Abstract
Among the greatest challenges facing AIDS vaccine development is the intrinsic diversity among circulating populations of HIV-1 in various geographical locations and the need to develop vaccines that can elicit enduring protective immunity to variant HIV-1 strains. While variation is observed in all of the viral proteins, the greatest diversity is localized to the viral envelope glycoproteins, evidently reflecting the predominant role of these proteins in eliciting host immune recognition and responses that result in progressive evolution of the envelope proteins during persistent infection. Interestingly, while envelope glycoprotein variation is widely assumed to be a major obstacle to AIDS vaccine development, there is very little experimental data in animal or human lentivirus systems addressing this critical issue. In this review, the state of vaccine development to address envelope diversity will be presented, focusing on the use of centralized and polyvalent sequence design as mechanisms to elicit broadly reactive immune responses.
Keywords: centralized, consensus, HIV-1, polyvalent, vaccine, viral diversity
In July 2008, there were 33.2 million people infected with HIV, with approximately 2.5 million new infections [1]. Throughout the world, individuals are infected with a diverse range of HIV isolates [1]. While diversity is an issue with many HIV gene products, the greatest amount of diversity is found in the envelope (Env) glyco-protein. The Env amino acid sequences can differ as much as 15% between isolates within a single clade and more than 35% between envelopes from different clades. This diversity is one of the major obstacles facing the development of AIDS vaccines. Therefore, an effective AIDS vaccine must overcome the challenges associated with HIV sequence diversity.
The development of a successful HIV/AIDS vaccine continues to be high priority for researchers. There have been many failures from the first clinical trials using Env-only-based vaccines to the more recent Vaxgen trial [2,3]. These vaccines failed to induce neutralizing antibody responses. Due to the genetic variability of the viral envelope proteins or the display of epitopes in the proper 3D structure, the virus can escape elicited neutralizing antibodies. In addition, there are difficulties in identifying immunogens and immunization platforms that consistently induce antibodies that can neutralize isolates from different clades. In light of the difficulties in eliciting neutralizing antibodies, the field switched its focus away from vaccines that induce humoral immunity toward immune responses that reduce viral load and transmission [4,5]. This shift was prompted by data showing that T-cell-mediated immunity was critical for resistance to lentiviral infection. However, the recent failure of the candidate vaccine from Merck, which was the first vaccine designed to elicit strong cellular immunity, was a tremendous setback for AIDS vaccine researchers. This adenovirus-5-based vaccine showed no protection against infection and, even more significantly, the vaccine appeared to increase the rate of HIV infection in individuals with prior immunity against the adenovirus vector. Even though this vaccine elicited HIV-specific immunogenicity in earlier studies, there were limited multifunctional responses.
Therefore, there is a new emphasis towards more basic scientific discovery to overcome the underlying obstacles that hinder the design of an effective prophylactic HIV vaccine. The ultimate goal is to enhance a preventive vaccine that could interrupt HIV transmission and/or substantially control disease progression. This refocused effort is addressing some fundamental issues associated with HIV infection and transmission to improve the basic understanding of the immune system’s response to natural infection and vaccination, to dissect the mechanisms of protection and to use this knowledge to identify and design effective immunogens and approaches toward manipulating the immune response for an improved vaccine. The use of appropriate animal models for understanding the pathogenesis and transmission of lentiviruses is necessary to construct effective HIV vaccine immunogens. There are many challenges for AIDS vaccine researchers, including the development of delivery mechanism to elicit robust B-cell and T-cell immunity in appropriate immune compartments and the use of appropriate animal models to understand the correlates of protection and vaccine assays to assess the induction of immunity. In this review, we focus on the issue of sequence diversity as an obstacle for the development of effective AIDS vaccines, with particular emphasis on the structure and sequence variability associated with the envelope glycoprotein.
HIV-1 diversity in AIDS vaccine development
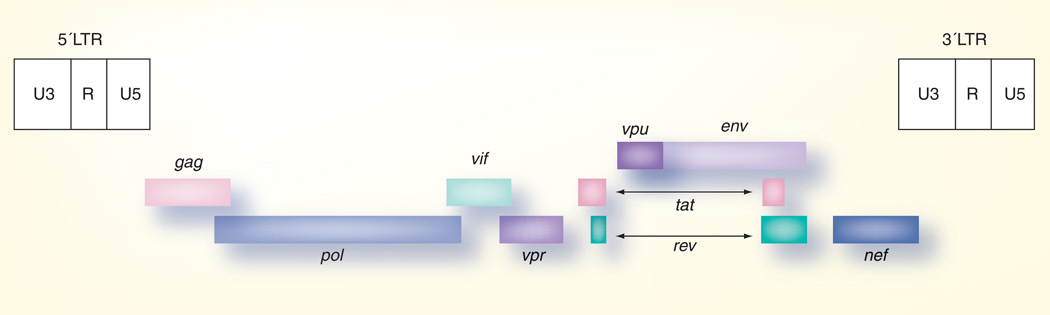
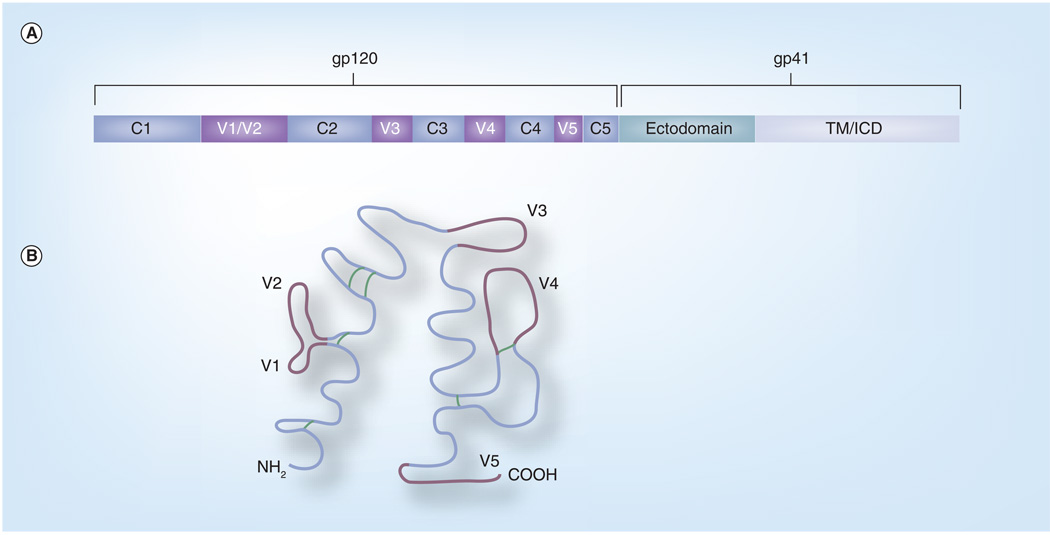
HIV-1 is a member of the family Retroviridae and the genus Lentiviridae [6]. The HIV-1 RNA genome encodes the essential retrovirus genes gag, pol and env, as well as the additional accessory/regulatory genes vif, vpr, vpu, rev, tat and nef (Figure 1) [6]. Envelope, located on the surface of viral particles, mediates binding to cellular receptors and entry into cells. The uncleaved envelope protein (Envgp160) is a highly glycosylated molecule that helps mask it from the immune system. Envgp160 is cleaved into Envgp120 and Envgp41 [7]. On the surface of the virion, envelope forms trimers, with Envgp120 on the outer surface bound to Envgp41 spanning the cellular membrane [8]. Envgp120 is composed of the constant regions C1–C5 and the variable regions V1–V5 (Figure 2).
Figure 1. Genomic organization of the HIV-1 proviral genome.
Structural and enzymatic proteins are encoded by the gag, pol and env genes. Regulatory gene products are encoded by the tat and rev genes and the major regulatory proteins are encoded by the vif, vpu, vpr and nef. The long terminal repeats (LTRs) are sites for initiation of viral RNA synthesis and necessary for proviral integration into host cell chromosomes.
Figure 2. Schematic of HIV-1 envelope.
(A) shows the division of Envgp160 into Envgp120 and Envgp41, as well as the regions of envelope and their location within Envgp120 and Envgp41. (B) A schematic of folded Envgp120. The constant regions are shown in blue with the variable regions shown in purple. Disulfide bonds are shown in green.
ICD: Intracytoplasmic domain.
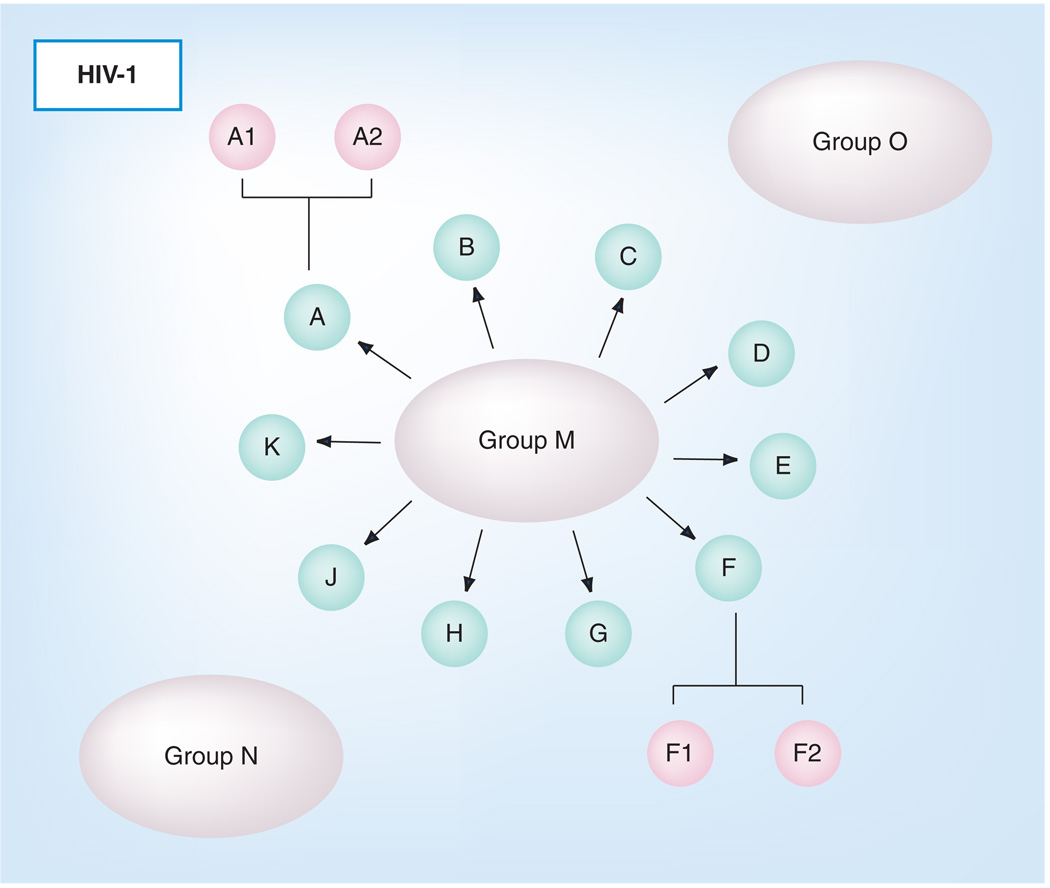
A central feature of lentiviruses in general, and HIV-1 in particular, is the propensity for these viruses to rapidly mutate and evolve in response to immunological or other selective pressures. While all of the viral genes are susceptible to mutation and protein variation, the envelope proteins are hotspots of sequence variation as they are primary targets for host immune responses, and Env has evolved to permit substantial sequence variability to confound immune recognition and control. Thus, HIV-1 populations at the individual and global levels exist as a heterogeneous mixture of genomic quasispecies unique to a patient or, on a larger scale, to a geographical area. With all of this diversity, it is reasonable to ask whether there is any commonality that can be identified and targeted for vaccine develop ment among diverse HIV-1 envelope species. Based on viral genetic distances and positions in phylogenetic trees, HIV-1 has been divided into three groups: M, O and N (Figure 3). Group M viruses are found globally and are largely responsible for the AIDS pandemic, while group O and N viruses are restricted to West Central Africa. Viral envelope sequences differ by up to 50% between these groups. Within group M, HIV-1 isolates have been further divided into distinct clades (designated A–K), which differ in envelope sequence by as much as 35% [9]. Envelope sequences can also differ by up to 15% between viruses in the same clade. There are also circulating recombinant forms (CRFs). CRFs are viral isolates that are the combination of two or more viral isolates from different clades that have been found in at least three epidemiologically independent individuals [10]. The combination of the high diversity and isolates within clades and between clades indicates that a vaccine based on a single isolate is unlikely to provide protection against all possible exposures to HIV-1 isolates. This point is further emphasized by studies that show protection can be elicited by vaccines against homologous challenge viruses, but not against heterologous challenges [11–13].
Figure 3. HIV-1 is divided into three groups: (M) main, (O) outlier and (N) non-M/non-O.
Groups M, O and N are shown as large ovals. Group M is further divided into clades, which are shown as small ovals arrows pointing from the group M oval. Clades A and F have also been further divided into A1/A2 and F1/F2.
Vaccine approaches to AIDS
The ultimate goal of an AIDS vaccine is to elicit potent cellular and humoral immune responses that will result in enduring, broadly protective immunity. While much effort has focused on elucidating the mechanisms and specificity of cellular immune responses, less is known about virus-specific antibody responses. It is well established that cellular immune responses mediate viral control during the primary infection, and antibody depletion studies in the SIV/monkey model also demonstrated a role for cellular immunity in maintaining the viral set-point in chronic infection [14]. On the other hand, passive protection experiments using human monoclonal antibodies resulted in protection against virus exposure in the HIV-1/chimp [15,16], SIV/monkey [17–20] and SHIV/monkey models [21–27], demonstrating the ability of antibody alone to mediate protection against pathogenic virus infection. Thus, the goal of an effective AIDS vaccine should be to elicit as robust and broadly reactive cellular and humoral immunity as possible, thereby maximizing the potential for protection from variant HIV-1 strains by different routes of exposure.
Initial trials using monomeric envelope proteins elicited high-titer antibody responses against the immunogen [28–35]. However, while these single-envelope vaccines have proven to be protective in both SIV and HIV nonhuman primate challenge models, protection was only observed when the envelope in the vaccine was precisely matched to the envelope on the challenge virus [11–13]. These results suggest that envelope diversity is an important obstacle in the development of an effective AIDS vaccine. This is supported by the fact that single-envelope vaccines have not been successful in humans, most likely due to the inability to preselect the challenge virus in humans and the tremendous diversity of HIV-1 isolates.
Recent vaccine studies using a polyvalent Env vaccine elicited broader immune responses, particularly neutralizing antibodies, but did not provide sterilizing protection against heterologous SHIV infection in pigtailed macaques [36]. Interestingly, animals in the multivalent vaccine group, but not those immunized with the monovalent vaccine, exhibited markedly lower levels of plasma virus than monkeys in the control group, suggesting superior cell-mediated responses induced by the polyvalent vaccine [36]. Therefore, it appears that immune responses capable of recognizing conserved epitopes within Env, with the concomitant ability to recognize these epitopes on more than one strain of HIV-1, may play an important role in inhibiting the viral spread in vivo. In addition to Env-stimulating classical cellular and neutralizing antibody responses, due to the surface exposure of Env on both virions and infected cells, antibodies can also support complement-mediated lysis of virions and antibody-dependent cellular cytotoxicity of infected cells, or contribute epitopes for MHC-I presentation and cytotoxicity. Despite a lack of reliable correlates for immune protection against HIV-1 infection, an effective vaccine against HIV or AIDS will most likely need to elicit high levels of cross-reactive neutralizing antibodies in combination with a robust cell-mediated response against multiple viral antigens. To this end, researchers have demonstrated a critical correlation of the maturation of immune responses to SIV, SHIV and other lentivirus envelope proteins and the development of protective immunity in animals inoculated with live-attenuated vaccine strains [20,37–43]. Thus, an effective AIDS vaccine may need to include a full-length, native envelope protein to enhance the breadth and maturation of both cellular and humoral immune responses.
Neutralizing antibodies, CD4+ and CD8+ T-cell responses and innate immunity have all been linked to protective efficacy. However, none of these immune parameters are universal. Vaccine studies performed in nonhuman primates indicate that protection from infection may be possible in humans [44–47]. Passively transferred neutralizing antibodies can protect against infection, but the elicitation of neutralizing antibodies has been difficult to achieve by vaccination. Live-attenuated SIV vaccines are able to protect nonhuman primates against subsequent infection [48–50]. Due to the possibility of reversion, live-attenuated HIV-1 vaccines are not suitable for human use. However, these types of vaccines do offer an effective mechanism for studying correlates of protection. Results from live-attenuated SIV infection studies indicate that cellular and humoral immunity elicited at the site of challenge can offer some level of protection from disease [48–50]. However, these responses were only protective when the challenge virus was homologous to the vaccine. These monkeys were not protected against heterologous challenge viruses, indicating that these vaccines elicited a restricted breadth of immunity [36,51,52]. Due to the vast diversity of HIV-1 isolates circulating in the population, it is unlikely that a vaccine based on a single viral isolate will be able to protect against human infection.
Centralized sequences
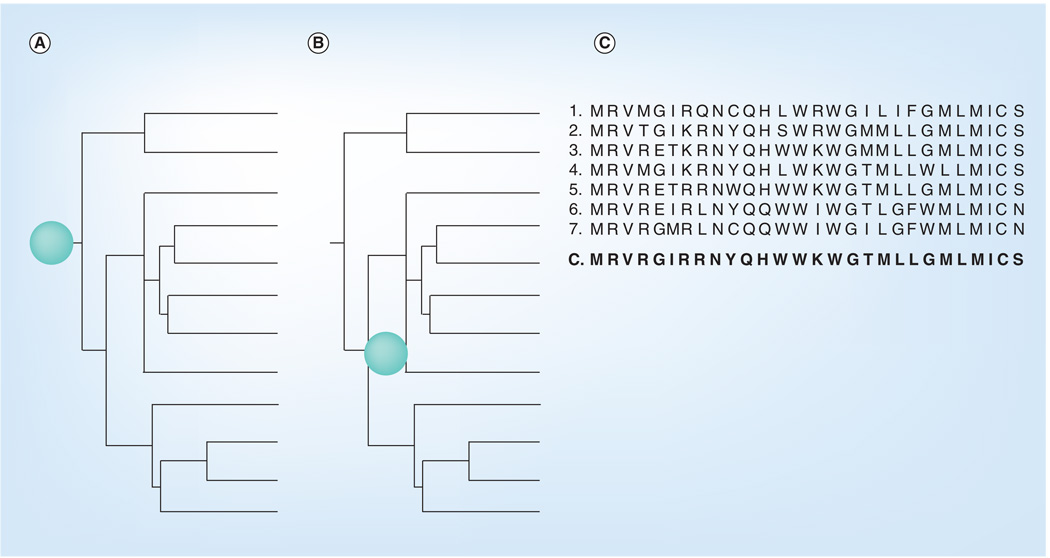
Centralized sequences are designed to limit the genetic distance between any given viral isolate and the vaccine strain. Vaccines using this approach can be designed against any protein. Three primary methods have been used to design centralized vaccines for HIV (Figure 4). Each method utilizes a given population of sequences that are based on a diverse set of parameters, such as regional location of viral isolation, the HIV clade, the year of isolation or the specific coreceptor usage. A phylogenic tree is then developed using primary viral protein sequences to develop one of two types of centralized sequences: center-of-the-tree or ancestral. Center-of-the-tree sequences are based upon generating a sequence that is equidistant to all points of the tree [9,53]. The ancestral approach is based upon the theoretical ancestor sequence that gave rise to all sequences of a particular tree [9,53]. In addition, the population of these sequences can also be aligned to make a consensus sequence by assigning the most common amino acid located at each position [9,53]. Generation of hybrid consensus sequences of the Envgp120 constant regions have been used in conjunction with the variable regions based upon a single viral isolate [54]. HIV vaccines based upon each of these centralized methods are in development.
Figure 4. Centralized vaccines.
Three methods of developing a centralized vaccine. (A) Ancestral vaccine. This method of centralization utilizes the theoretical ancestor that gave rise to the phylogenetic tree. The location of the ancestral vaccine is shown with a green dot. (B) Center of the tree. This method of centralization generates a sequence that is equidistant to all points of the phylogenetic tree. The approximate location of the vaccine sequence is shown with a green dot. (C) Consensus vaccine. A consensus sequence was generated from the seven sample envelope sequences by assigning the most common amino acid at each position in the amino acid sequence.
The development of centralized sequences is highly dependent on the quality of the starting sequence library. The center-of-the-tree and ancestral methods utilize a phylogenic tree constructed based on assumptions created to fill gaps generated by incomplete sampling of the viral population. If these assumptions are incorrect, it will alter the resulting centralized sequence to also be incorrect. The greatest degree of difficulty comes with the development of ancestral sequences, as they are the most heavily influenced by the assumptions used to generate a phylogenic tree. Sampling for the development of ancestral sequences also requires the inclusion of sequences from older samples. These samples are often difficult to obtain and are representative of only a small population of viruses, thus affecting the resulting sequence. A consensus sequence can also be biased by the sampling of the viral population. If this sampling only covers a small population, the resulting sequence will be unevenly weighted to that sample. Each centralized strategy is dependent on developing an unbiased and large database of sequences. Through the development of such databases, the short comings of these methods can be overcome.
Centralized HIV-1 envelope sequences
The three methods of developing centralized sequences are currently being used to overcome the challenges of viral diversity in HIV-1. The most common approach has been through the use of consensus sequences. However, any of the three designs may miss epitopes as possible in any given immunogen; therefore, combining several consensus sequences has advantages for covering as many epitopes within a diverse immunogen, such as the HIV-1 envelope. Recently, vaccine strategies utilizing consensus and ancestral Env epitopes have proven potent inducers of both humoral and cellular immunity [55–59]. The initial generation of centralized sequences was tested to ensure that these artificial genes were able to express functional products [55,60–64]. The majority of centralized sequences use codon optimization to enhance gene expression. This technique generates a sequence using the optimum codon for a given cell-type to maximize protein expression [65]. Codon optimized envelopes are cleaved into Envgp120 and Envgp41 at the surface, similar to native Env proteins [61,62]. Centralized envelopes form trimers that facilitate binding to CD4 and coreceptors, as well as establishing infection in vitro when pseudotyped onto viral backbones [55,61,62,64]. Other consensus HIV-1 proteins, such as Tat, Rev and Nef, have also been shown to express efficiently and be processed naturally, further demonstrating the utility of consensus sequences [63].
Centralized Env vaccines based upon geographical regions, specific group M clades, multiple clades and the entire group M have been investigated [54,55,60–64,66–74]. Regional centralized vaccines based upon viruses circulating in specific geographical regions of Africa [69,73] were designed to match the viral strains that caused over 70% of the infections within an area, such as Kenya [69]. This vaccine consisted of a consensus CAp24/MAp17, as well as cytotoxic T-lymphocyte epitopes derived from a clade A Env. Strong cellular immune responses were observed in murine studies, and immunogenicity was demonstrated in early clinical trials. A vaccine designed for Eastern and Central Africa consisted of a consensus clade A Tat, reverse transcriptase, Nef and Envgp41 [73]. This vaccine induced strong cellular responses to each of the above components; however, it did not elicit antibodies, which may limit its effectiveness in protective efficacy.
Vaccines based upon sequences from a single clade
Centralized vaccines targeted against clade B or C gene products elicit strong cellular and humoral immune responses [61–63]. A DNA vaccine based upon centralized clade C sequences expressing Tat, Rev and Nef elicited cellular immune responses that were equal to immunity elicited by corresponding vaccines based upon wild-type, primary sequences [63]. In addition, ancestral and consensus envelope-based vaccines for clade C are highly immunogenic, eliciting both strong cellular and humoral immune responses [62]. These centralized Env-based vaccines elicited antibodies that recognized an increased number of clade C isolates compared with vaccines composed of wild-type sequences. Consensus clade B Envs elicit an increased breadth of antibody recognition virus neutralization [61]. Interestingly, similar cellular responses were elicited by both clade B and C vaccine approaches [62]. These results indicate that vaccines based upon a centralized sequence can increase the breadth of immune responses, thereby allowing for an increase in the coverage of circulating viruses.
Vaccines based upon sequences from multiple clades
Centralized vaccines based upon the entire group M set of isolates strive to elicit immunity against various proteins in viral isolates across clades [54,55,64,72]. A group M consensus Env vaccine induced both cellular and humoral immune responses [54,55,64]. These vaccines induce immune responses that recognize both clade B and C cellular epitopes, as well as antibodies that neutralize isolates in both clades. Compared with vaccines using Envs derived from wild-type clade A, B and C isolates, the consensus M Env vaccine elicited higher-titer immune responses within each respective clade than vaccines based upon the wild-type sequences [54]. These results indicate that a group M consensus vaccine can generate both high-titer and broader immune responses that may elicit protection against current and emerging HIV isolates.
In contrast to a single consensus M Env strategy, a vaccine based upon Env sequences (consensus or ancestral) from multiple clades (A, B, C, F, G and H) is currently in preclinical testing [72]. This DNA vaccine is composed of rev, nef, tat and gag consensus sequences derived from clades A, B and C, and an ancestral sequence derived from clades F, G and H [72]. This vaccine was administered along with a second vaccine that encoded known cellular Env and Pol epitopes [72]. In mice, this vaccine strategy elicited strong cellular responses to Gag and Env that protected against murine leukemia virus with envelopes derived from clade A and B viruses. Overall, a multiclade centralized vaccine strategy elicits immune responses that recognize multiple HIV proteins from multiple clades. These vaccines demonstrate that group, clade and regional centralized vaccines are immunogenic, and that each type of vaccine is able to elicit increased coverage of viral isolates compared with wild-type based vaccines.
Vaccines based upon scrambled sequences
Consensus methods are also being used to deliver whole-genome vaccines. The synthetic scrambled antigen vaccine (SAVINE) is based on a weighted consensus genome of clades A, B, C and E [74]. The SAVINE vaccine is a novel use of centralized sequences. The vaccine is divided into three expression plasmids expressing scrambled peptides encompassing all the HIV gene products. SAVINE was designed using a consensus genome sequence. The HIV proteins were then divided into 30-mer peptides overlapping by 15 amino acids. These peptides were expressed as a single polypeptide from sequences that were randomly joined into a single cDNA. Due to the size of this cDNA, it was divided into three separate open reading frames (ORF). Each of the ORFs contained multiple small sections of various HIV proteins, thereby resulting in a single protein that presents multiple epitopes from every protein of the virus. The SAVINE vaccine is highly recognized by HIV-1-positive patient sera, demonstrating that the SAVINE vaccine can present epitopes, even though it does not encode any natural forms of HIV proteins [74]. The SAVINE vaccine is highly immunogenic, inducing strong cellular responses in mice [74]. The advantage of the SAVINE vaccine is that it can present epitopes from all of the HIV gene products without expressing a complete HIV protein.
Polyvalent vaccines
An alternative strategy to centralized sequences is the use of mixtures of the same immunogen from different isolates. These polyvalent vaccines aim to increase the coverage of the vaccine by combining multiple components into a single vaccine. This strategy has been used successfully against infectious disease pathogens, such as pneumococcus, poliovirus and influenza virus [75]. Multiple methods of making a polyvalent HIV vaccine have been used (Figure 5). One method of making a polyvalent vaccine is to include multiple components of the virus together in the vaccine. These methods are not designed to increase the coverage of viral isolates, but instead to increase the breadth of the immune responses against a single isolate by eliciting immune responses to multiple components. The second method is to use antigens from multiple viral isolates. These vaccines can be developed using either single antigens or multiple antigens from multiple isolates. In this way, a single vaccine can employ both methods of making a polyvalent vaccine.
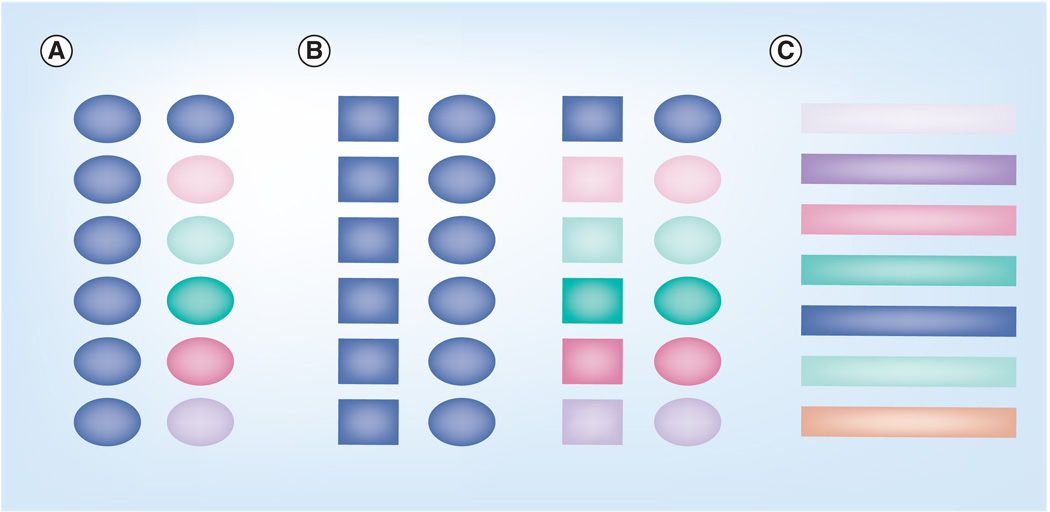
Figure 5. Polyvalent vaccines.
Three main methods of developing a polyvalent vaccine. (A) Single-component vaccine. These polyvalent vaccines are composed of a single repeated component shown as an oval. These vaccines can be based on a single clade shown in blue or multiple clades as demonstrated by multiple colors. (B) Multiple-component vaccine. These polyvalent vaccines are composed of two or more target proteins and can also be based on single or multiple clades. One antigen is shown as a square with the second antigen being shown as an oval. (C) Polyvalent peptide vaccines. These vaccines are designed to present the most common cellular epitopes from a population of sequences to the immune system. The different colors represent different epitopes.
Polyvalent vaccines based upon sequences representing a single protein within a single clade
The simplest polyvalent vaccines are those consisting of multiple repeats of a single protein. The Env immunogen has been the main focus of this type of polyvalent vaccine due to its high sequence diversity, using envelopes from a single clade or from multiple clades. Polyvalent vaccines containing either four or five clade B envelopes protected vaccinated monkeys against SHIVDH12 infection and neutralized a greater number of viral isolates than the monovalent vaccines, but this increased recognition was primarily restricted to the viral isolates included in the vaccines [36]. These results indicate that, while a polyvalent vaccine can increase the breadth of a vaccine, this breadth may be limited. Both the monovalent and polyvalent vaccine groups containing the challenge strain elicited greater protection than either the monovalent or polyvalent vaccine, which did not contain the SHIVDH12 Env. These results indicate a direct link of neutralizing antibody titers to protection against SHIVDH12 infection [36]. In addition to neutralizing antibody, other immune components contributed to protection. Monkeys vaccinated with the polyvalent vaccine without the SHIVDH12 Env had lower plasma viremia than monkeys vaccinated with the matched monovalent vaccine [36]. These results, along with other studies [76], indicate that polyvalent Env vaccines elicit broader cellular immune responses that partially control viral infection. Even a polyvalent vaccine composed of 14 Envs induced stronger cellular and humoral responses than a monovalent vaccine [76]. Interestingly, even though this vaccine was composed of exclusively clade B envelopes, it generated cellular immune responses that recognized epitopes from clades C and A/E viruses [76]. Therefore, a polyvalent vaccine composed of Envs from a single clade can induce broader humoral and cellular immune responses than a monovalent vaccine, and these responses may also recognize viral isolates or epitopes from different clades.
Polyvalent vaccines based upon sequences from different clades
To address the worldwide diversity of HIV, polyvalent vaccines composed of envelopes from different clades are in preclinical testing. A majority of these studies use polyvalent vaccines composed of isolates from clades A, B and C [77–79] and one study used isolates from clades A, B, C, D and E [80]. These multiclade polyvalent vaccines induce broader neutralizing antibodies than monovalent vaccines against each clade [77–80]. The multiclade vaccines also increase the breadth of cellular immunity compared with a monovalent vaccine [78,79]. Envelope-only vaccines using either multiclade or single-clade polyvalent strategies increase the breadth of both cellular and humoral immunity against Env. This approach may be feasible for other HIV antigens or immunogens from other viruses with a high degree of diversity.
Polyvalent vaccines based upon sequences representing multiple viral proteins
To target a wider range of HIV proteins, polyvalent vaccines incorporating multiple Env isolates, as well as other viral proteins, have been constructed to address the Env sequence diversity while combining these immunogens with other viral targets. These vaccines increase the breadth of cellular and humoral immune responses to envelope, similar to the increase observed with Env-only polyvalent vaccines [81–89]. In addition, these vaccines elicited robust immune responses against Gag epitopes [81–89] and several of these vaccines protected monkeys against SHIV infections [85,86]. One of the limitations of these challenge studies was the use of a challenge strain that was homologous to one of the Env isolates within the polyvalent vaccine [85,86]; therefore, it is unclear whether these vaccines will protect against a truly heterologous virus. A recent clinical Phase I trial using a polyvalent vaccine incorporating Env from clades A, B, C and E, as well as a Gag p24 from clade C, induced strong cellular immune responses against A, B, C and E envelopes, as well as Gag epitopes [89]. Antibody binding and neutralization were observed against a wide range of viral isolates from clades A, B, C, D and E [89]. These encouraging results indicate that a polyvalent HIV-1 vaccine is a viable vaccine strategy for humans.
Polyvalent vaccines based upon sequences to increase the breadth of cellular immunity
A modified polyvalent vaccine, designed for optimum coverage of potential T-cell epitopes, utilizes a large population of sequences similar to those used in centralized sequence design [90]. These populations are used to generate mosaic sequences by using recombination events designed to generate a sequence, which provides optimum coverage of the most common ninemer epitopes for a given target protein. These mosaic sequences exclude any rare ninemer epitopes, so only the most likely epitopes are included in the final product. These computationally derived mosaic vaccines were compared with both polyvalent vaccines, using natural strains, and consensus vaccines. The computational data indicate that the optimized polyvalent mosaics will elicit broader coverage of potential T-cell epitopes than either natural polyvalent or consensus vaccines [90], but these assumptions remain to be tested in animal models to determine if the computational increase in T-cell epitope coverage will result in increased protection or immunological recognition. This study does demonstrate a new method of designing future vaccines through the use of computational methods.
Another modified polyvalent strategy designed to target multiple clades utilizes sequences from 21 clade A, 128 clade B, 51 clade C, 17 clade D and 33 clade E isolates to generate 96 lipidated and 80 nonlipidated peptides [91]. These peptides represent the five hypervariable regions of the HIV-1 gp120 and the two variable epitopes in HIV-1 Gag [91]. The peptides were combined and delivered subcutaneously to Cynomolgus macaques and humanized HLA-A2.1 mice. The vaccine elicited strong CD4+ and CD8+ T-cell responses that were directed to isolates from clades A–E demonstrating that a multiclade polyvalent vaccine can elicit cellular immune responses to viruses from each of the clades included in the vaccine. Even though this vaccine was designed to elicit mostly cellular immune responses, moderate humoral responses were also observed. Low levels of binding antibodies were observed against the vaccine peptides, as well as primary Env isolates from clades A–E. Moderate neutralization was observed against T-cell line adapted virus, with weak neutralization observed against a few primary isolates [91]. Viral challenges were completed in humanized mice against a recombinant vaccinia virus expressing HIV-1IIIB Gag/Pol/Env proteins. Vaccinated mice had a two- to threefold reduction in viral titer demonstrating control and clearance of infection [91]. It is unclear if this challenge model can be correlated to nonhuman primate challenge systems. Together, all of the polyvalent vaccines demonstrate that there is not a loss of immunogenicity when additional HIV targets are added to the vaccine. This is important, as it is unlikely that a vaccine that focuses on only one target of HIV will be successful in the long-term due to the high mutation rates of HIV.
HIV-1 clinical trials
The recent apparent failure of HIV-1 vaccine trials has led many to question our ability to develop an effective vaccine. The results of these trials have been extensively reviewed elsewhere and will only be mentioned briefly as they apply to the development of broadly reactive vaccines [92,93]. The vaccines tested in these trials were designed using single isolates. Therefore, they were only able to generate limited reactivity against heterologous viruses. These vaccines were able to protect against homologous infections in nonhuman primates, but proved to be ineffective against human infections. This is not be because the vaccines are not inducing strong immune responses in humans, but is probably due to the fact that the infecting viruses are no longer homologous to the vaccine. These studies do not indicate the impossibility of developing an HIV-1 vaccine, but instead highlight the need for broadly reactive vaccines. These vaccines are more likely to be able to protect against heterologous infections in both nonhuman primates and humans. Therefore, further work is needed to continue the development of these broadly reactive vaccine strategies and to bring these strategies forward into clinical trials.
Summary & conclusion
One of the major obstacles in the development of an effective HIV-1 vaccine is overcoming the vast diversity of HIV-1 isolates. An effective vaccine must be able to protect against a wide population of viral isolates. Two strategies have been discussed in this article to address this issue: the use of centralized and polyvalent sequence vaccines. The centralized vaccines are designed to minimize the genetic distance between the vaccine strain and the infecting virus. Three methods of developing centralized vaccines are ancestral, center-of-the-tree and consensus. Each of these methods generates an artificial sequence that is then used to produce the vaccine of interest. Centralized vaccines produce proteins that are similar to native proteins [55,60–64]. Polyvalent vaccines use multiple components to generate better coverage of the viral population. Polyvalent vaccines are based on repeats of either a single or multiple antigens. These vaccines are based on a single genotype/clade or multiple clades. Both centralized and polyvalent vaccines are immunogenic and generate a greater breadth of humoral immunity than monovalent vaccines [36,54,77–79,81–89]. Vaccines using these strategies protect against homologous viral challenges [36,76,85,86]. The use of polyvalent and centralized vaccines offers an exciting method for addressing the issue of diversity in HIV-1 protein sequences.
Expert commentary
Some 25 years following the identification of HIV as the causative agent of AIDS, the correlate(s) of protection against viral infection are still unknown and AIDS researchers continue to debate the most effective vaccine strategies to prevent infection or the onset of disease. Previous AIDS vaccines have been designed using a variety of HIV antigens, but most include a component based upon the gag, pol and/or env genes. Two of the major obstacles in developing an effective HIV-1 vaccine are mimicking the 3D trimeric structure of Env and overcoming the vast genetic diversity of Env in emerging HIV-1 isolates.
Early trials using monomeric envelope proteins, while eliciting high-titer antibody responses against the immunogen, failed to elicit antibodies capable of neutralizing HIV-1 in vitro [28–35]. Thus, while neutralizing antibody responses appear to be an important component of an AIDS vaccine [31,59,94–97], they tend to recognize highly conformational epitopes that may not be immunogenic in vivo. The inability of these monomeric envelope proteins to elicit neutralizing antibody responses is probably due to differences in structure between monomeric forms of Envgp120 and oligomeric forms of envelope as they are expressed on the surface of a virus particle. To address the need for conformation in eliciting neutralizing antibody responses, recent studies have attempted to use soluble, truncated envelope trimers as immunogens. These trials led to disappointing results and suggest that the form of Env on the virus particle is structurally different from the soluble forms currently being produced by manufactures. While single-envelope vaccines have proven protective in both SIV and HIV nonhuman primate challenge models, protection was only observed when the envelope in the vaccine was precisely matched to the envelope on the challenge virus [11–13]. The use of inactivated virus, pseudovirions or virus-like particles [98,99] appear to be more effective at generating antibodies that recognize conformational epitopes on Env compared with soluble monomers or trimers of Env; however, neutralizing antibodies are directed to both cellular proteins embedded in the viral membrane, as well as trimeric Env spikes (Ross TM, Personal Observations) [98]. These results suggest that the structure of the Env molecule is an important determinant in the elicitation of protective immune responses.
Centralized gene strategies based upon computer models (consensus, ancestor and center-of-the-tree) have been used to construct HIV-1 gene sequences [54,55,60–64,66–74]. Each of these designs has advantages for vaccine development. Vaccine strategies utilizing consensus envelopes induce IFN-γ-producing T cells (Ross TM, Personal Observation) and cytotoxic T-cell activity [55–59]. Consensus sequences minimize the degree of sequence dissimilarity between vaccine immunogens and circulating virus strains by creating artificial sequences based upon the most common amino acid in each position in an alignment. Consensus sequences are arguably the most representative of current circulating viral populations. However, any of the three designs may miss epitopes in any given immunogen, therefore combining several consensus sequences has advantages for covering as many epitopes within a diverse immunogen, such as the HIV-1 envelope. However, focusing vaccine responses on epitopes that have escaped and are rare in the current population of viral isolates may be a disadvantage for an effective vaccine. No matter how potent the response to a particular epitope in Env, it is not advantageous if the vaccine responses are focused on epitopes that are retained only in a small percentage of HIV-1 isolates [100,101]. The use of centralized, consensus envelope immunogens may elicit broadly reactive anti-HIV-1 immunity. Expressing these consensus Envs on the surface of a particle in a native form is an attractive approach to combat both the structural and diversity issues associated with Env vaccine design. These results suggest that envelope structure and diversity are important obstacles in the development of an effective AIDS vaccine. This is supported by the fact that single envelope vaccines have not been successful in humans, most likely due to the inability to preselect the challenge virus in humans and the tremendous diversity of HIV-1 isolates.
This review also examined the use of polyvalent vaccine strategies to overcome viral sequence diversity. Polyvalent Env vaccines elicit broader immune responses, particularly neutralizing antibodies, compared with monomeric Env vaccines, but do not provide sterilizing protection against heterologous SHIV challenge [36]. Interestingly, animals in the multivalent vaccine group, but not those immunized with the monovalent vaccine, exhibited markedly lower levels of plasma virus than monkeys in the control group, suggesting superior cell-mediated responses induced by the polyvalent vaccine [36]. Therefore, it appears that immune responses capable of recognizing conserved epitopes within Env, with the concomitant ability to recognize these epitopes on more than one strain of HIV-1, may play an important role in inhibiting the viral spread in vivo. Our group and others have observed that a mixture of Envs elicit antibodies that recognize viruses from different clades [36,76–89]. A mixture of Envs from the same clade expressed from the surface of a virus-like particle elicited broadly reactive cellular responses as effectively as a single consensus Env from the same clade (Ross TM, Personal Observation). The use of a single immunogen, such as a consensus sequence, that elicits the same broadly reactive immunity as a polyvalent vaccine has advantages for vaccine development, since only one protein will need to be produced, characterized and receive approval by the US FDA for clinical trials and future licensing.
The use of centralized or polyvalent sequences may be an effective vaccine strategy against other infectious disease pathogens with genetic viral diversity, such as emerging and circulating influenza and dengue viruses. Each year, genetically distinct isolates evolve faster than vaccine manufactures can update and generate new vaccines. Centralized, sequence-based vaccines may effectively elicit immune responses that can protect individuals against future viral isolates. Based upon their potential, the centralized and polyvalent sequence strategies are strong candidates for human vaccine use against several disease pathogens.
Five-year view
Vaccine manufacturers are using a variety of different techniques to address the issues of viral sequence diversity. Over the next 5 years, these techniques will be combined and compared with develop the best method for developing a broadly reactive AIDS vaccine. Initial immunogenicity studies have shown that consensus and polyvalent vaccines can elicit strong immune responses. Further work will be needed to determine the optimal sequences, structure and delivery system for these vaccines. DNA, viral vectors, virus-like particles and liposomes carrying polyvalent and/or centralized antigens will all be tested separately and in combination. Computationally designed vaccines will also become more important as initial animal studies are completed using these vaccines. Broadly reactive vaccines will move further into nonhuman primate studies using heterologous viral challenge models to evaluate the broadest protection against viruses from different HIV clades. Centralized and polyvalent vaccines will also move further into human clinical trials, with these results setting the baseline for the use of these vaccine approaches in the future.
Key issues
HIV-1 antigens contain a wide diversity in sequences.
HIV sequence diversity is a challenge for AIDS vaccine manufacturers.
Centralized and polyvalent vaccines are two strategies to address HIV sequence diversity.
There are three primary methods to develop a centralized vaccine: ancestral, center-of-the-tree and consensus.
Centralized vaccines can be based on the region from where the virus was isolated, the genetic clade or the HIV-1 group.
Polyvalent vaccines can be developed using multiple repeats of a single protein or repeats of multiple proteins.
Both centralized and polyvalent vaccines elicit immune responses with greater breadth of immunity than vaccines based upon a single isolate.
Both strategies protect against homologous viral challenges, but it is unclear if these strategies can protect against heterologous viral isolates.
Future work will determine the best method for developing a broadly reactive immune response that will protect against HIV-1 infection.
Acknowledgements
The authors thank Hermancia S Eugene and Jodi K Craigo for helpful discussions and Dilhari DeAlmeida and Corey Crevar for technical assistance.
Footnotes
Financial & competing interests disclosure
SP McBurney received a Pittsburgh AIDS Research Training fellowship AI065380-02 from the NIH/NIAID. TM Ross is supported by the research grant award R01AI068507 from NIH/NIAID. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Sean P McBurney, University of Pittsburgh, School of Medicine, Center for Vaccine Research, Program in Molecular Virology and Microbiology, 9051 Biomedical Science Tower 3, 3501 Fifth Avenue, Pittsburgh, PA 15261, USA, Tel.: +1 412 383 9605, Fax: +1 412 624 4440, spm10@pitt.edu.
Ted M Ross, University of Pittsburgh, School of Medicine, Center for Vaccine Research, Department of Microbiology and Molecular Genetics, 9047 Biomedical Science Tower 3, 3501 Fifth Avenue, Pittsburgh, PA 15261, USA, Tel.: +1 412 648 8666, Fax: +1 412 624 4440, tmr15@pitt.edu.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1. Joint United Nations Programme on HIV/AIDS, World Health Organization. AIDS epidemic update report. 2007 • Report of the number of new and current infections during 2007, as well as the number of deaths caused by HIV in 2007.
- 2.Letvin NL. Progress and obstacles in the development of an AIDS vaccine. Nature Rev. Immunol. 2006;6(12):930–939. doi: 10.1038/nri1959. [DOI] [PubMed] [Google Scholar]
- 3.Sekaly R-P. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 2008;205(1):7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMichael AJ. HIV VACCINES. Annu. Rev. Immunol. 2006;24(1):227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 5.Thorner AR, Barouch DH. HIV-1 vaccine development: progress and prospects. Curr. Infect. Dis. Rep. 2007;(9):71–75. doi: 10.1007/s11908-007-0025-0. [DOI] [PubMed] [Google Scholar]
- 6.Freed EO, Martin MA. In: Fields Virology. Howley PM, Knipe DM, editors. PA, USA: Lippincott Williams and Wilkins; 2001. [Google Scholar]
- 7.Hallenberger S, Bosch V, Angliker H, et al. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360(6402):358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 8.Prabakaran P, Dimitrov AS, Fouts TR, Dimitrov DS, KuanTeh J. Advances in Pharmacology. MA, USA: Academic Press; 2007. Structure and function of the HIV envelope glycoprotein as entry mediator, vaccine immunogen, and target for inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaschen B, Taylor J, Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 10.Robertson DL, Anderson JP, Bradac JA, et al. A reference guide to HIV-1 classification. In: Korber B, Kuiken C, Foley B, et al., editors. Human Retroviruses and AIDS. 1999. pp. 492–505. [Google Scholar]
- 11.Johnston MI. Progress in HIV vaccine development. Curr. Opin. Pharmacol. 2001;1:504–510. doi: 10.1016/s1471-4892(01)00086-8. [DOI] [PubMed] [Google Scholar]
- 12.Berman P, Gregory T, Riddle L, et al. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature. 1990;345:622–625. doi: 10.1038/345622a0. [DOI] [PubMed] [Google Scholar]
- 13.Polacino P. Limited breadth of protetive immunity elicited by SIVmne gp160 vaccines in combination immunization regimen. J. Virol. 1999;73:618–630. doi: 10.1128/jvi.73.1.618-630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barry AP, Silvestri G, Safrit JT, et al. Depletion of CD8+ cells in sooty mangabey monkeys naturally infected with simian immunodeficiency virus reveals limited role for immune control of virus replication in a natural host species. J. Immunol. 2007;178(12):8002–8012. doi: 10.4049/jimmunol.178.12.8002. [DOI] [PubMed] [Google Scholar]
- 15.Murthy KK, Cobb EK, Rouse SR, et al. Active and passive immunization against HIV type 1 infection in chimpanzees. AIDS Res. Hum. Retroviruses. 1998;14 Suppl. 3:S271–S276. [PubMed] [Google Scholar]
- 16.Conley AJ, Kessler JA, II, Boots LJ, et al. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J. Virol. 1996;70(10):6751–6758. doi: 10.1128/jvi.70.10.6751-6758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clements JE, Montelaro RC, Zink MC, et al. Cross-protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J. Virol. 1995;69(5):2737–2744. doi: 10.1128/jvi.69.5.2737-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner M, Rosenthal A, Jennings M, et al. Passive immunization of rhesus macaques against SIV infection and disease. AIDS Res. Hum. Retroviruses. 1995;11(7):843–854. doi: 10.1089/aid.1995.11.843. [DOI] [PubMed] [Google Scholar]
- 19.Haigwood NL, Watson A, Sutton WF, et al. Passive immune globulin therapy in the SIV/macaque model: early intervention can alter disease profile. Immunol. Lett. 1996;51(1–2):107–114. doi: 10.1016/0165-2478(96)02563-1. [DOI] [PubMed] [Google Scholar]
- 20.Van Rompay KK, Berardi CJ, Dillard-Telm S, et al. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J. Infect. Dis. 1998;177(5):1247–1259. doi: 10.1086/515270. [DOI] [PubMed] [Google Scholar]
- 21.Mascola JR, Lewis MG, Stiegler G, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 1999;73(5):4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascola JR, Stiegler G, Vancott TC, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 23.Baba TW, Liska V, Hofmann-Lehmann R, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian–human immunodeficiency virus infection. Nat. Med. 2000;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 24.Foresman L, Jia F, Li Z, et al. Neutralizing antibodies administered before, but not after, virulent SHIV prevent infection in macaques. AIDS Res. Hum. Retroviruses. 1998;14(12):1035–1043. doi: 10.1089/aid.1998.14.1035. [DOI] [PubMed] [Google Scholar]
- 25.Li A, Baba TW, Sodroski J, et al. Synergistic neutralization of a chimeric SIV/HIV type 1 virus with combinations of human anti-HIV type 1 envelope monoclonal antibodies or hyperimmune globulins. AIDS Res. Hum. Retroviruses. 1997;13(8):647–656. doi: 10.1089/aid.1997.13.647. [DOI] [PubMed] [Google Scholar]
- 26.Li A, Katinger H, Posner MR, et al. Synergistic neutralization of simian–human immunodeficiency virus SHIV-vpu+ by triple and quadruple combinations of human monoclonal antibodies and high-titer anti-human immunodeficiency virus type 1 immunoglobulins. J. Virol. 1998;72(4):3235–3240. doi: 10.1128/jvi.72.4.3235-3240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata R, Igarashi T, Haigwood N, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 1999;5(2):204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 28.Barnett SW. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine. 1997;15:869–873. doi: 10.1016/s0264-410x(96)00264-2. [DOI] [PubMed] [Google Scholar]
- 29.Burton DR, Montefiori DC. The antibody response in HIV-1 infection. AIDS. 1997;11:S87–S98. [PubMed] [Google Scholar]
- 30.Graham BS. Determinants of antibody response after recombinant gp160 boosting in vaccinia-naive volunteers preimed with gp160-recombinant vaccinina virus. J. Infect. Dis. 1994;170:782–786. doi: 10.1093/infdis/170.4.782. [DOI] [PubMed] [Google Scholar]
- 31.Mascola JR, Snyder SW, Weislow OS, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against labratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 32.Mascola JR. Protection of macaques against vaginal transmission of pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 33.Parren PW. Neutralization of HIV-1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of eptiope specificity. J. Virol. 1998;73:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Cott TC. Antibodies with specificity to native gp120 and neutralizaion activity against primary HIV-1 isolates elciited by immunization with oligomeric gp160. J. Virol. 1997;71:4319–4330. doi: 10.1128/jvi.71.6.4319-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X. Characterization of stable, soluble trimer contaiing complete ectodomains of HIV-1 envelope glycoprotein. J. Virol. 2000;74:5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho MW. Polyvalent envelope glycoprotein vaccine elicits a broader neutralizing antibody response but is unable to provide sterilizing protection against heterologous Simian/human immunodeficiency virus infection in pigtailed macaques. J Virol. 2001;75:2224–2234. doi: 10.1128/JVI.75.5.2224-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole KS, Rowles JL, Murphey-Corb M, et al. A model for the maturation of protective antibody responses to SIV envelope proteins in experimentally immunized monkeys. J. Med. Primatol. 1997;26(1–2):51–58. doi: 10.1111/j.1600-0684.1997.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 38.Cole KS, Murphey-Corb M, Narayan O, et al. Common themes of antibody maturation to simian immunodeficiency virus, simian–human immunodeficiency virus, and human immunodeficiency virus type 1 infections. J. Virol. 1998;72(10):7852–7859. doi: 10.1128/jvi.72.10.7852-7859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuller DH, Simpson L, Cole KS, et al. Gene gun-based nucleic acid immunization alone or in combination with recombinant vaccinia vectors suppresses virus burden in rhesus macaques challenged with a heterologous SIV. Immunol. Cell Biol. 1997;75(4):389–396. doi: 10.1038/icb.1997.61. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RP, Lifson JD, Czajak SC, et al. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J. Virol. 1999;73(6):4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond SA, Cook SJ, Lichtenstein DL, Issel CJ, Montelaro RC. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J. Virol. 1997;71(5):3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montelaro RC, Cole KS, Hammond SA. Maturation of immune responses to lentivirus infection: implications for AIDS vaccine development. AIDS Res. Hum. Retroviruses. 1998;14 Suppl. 3:S255–S259. [PubMed] [Google Scholar]
- 43.Van Rompay K, Greenier JL, Cole KS, et al. Immunization of newborn rhesus macaques with simian immunodeficiency virus (SIV) vaccines prolongs survival after oral challenge with virulent SIVmac251. J. Virol. 2003;77:179–190. doi: 10.1128/JVI.77.1.179-190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emini EA, Schleif WA, Nunberg JH, et al. Prevention of HIV-1 infection in chimpanzees by gpl20 V3 domain-specific monoclonal antibody. Nature. 1992;355(6362):728. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- 45.Holl V, Peressin M, Decoville T, et al. Nonneutralizing antibodies are able to inhibit human immunodeficiency virus type 1 replication in macrophages and immature dendritic cells. J. Virol. 2006;80(12):6177–6181. doi: 10.1128/JVI.02625-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mascola JR, Lewis MG, Stiegler G, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6pd by passive transfer of neutralizing antibodies. J. Virol. 1999;73(5):4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manrique M, Micewicz E, Kozlowski PA, et al. DNA-MVA Vaccine protection after X4 SHIV challenge in macaques correlates with day-of-challenge antiviral CD4+ cell-mediated immunity levels and postchallenge preservation of CD4+ T cell memory. AIDS Res. Hum. Retroviruses. 2008;24(3):505–519. doi: 10.1089/aid.2007.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsson C, Makitalo B, Thorstensson R, et al. Live attenuated simian immunodeficiency virus (SIV)mac in macaques can induce protection against mucosal infection with SIVsm. AIDS. 1998;12(17):2261–2270. doi: 10.1097/00002030-199817000-00006. [DOI] [PubMed] [Google Scholar]
- 49.Wyand MS, Manson K, Montefiori DC, et al. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 1999;73(10):8356–8363. doi: 10.1128/jvi.73.10.8356-8363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyand MS, Manson KH, Garcia-Moll M, Montefiori D, Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 1996;70(6):3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu R, Srivastava IK, Kuller L, et al. Immunization with HIV-1 SF162-derived envelope gp140 proteins does not protect macaques from heterologous simian–human immunodeficiency virus SHIV89.6P infection. Virology. 2006;349(2):276–289. doi: 10.1016/j.virol.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 52.Someya K, Cecilia D, Ami Y, et al. Vaccination of rhesus macaques with recombinant Mycobacterium bovis bacillus Calmette–Guerin env V3 elicits neutralizing antibody-mediated protection against simian–human immunodeficiency virus with a homologous but not a heterologous V3 motif. J. Virol. 2005;79(3):1452–1462. doi: 10.1128/JVI.79.3.1452-1462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nickle DC, Jensen MA, Gottlieb GS, et al. Consensus and ancestral state HIV vaccines. Science. 2003;299(5612):1515–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 54. Weaver EA, Lu Z, Camacho ZT, et al. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group M consensus env immunogen. J. Virol. 2006;80(14):6745–6756. doi: 10.1128/JVI.02484-05. •• Report of the possibility of generating a larger breadth of immunity across multiple clades using a ConM immunogen.
- 55.Gao F, Weaver EA, Lu Z, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J. Virol. 2005;79(2):1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuiken CL, Foley B, Freed E, et al., editors. Theoretical Biology and Biophysics Group. HIV Sequence Compendium. NM, LA-UR 03-3564, USA: Los Alamos National Laboratory; 2002. [Google Scholar]
- 57.Zhan X. Minor components of a multi-envelope HIV vaccine are recognized by type-specific T-helper cells. Vaccine. 2004;22:1206–1213. doi: 10.1016/j.vaccine.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 58.Yusim K, Kesmir C, Gaschen B. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 2002;76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doria-Rose NA, Learn GH, Rodrigo AG, et al. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 2005;79(17):11214–11224. doi: 10.1128/JVI.79.17.11214-11224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kothe DL, Decker JM, Li Y, et al. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology. 2007;360(1):218–234. doi: 10.1016/j.virol.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kothe DL, Li Y, Decker JM, et al. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology. 2006;352(2):438–449. doi: 10.1016/j.virol.2006.05.011. • One of the first reports to demonstrate that a consensus envelope protein is as functional as a wild-type envelope protein.
- 63. Scriba TJ, zur Megede J, Glashoff RH, et al. Functionally-inactive and immunogenic Tat, Rev and Nef DNA vaccines derived from sub-Saharan subtype C human immunodeficiency virus type 1 consensus sequences. Vaccine. 2005;23(9):1158. doi: 10.1016/j.vaccine.2004.08.026. • First report that consensus Tat, Rev and Nef were processed as well as and in the same manner as native proteins.
- 64.Liao H-X, Sutherland LL, Xia SM, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353(2):268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davies MN, Flower DR. Harnessing bioinformatics to discover new vaccines. Drug. Discov. Today. 2007;12(9–10):389–395. doi: 10.1016/j.drudis.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 66.Burgers WA, van Harmelen JH, Shephard E, et al. Design and preclinical evaluation of a multigene human immunodeficiency virus type 1 subtype C DNA vaccine for clinical trial. J. Gen. Virol. 2006;87(2):399–410. doi: 10.1099/vir.0.81379-0. [DOI] [PubMed] [Google Scholar]
- 67.De Groot AS, Bishop EA, Khan B, et al. Engineering immunogenic consensus T-helper epitopes for a cross-clade HIV vaccine. Methods. 2004;34(4):476. doi: 10.1016/j.ymeth.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 68.Hamano T, Sawanpanyalert P, Yanai H, et al. Determination of HIV type 1 CRF01_AE gag p17 and env-V3 consensus sequences for HIV/AIDS vaccine design. AIDS Res. Hum. Retroviruses. 2004;20(3):337–340. doi: 10.1089/088922204322996572. [DOI] [PubMed] [Google Scholar]
- 69.Hanke T, McMichael AJ, Mwau M, et al. Development of a DNA-MVA/HIVA vaccine for Kenya. Vaccine. 2002;20(15):1995. doi: 10.1016/s0264-410x(02)00085-3. [DOI] [PubMed] [Google Scholar]
- 70.Krohn K, Stanescu I, Blazevic V, et al. A DNA HIV-1 vaccine based on a fusion gene expressing non-structural and structural genes of consensus sequence of the A–C subtypes and the ancestor sequence of the F–H subtypes. Preclinical and clinical studies. Microbes Infect. 2005;7(14):1405. doi: 10.1016/j.micinf.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 71.Kumar S, Aggarwal P, Vajpayee M, Pandey RM, Seth P. Development of a candidate DNA/MVA HIV-1 subtype C vaccine for India. Vaccine. 2006;24(14):2585. doi: 10.1016/j.vaccine.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 72. Malm M, Rollman E, Ustav M, et al. Cross-clade protection induced by human immunodeficiency virus-1 DNA immunogens expressing consensus sequences of multiple genes and epitopes from subtypes A, B, C, and FGH. Viral Immunol. 2005;18(4):678–688. doi: 10.1089/vim.2005.18.678. •• First clinical trial of consensus immunogens.
- 73.Nkolola J, Wee EG-T, Im E-J. Engineering RENTA, a DNA prime–MVA boost HIV vaccine tailored for Eastern and Central Africa. Gene Ther. 2004;11(13):1069–1080. doi: 10.1038/sj.gt.3302241. [DOI] [PubMed] [Google Scholar]
- 74. Thomson SA, Jaramillo AB, Shoobridge M, et al. Development of a synthetic consensus sequence scrambled antigen HIV-1 vaccine designed for global use. Vaccine. 2005;23(38):4647. doi: 10.1016/j.vaccine.2005.04.045. • Centralized immunogen that can present epitopes from all regions of the HIV genome.
- 75.Slobod K, Coleclough C, Bonsignori M, et al. HIV vaccine rationale, design and testing. Curr. HIV Res. 2005;3(2):107–112. doi: 10.2174/1570162053506928. [DOI] [PubMed] [Google Scholar]
- 76. Azizi A, Anderson D, Ghorbani M, Gee K, Diaz-Mitoma F. Immunogenicity of a polyvalent HIV-1 candidate vaccine based on fourteen wild type gp120 proteins in golden hamsters. BMC Immunol. 2006;7(1):25. doi: 10.1186/1471-2172-7-25. • Demonstration of the ability of a polyvalent vaccine to protect against viral challenge.
- 77.Chakrabarti BK, Ling X, Yang ZY, et al. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine. 2005;23(26):3434. doi: 10.1016/j.vaccine.2005.01.099. [DOI] [PubMed] [Google Scholar]
- 78.Ljungberg K, Rollman E, Eriksson L, Hinkula J, Wahren B. Enhanced immune responses after DNA vaccination with combined envelope genes from different HIV-1 subtypes. Virology. 2002;302(1):44. doi: 10.1006/viro.2002.1547. [DOI] [PubMed] [Google Scholar]
- 79.Rollman E, Hinkula J, Arteaga J, et al. Multi-subtype gp160 DNA immunization induces broadly neutralizing anti-HIV antibodies. Gene Ther. 2004;11(14):1146–1154. doi: 10.1038/sj.gt.3302275. [DOI] [PubMed] [Google Scholar]
- 80.Wang S, Pal R, Mascola JR, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350(1):34–47. doi: 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 81.Brave A, Boberg A, Gudmundsdotter L, et al. A new multi-clade DNA prime/recombinant MVA Boost vaccine induces broad and high levels of HIV-1-specific CD8+ T-cell and humoral responses in mice. Mol. Ther. 2007;15(9):1724–1733. doi: 10.1038/sj.mt.6300235. [DOI] [PubMed] [Google Scholar]
- 82.Brave A, Ljungberg K, Boberg A, et al. Multigene/multisubtype HIV-1 vaccine induces potent cellular and humoral immune responses by needle-free intradermal delivery. Mol. Ther. 2005;12(6):1197. doi: 10.1016/j.ymthe.2005.06.473. [DOI] [PubMed] [Google Scholar]
- 83.Cristillo AD, Wang S, Caskey MS, et al. Preclinical evaluation of cellular immune responses elicited by a polyvalent DNA prime/protein boost HIV-1 vaccine. Virology. 2006;346(1):151. doi: 10.1016/j.virol.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 84.Kong W-P, Huang Y, Yang Z-Y, et al. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J. Virol. 2003;77(23):12764–12772. doi: 10.1128/JVI.77.23.12764-12772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pal R, Wang S, Kalyanaraman VS, et al. Polyvalent DNA prime and envelope protein boost HIV-1 vaccine elicits humoral and cellular responses and controls plasma viremia in rhesus macaques following rectal challenge with an R5 SHIV isolate. J. Med. Primatol. 2005;34(5–6):226–236. doi: 10.1111/j.1600-0684.2005.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pal R, Wang S, Kalyanaraman VS, et al. Immunization of rhesus macaques with a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine elicits protective antibody response against simian human immunodeficiency virus of R5 phenotype. Virology. 2006;348(2):341–353. doi: 10.1016/j.virol.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 87.Pal R, Yu Q, Wang S, et al. Definitive toxicology and biodistribution study of a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 (HIV-1) vaccine in rabbits. Vaccine. 2006;24(8):1225. doi: 10.1016/j.vaccine.2005.07.112. [DOI] [PubMed] [Google Scholar]
- 88.Seaman MS, Xu L, Beaudry K, et al. multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J. Virol. 2005;79(5):2956–2963. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang S, Kennedy JS, West K, et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime–protein boost HIV-1 vaccine in healthy human volunteers. Vaccine. 2008;26(8):1098–1110. doi: 10.1016/j.vaccine.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fischer W, Perkins S, Theiler J, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 2007;13(1):100–106. doi: 10.1038/nm1461. •• Computational design of a polyvalent vaccine to cover the most common epitopes in HIV-1.
- 91. Azizi A, Anderson DE, Torres JV, et al. Induction of broad cross-subtype-specific HIV-1 immune responses by a novel multivalent HIV-1 peptide vaccine in cynomolgus macaques. J. Immunol. 2008;180(4):2174–2186. doi: 10.4049/jimmunol.180.4.2174. •• Computational design of polyvalent peptide vaccine.
- 92.Hanke T. STEP trial and HIV-1 vaccines inducing T-cell responses. Expert Rev. Vaccines. 2008;7(3):303–309. doi: 10.1586/14760584.7.3.303. [DOI] [PubMed] [Google Scholar]
- 93.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 2008;14(6):617–621. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cole K, Rowles J, Jagerski B, et al. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J. Virol. 1997;71:5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dorner F, Barrett PN. Vaccine technology: looking to the future. Ann. Med. 1999;31(1):51–60. doi: 10.3109/07853899909019262. [DOI] [PubMed] [Google Scholar]
- 96.Graham B. Clinical trials of HIV vaccines. Annu. Rev. Med. 2002;53:207–221. doi: 10.1146/annurev.med.53.082901.104035. [DOI] [PubMed] [Google Scholar]
- 97.Mascola JR. Defining the protective antibody response for HIV-1. Curr. Mol. Med. 2003;3:209–216. doi: 10.2174/1566524033479799. [DOI] [PubMed] [Google Scholar]
- 98.Crooks ET, Moore PL, Franti M, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366(2):245–262. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McBurney SP, Young KR, Ross TM. Membrane embedded HIV-1 envelope on the surface of a virus-like particle elicits broader immune responses than soluble envelopes. Virology. 2007;358(2):334–346. doi: 10.1016/j.virol.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 100.Gaschen B. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 101.Gao F. Consensus and ancestral state of HIV vaccines. Science. 2003;299:1515–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]