Abstract
Purpose
We have noted that hypermethylation at GSTP1 in the preoperative serum of men with localized prostate cancer predicts early prostate specific antigen failure following surgical treatment. In this study we investigated the hypermethylation profile of several genes in the serum of men with localized and hormone refractory prostate cancer.
Materials and Methods
We assayed the serum of 192 men with clinically localized prostate cancer and 18 with hormone refractory metastatic disease. A total of 35 serum samples from patients with negative prostate biopsy served as a negative control. CpG Island hypermethylation status of certain genes was assessed, including MDR1, EDNRB, CD44, NEP, PTGS2, RASSF1A, RAR-β and ESR1. The results of hypermethylation at GSTP1 were included from a previous study.
Results
CpG island hypermethylation at MDR1 was positive in 38.2% of cases without PSA recurrence and in 16.1% of those with biochemical recurrence after radical prostatectomy. DNA hypermethylation at the remaining 7 gene loci was not detected in the serum of patients with localized prostate cancer. In serum from metastatic prostate cancer cases CpG island hypermethylation was detected at MDR1 in 15 (83.3%), EDNRB in 9 (50%), RAR-β in 7 (38.9%), GTSP1 in 5 (27.8%) and NEP or RASSF1A in 3 (16.7%). CpG island hypermethylation at CD44, PTGS2 or ESR was not detected in any samples. All histologically normal cases were negative for CpG island hypermethylation.
Conclusions
DNA hypermethylation at MDR1 was detected in cases of localized prostate cancer. CpG island hypermethylation at several gene loci was detected in men with advanced disease. No single gene was consistently observed to be hypermethylated in men with hormone refractory disease. These results suggest that the CpG island hypermethylation status of a defined panel of genes may be a useful biomarker in men with hormone refractory prostate cancer.
Keywords: prostate, prostatic neoplasms, tumor markers, biological, glutathione S-transferase pi, neoplasm recurrence, local
Prostate cancer remains the leading diagnosed solid organ malignancy in men in the United States and Western Europe. Primary therapy in the form of radical prostatectomy is one of the principal treatment modalities available for men with clinically localized disease. Approximately two-thirds of patients treated with surgery with curative intent remain disease-free 15 years following radical prostatectomy. Men who experience early PSA recurrence, particularly within the first 2 years after surgery, are likely to have metastatic lesions and they are prone to die of prostate cancer.
Clinically localized prostate cancer, which may be cured with radical prostatectomy, depends on androgens for growth and survival. However, certain prostate cancers become hormone refractory and these patients are likely to die of prostate cancer. How hormone independent prostate cancer arises is still a matter of debate and multiple theories exist.
DNA hypermethylation at various gene sites has been noted frequently in human cancers.1 Dinucleotides containing cytosine and guanine (CpG) in the DNA sequence are prone to DNA methyltransferases, which catalyze the transfer of a methyl group to the cytosine ring, yielding 5-methyl cytosine. Clusters of CpG dinucleotides, called CpG islands, are found in about 60% of all genes in the humane genome. Hypermethylation of promoter associated CpG islands is thought to inhibit transcriptional activity of the downstream gene, leading to a loss of the production of mRNA and the corresponding protein. CpG island hypermethylation in the regulatory region of the π-class GSTP1 gene has been described as the most prevalent somatic abnormality in prostate cancer.1 Somatic GSTP1 CpG island hypermethylation has been observed in greater than 90% of prostate cancer tissue but not in histologically normal prostate cancer tissue.1 Furthermore, multigene hypermethylation analysis has improved the sensitivity and specificity of epigenetic tests in the diagnosis and prognosis of prostate cancer.1,2
Recently analysis of the bodily fluids of patients with prostate cancer revealed CpG island hypermethylation as a novel biomarker for prostate cancer.3,4 Only a few groups have investigated CpG island hypermethylation in the serum or plasma of patients with prostate cancer.3,4 We examined GSTP1 hypermethylation in serum of men with localized and metastatic hormone refractory prostate cancer, and found that GSTP1 hypermethylation is the strongest independent predictor of early biochemical recurrence following surgical monotherapy for localized prostate cancer.5
Today no established pretreatment laboratory test exists to distinguish men with localized and metastatic hormone refractory prostate cancer. We examined a CpG island hypermethylation profile of multiple genes in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. All gene loci were chosen because they were shown to be of importance for determining the diagnosis or progression of prostate cancer in cancerous tissue samples in previous studies.1,2,6–8
MATERIALS AND METHODS
Sample Collection and DNA Isolation
All serum samples of 192 patients who underwent radical prostatectomy were collected before or at least 4 months following prostate biopsy and stored at −80C. All serum samples were stored on ice between the blood draw and further processing. Clotting of serum samples was allowed for 30 minutes on ice before centrifugation at 1,800 × gravity per minute for 10 minutes. The study cohort consisted of 192 men who underwent radical prostatectomy at The Johns Hopkins Hospital for localized prostate cancer (table 1). All patients were examined for CpG island hypermethylation at GSTP1 in a previous study and the results were included in the data analysis (table 2).5
Table 1.
Characteristics of 192 men with clinically localized prostate cancer treated with radical prostatectomy
| No. pts | 192 |
| Mean age (range) | 58.9(40–71) |
| Mean ng/ml, preop PSA (range) | 7.68 (0.9–38) |
| No. pathological Gleason score (%):* | |
| 5–6 | 89 (46) |
| 7 | 77 (40) |
| 8–10 | 26 (14) |
| No. pathological stage (%): | |
| Organ confined† | 104 (54) |
| Nonorgan confined | 87 (45) |
| Extraprostatic extension | 86 (45) |
| Seminal vesicle invasion | 18 (9) |
| Pos surgical margins | 37 (19) |
| No. pos pathological lymph node (%) | 7 (4) |
| Followup of disease-free men (yrs): | |
| Median | 3 |
| Mean (range) | 4 (1–14) |
| Yrs to PSA recurrence: | |
| Median | 2 |
| Mean (range) | 3 (1–7) |
Unknown in 1 patient.
Unknown in 2 patients.
Table 2.
CpG island hypermethylation frequency at GSTP1, RAR-β, MDR1 and EDNRB in serum samples of patients with prostate cancer at multiple disease stages
| No. Pts (% methylation) |
||||
|---|---|---|---|---|
| Disease Progression | GSTP1* | MDR1 | EDNRB | RAR-β |
| Neg biopsy | 35 (0.0) | 35 (0.0) | 35 (0.0) | 35 (0.0) |
| Prostate Ca: | ||||
| No recurrence | 136 (5.2) | 136 (38.2) | 136 (0.0) | 136 (0.0) |
| Recurrence | 56 (19.6) | 56 (16.1) | 56 (0.0) | 56 (0.0) |
| Metastasis | 18 (27.8) | 18 (88.9) | 18 (50.0) | 18 (38.9) |
| Chi-square (p <0.0001) | 18.6 | 15.4 | 29.7 | 23.4 |
Results included from a previous study.5
DNA was extracted from 600 μl archived serum from each patient using a Qiagen® Blood Mini Kit and diluted in a total volume of 25 μl. A total of 35 men without evidence of cancer on prostate biopsy served as a control population. In that group mean age was 60.1 years (range 40 to 79) and mean PSA was 2.3 ng/ml (range 0.1 to 4.0).
Biopsy was performed because of abnormal digital rectal examination. Histological examination revealed benign prostatic hyperplasia combined with prostate inflammation. Serum from 18 men with hormone refractory metastatic prostate cancer was also examined. These samples were collected before treatment with suramin in a clinical trial after established hormone refractory prostate cancer, which had been treated with different courses of hormonal ablation therapy. Unfortunately no information on the site of metastasis was available, eg bone or lymph nodes.
In men undergoing surgery for localized prostate cancer routine PSA followup consisted of assessment 3 months postoperatively and annually thereafter. All men studied had undetectable serum PSA 3 months after radical prostatectomy. All studies were performed with the approval of the institutional review board, and with Health Insurance Portability and Accountability Act compliance.
Quantitative Real-Time PCR
All samples from histologically benign prostatic tissue, clinically localized prostate cancer and hormone refractory metastatic prostate cancer were examined for CpG island hypermethylation at all 8 gene loci. We used a previously described quantitative real-time PCR method5 to detect GSTP1 CpG island hypermethylation. This method allows the quantification of the amount of total DNA and methylated DNA in each sample and it is able to detect as few as 1 methylated allele. Briefly, DNA extracted from serum samples was subjected to RE digestion with HpaII. HpaII cuts the sequence CCGG but does not cut the methylated form of this sequence, C5mCGG. If the CpG island does not contain C5mCGG, the DNA is cut and no product can be detected after PCR amplification.
Typically 4 μl DNA were incubated at 37C for 5 hours with 60 U RE, 2 μl 10 × RE buffer and the appropriate amount of deoxyribonuclease-free water to achieve a final reaction volume of 20 μl. To ensure complete digestion an additional 30 U RE, 1 μl RE buffer and 6 μl deoxyribonuclease-free water were added to each reaction and incubated at 37C overnight. As individual sample controls, PCR amplification was performed on undigested DNA and DNA digested with the RE MspI using the protocol described. MspI cuts the unmethylated and methylated sequences and, thus, no PCR product should be detected.
All PCR reactions were done in an iCycler® real-time thermal cycler using a specific primer sets that brackets at least 3 HpaII/MspI recognition sites (table 3).2 PCR conditions were set as 95C for 15 minutes, followed by 45 cycles of 94C for 30 seconds, 60C for 30 seconds and 72C for 45 seconds. Calibration curve analysis was performed after each PCR. PCR was accomplished using a 25 μl reaction mixture consisting of 4 μl template DNA, 100 nM forward and reverse primer, and 12.5 μl 2 × SYBR® Green PCR Master Mix. In each reaction set RE treated universally methylated DNA served as the positive control, and RE treated white blood cell DNA and a water blank served as negative controls. Table 4 lists primer sequences.
Table 3.
CpG island hypermethylation frequency in serum samples of patients with hormone refractory metastatic prostate cancer
| Gene Locus | No. Methylation (%) |
|---|---|
| Overall | 18 |
| MDR1 | 15 (83.3) |
| EDNRB | 9 (50) |
| RAR-β | 7 (38.9) |
| GSTP1* | 5 (27.8) |
| NEP | 3 (16.7) |
| RASSF1a | 3 (16.7) |
No ESR, CD44 or PTGS2 hypermethylation.
Results included from a previous study.5
Table 4.
Primer sequences used to amplify restriction enzyme treated DNA at various genes in real-time methylation specific PCR reactions
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GSTP15 | GACCTGGGAAAGAGGGAAAG | ACTCACTGGTGGCGAAGACT |
| MDR1 | AGCATCTCCACGAAGGCAGAGTT | AAACCGTAGTGGCACTGGACCAT |
| EDNRB | CGCGATTGAACTCGAAAGAC | ATTCTCGCAGCGTTTCGT |
| CD44 | GTGCCACCAAAACTTGTCCA | TCCAACATCCCTGTGAAACC |
| NEP | GACACATCCCGAAACATGAG | GGGGTCACGGTGAGTCTCT |
| PTGS2 | AGGAGGTCAGAGCGGAAACT | CTGGGTTTCCGATTTTCTCA |
| RASSF1a | CCTTCCTTCCCTCCTTCGT | ACCTCAAGATCACGGTCCAG |
| RAR-β | AATGCTTGCAAAAAGCCTTC | AGAAGTTGGTGCTCAACGTG |
| ESR1 | AGGGCAGAAGGCTCAGAAA | ATGAGCTCGGGAGACCAGTA |
Each reaction was performed in duplicate. The relative levels of methylated DNA in each sample from patients with metastasis were calculated and they are described as an NIM.1 The NIM scale from white—no methylation detected to black—greater than 99% of input DNA methylated was designed using Microsoft® Visual Basic.
Statistical Analysis
All samples tested for CpG island hypermethylation at MDR1, RAR-β and EDNRB in the disease progression groups were analyzed. The results of GSTP1 hypermethylation were included from our previous investigation.5 The percent of patients with DNA hypermethylation were compared in each group. Because the 4 groups represented worsening disease categories, the percent with DNA hypermethylation was evaluated for a linear trend, ie whether the percent that was methylated increased with each worsening category. They were compared using the Mantel-Haenszel chi-square test for linear trend. All statistical tests were done using SAS®, version 8.0 software.
RESULTS
CpG Island Hypermethylation at Various Gene Loci in Men Without Evidence of Prostate Cancer and in Men With Clinically Localized Prostate Cancer
All samples from men without evidence of prostate cancer in the prostate biopsy were negative for CpG island hypermethylation at all gene loci, including EDNRB, CD44, NEP, PTGS2, RASSF1A, RAR-β, MDR1 and ESR1. All samples from men without evidence of prostate cancer were negative for GSTP1 methylation in our previous study.5
CpG island hypermethylation at MDR1 was positive in 38.2% of cases (52 of 136) with localized prostate cancer but without PSA recurrence following surgical monotherapy and in 16.1% (9 of 56) with biochemical recurrence after radical prostatectomy. No CpG island hypermethylation at the remaining 7 gene loci (EDNRB, CD44, NEP, PTGS2, RASSF1A, RAR-β and ESR1) was detected in cases of localized prostate cancer with and without PSA recurrence, respectively, following surgical monotherapy.
Of the 192 men in our study 56 (29%) experienced PSA recurrence during the study period. Median time to PSA recurrence was 2 years (mean 4, range 1 to 14). Median followup in men who were free of disease at last followup was 3 years (mean 3, range 1 to 7).
CpG Island Hypermethylation in Men With Hormone Refractory Metastatic Prostate Cancer
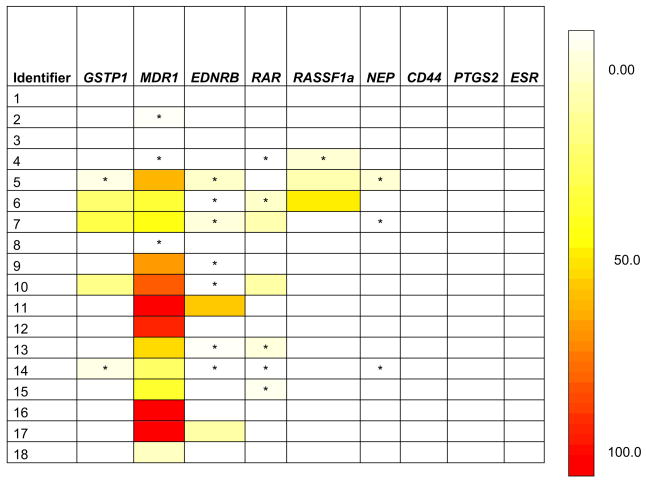
CpG island hypermethylation at various gene loci was positive in 15 of 18 cases (83.3%) (see figure and table 3). CpG island hypermethylation was most common at MDR1 (83.3% of cases), EDNRB (50%), RAR-β (38.9%) and GSTP1 (27.8%). Even when there was no hypermethylation at all 7 gene loci in localized prostate cancer, the chance to detect hypermethylation was more prevalent in hormone refractory metastatic prostate cancer.
NIM in serum samples of patients with hormone refractory metastatic prostate cancer. Results from CpG island hypermethylation at GSTP1 were included from previous study.5 Asterisk indicates small but detectable NIM.
CpG Island Hypermethylation at MDR1, RAR-β and EDNRB in Disease Progression Groups
Table 2 shows the results of CpG island hypermethylation at MDR1, RAR-β and EDNRB among worsening disease categories. The results of GSTP1 hypermethylation were included from our earlier study.5 These results may suggest that EDNRB methylation shows the strongest correlation with disease progression. However, due to the small number in all other groups, ie 3 groups had 0 patients with methylation, these results must be interpreted with care even when they were clearly statistically significant.
DISCUSSION
Aberrant CpG island hypermethylation is a hallmark of cancer, and its role as a diagnostic and prognostic marker is well established for primary and metastatic prostate cancer tissue.1,2 To further investigate its diagnostic and prognostic usefulness we studied the CpG island hypermethylation at 8 gene loci in serum samples of men with localized and metastatic prostate cancer.
To our knowledge this study is the largest study of CpG island hypermethylation of a gene panel in the serum of patients with prostate cancer to date. We noted that CpG island hypermethylation at various gene loci could be detected in serum samples from patients with worsening disease categories. Interestingly the hypermethylation status at several gene loci correlated with disease progression, although these results must be interpreted with care. These results suggest that the CpG island hypermethylation status of a defined panel of genes may be a useful biomarker in men with hormone refractory prostate cancer. Furthermore, CpG island hypermethylation at MDR1 was also observed in the serum samples of 38.8% of cases without PSA recurrence and in 15.5% with biochemical recurrence after radical prostatectomy.
Only a few other groups have examined the DNA methylation of various genes in plasma and serum samples as biomarkers for a number of human cancers.9 Recently Wallner et al reported that serum CpG island hypermethylation at certain gene loci was an independent prognostic marker in colorectal cancer.10 Similarly 2 reports described the use of methylated DNA in the serum of patients with melanoma and its predictive usefulness.11,12 These articles suggest that methylated DNA released into the circulation has potential for tumor detection and as a prognostic indicator.
In men with prostate cancer Goessl et al noted GSTP1 hypermethylation in the plasma of 56% with pT2-3 disease and of 93% with pT4M+ prostate cancer.3 Jeronimo et al detected GSTP1 serum hypermethylation in 32% of patients with nonpalpable, clinically localized prostate cancer.4 However, no correlations with diagnostic or prognostic variables were reported.
In our previous study we investigated the role of CpG island hypermethylation at GSTP1 in serum samples of men with localized prostate cancer undergoing surgical monotherapy.5 In that study we observed that CpG island hypermethylation at GSTP1 predicted early PSA recurrence following radical prostatectomy. Of all men with clinically localized disease those with a positive test for DNA GSTP1 CpG island hypermethylation were at 4.4 times greater risk for biochemical relapse than those with a negative test. Surprisingly of the previously defined prognostic features associated with PSA recurrence preoperative GSTP1 CpG island hypermethylation was the single most powerful predictor of PSA recurrence.5 In addition, GSTP1 hypermethylation was not significantly associated with any established variables.
Recently 2 studies described CpG island hypermethylation in the serum of patients with prostate cancer. 1) Reibenwein et al observed CpG island hypermethylation at GSTP1, AR and 14-3-3 σ.13 Using conventional methylation specific PCR they detected GSTP1 hypermethylation in 32.2% of patients with hormone refractory prostate cancer and in 21.4% of patients with presumably localized prostate cancer who were undergoing radiation. Furthermore, they found an association between GSTP1 hypermethylation and a higher Gleason sum, ie greater than 8. However, it remains unclear whether the reported Gleason sum was related to the biopsy Gleason sum or to the pathological Gleason sum of the surgical specimen. Keeping in mind that 34% to 55% of cases with a biopsy Gleason sum of greater than 8 are over graded, these results must be interpreted with caution.14 To our knowledge the role of CpG island hypermethylation at AR and 14-3-3 σ for detecting prostate cancer from serum samples cannot be foreseen today and it requires further investigation.
2) Chuang et al reported GSTP1 hypermethylation status in 36 patients with prostate cancer using a restriction enzyme based, real-time PCR approach.15 They found CpG island hypermethylation in 11 of 36 plasma samples of patients with prostate cancer but did not report a correlation with diagnostic or prognostic variables. Unfortunately Chuang et al provided almost no information on sample collection and processing.
As reported previously, GSTP1 CpG island hypermethylation in the serum of patients with prostate cancer was detected in 27.8% of those with metastatic prostate cancer compared to only 12% of those with localized prostate cancer.5 Although GSTP1 CpG island hypermethylation is thought to be an early event during prostate carcinogenesis, it has also been detected in advanced prostate cancer and metastatic lesions.1 Interestingly there is a statistically significant difference among each disease progression group. This seems to indicate that GSTP1 hypermethylation in serum is also a marker of progressive disease. A quantitative approach may also improve the diagnostic usefulness of the test (see figure).
Likewise we detected a statistically significant difference for CpG Island hypermethylation at MDR1, RAR-β and EDNRB among the disease progression groups. For instance, we detected CpG island hypermethylation at MDR1 in the serum samples of 83% of hormone refractory cases of prostate cancer. This may imply a role as a test for hormone refractory, metastatic prostate cancer. However, we also detected that CpG island hypermethylation at MDR1 was positive in 39% and 16% of cases without and with PSA recurrence, respectively. Thus, the true nature of CpG island hypermethylation at MDR1 in the serum of patients with prostate cancer will remain unknown until further investigations.
Previous studies of MDR1 hypermethylation status also showed controversial findings in prostate cancer and benign prostate tissues.1,2,16 These results leave room for future studies to evaluate the role of MDR1 hypermethylation in prostate cancer. Prostate cancer tissue studies using laser capture microdissection looking at multiple disease stages are warranted. Maybe these investigations can shed new light on the contrasting findings published today. Possibly an unidentified member of the adenosine triphosphate binding cassette transporter gene superfamily is stress induced in benign prostatic hyperplasia and prostate cancer, thus, shutting down P-glycoprotein expression.2
According to the results of studies of prostatic tissue on CpG island hypermethylation there is little doubt about the diagnostic and prognostic usefulness of RAR-β and EDNRB hypermethylation.1,7,17 Although hypermethylation at RAR-β was reported in patients with localized prostate cancer and prostate cancer precursor lesions, we did not detect it in the serum of patients with localized disease.18
Interestingly CpG island hypermethylation at PTGS2 and CD44 was not detected in samples of localized or metastatic prostate cancer. These findings warrant further discussion since methylation status at the 2 genes in prostate cancer tissue is associated with higher pathological Gleason sum disease recurrence.1,6,17 Additionally, CpG island hypermethylation at NEP and RASSF1a was only detected in 17% of metastatic cases. Finally, CpG island hypermethylation was not detectable at ESR. Future studies must corroborate these findings and provide further information on whether testing DNA hypermethylation in bodily fluids at any of these gene loci may improve diagnostic sensitivity or specificity for prostate cancer. Furthermore, future studies must evaluate the change in DNA markers during different disease stages of prostate cancer, ie before diagnosis, and before surgery, hormonal ablation and chemotherapy.
A pitfall of our study warrants further discussion. We used serum from patients with a single negative prostate biopsy as negative controls. In the absence of alternatives, eg cystoprostatectomy specimens with a histologically negative prostate, which still may be positive for CpG island hypermethylation due to bladder cancer, we had to use this cohort as a control cohort. The ideal negative control samples would include serum samples from patients after autopsy without histological proof of any malignancy.
Other DNA alterations, such as mutations or amplifications, frequently occur at multiple sites in a given gene in individual cancers. In contrast, CpG island hypermethylation occurs over the same region (promoter region) of genes, thus, simplifying the design of such a test. Additionally, DNA based biomarkers reveal advantages over RNA or protein markers. DNA is more stable than RNA and easier to handle than protein. Despite the use of aberrantly methylated DNA in serum of prostate cancer certain points must be addressed in future studies. It is mandatory to standardize sample collection, DNA isolation and modification, and PCR assays. High throughput tests for CpG island hypermethylation, such as COMPARE-MS (combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes)16 or luminometric methylation assay,19 have been reported and they simplify detection. Furthermore, test reproducibility, sensitivity, specificity and efficacy compared to other markers must be considered. Finally, the role of circulating methylated DNA as a therapeutic target for novel treatment options warrants further investigations. Nevertheless, the detection of DNA hypermethylation in serum or plasma samples is technically possible and it may improve current screening and detection methods for prostate cancer. Including CpG island hypermethylation tests of urine samples from cases suspicious for prostate cancer may increase diagnostic sensitivity and specificity.20
CONCLUSIONS
We noted that serum DNA hypermethylation at various gene loci may be a useful biomarker in men with hormone refractory prostate cancer and in worsening disease progression groups. If corroborated by other prospective studies, DNA hypermethylation in the serum samples of patients with prostate cancer may be helpful for identifying men who are most likely to have hormone refractory prostate cancer and, thus, most likely to benefit from early multimodal therapy.
Acknowledgments
Supported by National Institutes of Health/National Cancer Institute SPORE Grant P50CA58236, Early Detection Research Network Grant U01-CA86323 and the American Foundation for Urologic Diseases/American Urological Association Education and Research, Inc.® program (GSP).
Abbreviations and Acronyms
- CD44
CD44 molecule (Indian blood group)
- EDNRB
prostate cancer endothelin receptor type B
- ESR1
estrogen receptor 1
- GSTP1
glutathione-S-transferase-π
- MDR1
multi-drug resistance-1
- NEP
neuroepithelial tyrosine kinase
- NIM
normalized index of methylation
- PCA
endothelin receptor type B
- PCR
polymerase chain reaction
- PSA
prostate specific antigen
- PTGS2
prostaglandin-endoperoxide synthase 2 or cyclooxygenase 2
- RAR-β
retinoic acid receptor β
- RASSF1a
Ras association domain family 1 isoform A
- RE
restriction enzyme
- TSG
tumor suppressor gene
References
- 1.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 2.Bastian PJ, Ellinger J, Wellmann A, Wernert N, Heukamp LC, Muller SC, et al. Diagnostic and prognostic information in prostate cancer with the help of a small set of hypermethylated gene loci. Clin Cancer Res. 2005;11:4097. doi: 10.1158/1078-0432.CCR-04-1832. [DOI] [PubMed] [Google Scholar]
- 3.Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, et al. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000;60:5941. [PubMed] [Google Scholar]
- 4.Jeronimo C, Usadel H, Henrique R, Silva C, Oliveira J, Lopes C, et al. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 5.Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, et al. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res. 2005;11:4037. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- 6.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55:199. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 7.Jeronimo C, Henrique R, Hoque MO, Ribeiro FR, Oliveira J, Fonseca D, et al. Quantitative RARbeta2 hypermethylation: a promising prostate cancer marker. Clin Cancer Res. 2004;10:4010. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 8.Usmani BA, Shen R, Janeczko M, Papandreou CN, Lee WH, Nelson WG, et al. Methylation of the neutral endopeptidase gene promoter in human prostate cancers. Clin Cancer Res. 2000;6:1664. [PubMed] [Google Scholar]
- 9.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 10.Wallner M, Herbst A, Behrens A, Crispin A, Stieber P, Goke B, et al. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res. 2006;12:7347. doi: 10.1158/1078-0432.CCR-06-1264. [DOI] [PubMed] [Google Scholar]
- 11.Koyanagi K, Mori T, O’Day SJ, Martinez SR, Wang HJ, Hoon DS. Association of circulating tumor cells with serum tumor-related methylated DNA in peripheral blood of melanoma patients. Cancer Res. 2006;66:6111. doi: 10.1158/0008-5472.CAN-05-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori T, O’Day SJ, Umetani N, Martinez SR, Kitago M, Koyanagi K, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol. 2005;23:9351. doi: 10.1200/JCO.2005.02.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reibenwein J, Pils D, Horak P, Tomicek B, Goldner G, Worel N, et al. Promoter hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67:427. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 14.Bastian PJ, Gonzalgo ML, Aronson WJ, Terris MK, Kane CJ, Amling CL, et al. Clinical and pathologic outcome after radical prostatectomy for prostate cancer patients with a preoperative Gleason sum of 8 to 10. Cancer. 2006;107:1265. doi: 10.1002/cncr.22116. [DOI] [PubMed] [Google Scholar]
- 15.Chuang CK, Chu DC, Tzou RD, Liou SI, Chia JH, Sun CF. Hypermethylation of the CpG islands in the promoter region flanking GSTP1 gene is a potential plasma DNA bio-marker for detecting prostate carcinoma. Cancer Detect Prev. 2007;31:59. doi: 10.1016/j.cdp.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res. 2006;34:e19. doi: 10.1093/nar/gnj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastian PJ, Ellinger J, Heukamp LC, Kahl P, Muller SC, von Rucker A. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur Urol. 2007;51:665. doi: 10.1016/j.eururo.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Woodson K, Gillespie J, Hanson J, Emmert-Buck M, Phillips JM, Linehan WM, et al. Heterogeneous gene methylation patterns among pre-invasive and cancerous lesions of the prostate: a histopathologic study of whole mount prostate specimens. Prostate. 2004;60:25. doi: 10.1002/pros.20013. [DOI] [PubMed] [Google Scholar]
- 19.Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, et al. LUMA (LUminometric Methylation Assay)—a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Roupret M, Hupertan V, Yates DR, Catto JW, Rehman I, Meuth M, et al. Molecular detection of localized prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. Clin Cancer Res. 2007;13:1720. doi: 10.1158/1078-0432.CCR-06-2467. [DOI] [PubMed] [Google Scholar]