundolo V, Lévy F. J Exp Med. 1999;189:895–905. doi: 10.1084/jem.189.6.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm U, Güthlein L, Klamp T, Ozbek K, Schaub A, Futterer A, Pfeffer K, Howard J C. J Immunol. 1998;161:6715–6723. [PubMed] [Google Scholar]
- 17.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 18.Colicelli J, Nicolette C, Birchmeier C, Rodgers L, Riggs M, Wigler M. Proc Natl Acad Sci USA. 1991;88:2913–2917. doi: 10.1073/pnas.88.7.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn B, Stearns T, Botstein D. Nature (London) 1993;362:563–565. doi: 10.1038/362563a0. [DOI] [PubMed] [Google Scholar]
- 20.Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel P-M. J Mol Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M, Zantopf D, Kraft R, Kostka S, Preissner R, Kloetzel P-M. J Mol Biol. 1999;288:117–128. doi: 10.1006/jmbi.1999.2660. [DOI] [PubMed] [Google Scholar]
- 22.Frishman D, Argos P. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]