Abstract
Structural and biochemical studies have begun to elucidate the pathway of RNA silencing that leads to the formation of the RISC complex. The outstanding feature of this pathway is the precise recognition and processing of double-stranded RNA. We present a review of recent structures that illustrate the molecular mechanisms contributing to these two related functions, highlighting models of Drosha, Dicer, and RISC function.
Keywords: PAZ, PIWI, Argonaute, dsRBD, RNaseIII
1. Introduction: Structural biology of the core pathway of RNAi
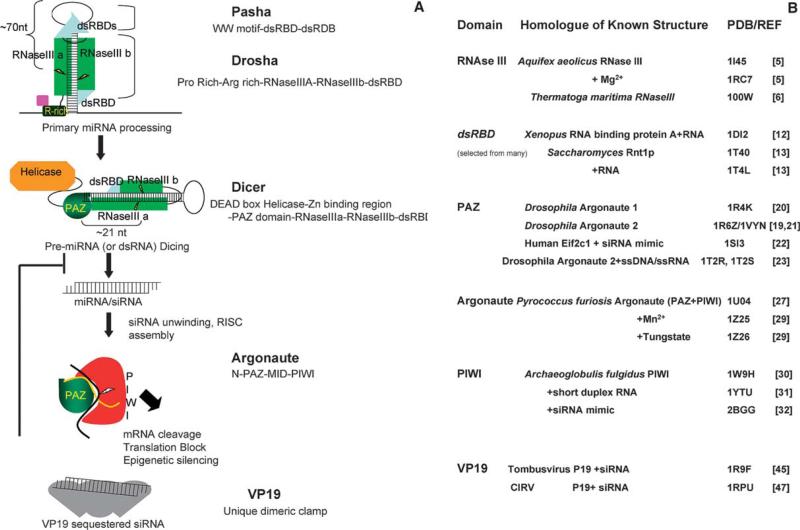
The pathways of RNA silencing [1] have at their core a set of reactions to generate siRNAs and miRNAs (∼21 nucleotide duplex RNA with 2-nucleotide 3′ overhangs) from dsRNA and endogenous transcripts containing long hairpin structures, respectively, and load them into RISC complexes, which effect a variety of silencing mechanisms [1,2]. Fig. 1 illustrates the heart of this pathway: miRNAs are transcribed as primary miRNA (pri-miRNAs) of several hundred nucleotides (nt) which are processed into stem-loop pre-miRNAs of ∼70 nt by Drosha prior to nuclear export [3]. Dicer(s) further cleave these structures into miRNAs of ∼22 nt, and processes dsRNAs into siRNAs of the same length. Once a siRNA/miRNA is unwound and loaded into an Argonaute protein (see [2] for RISC-loading details), the core of the RNAi effector complex (RISC) is formed. This review highlights the structural biology of this pathway, with an emphasis on the domains that contribute to the unique recognition and precise processing required in generating a RNA guide strand. The RNase activities of Dicer, Drosha, and RISC will be considered (see also review by Hammond, this issue). Specific modes of RNA recognition are critical to the pathway. Structures have revealed the basis of 3′ overhang recognition by PAZ, 5′ end recognition by PIWI, and of siRNA binding by a unique viral suppressor of RNAi (see Fig. 1B for a list of these structures, and references).
Fig 1.
A “Domain-centric” view of RNAi. (A) The conserved pathways of RNA silencing. The domain structure of each protein in (hypothetical) interaction with its RNA is shown. For clarity, the second column lists domains in order N- to C-terminal. Figures are not to scale. In brief, Drosha, an RNase III enzyme, and its obligate binding partner, Pasha recognize pri-mRNA loops, and cut these into ∼70 nt hairpin pre-miRNAs. Dicer utilizes a PAZ domain to sense the 3′ 2-nt overhang created, and further processes these, and dsRNAs into miRNAs and siRNAs. Argonaute binds the 5′ end of guide RNAs via its PIWI domain, and the 3′ end via a PAZ domain, yielding RISCs that effect RNA silencing through several mechanisms. A Viral protein, VP19 can suppress RNA silencing by sequestering siRNAs. (B) A summary of known siRNA structural biology. Listed by domain are solved structures, their protein/organism of origin, and ligands, where applicable. Also shown are PDB codes.
2. The RNase III domain: Drosha and Dicer
Drosha is composed of variable, long N-terminal regions, two RNase III domains, and a ‘traditional’ double-stranded RNA binding domain (dsRBD) [4]. Dicer typically contains an N-terminal RNA helicase domain, a PAZ domain, two RNase III domains, and a dsRBD [4]. Although the structure of neither Drosha nor Dicer is known, structural and biochemical characterization of homologues in the RNase III enzyme family has led to a model for the mechanism of both.
The structure of the endonuclease domain of RNase III from Aquifex aeolicus [5], and more recently, the unliganded full-length Aquifex and Thermatoga RNase III [6] and mutant Aquifex RNase III bound to dsRNA in a non-catalytic assembly [6] have been determined. The Aquifex RNase domain is a novel all-α globular fold of 7-α helices and one 310 helix. A tight dimer with more than 120 hydrophobic interactions mediated by side chains from 4 helices forms a deep cleft (50Å wide by 20Å deep). The base of this cleft is generated by two antiparallel helices, and is electrostatically basic. At each end of the valley, clusters of six acidic side chains were proposed to form a dual active site. Therefore, both active centers were expected to have double-stranded RNAse activity, for a total of four phosphodiester bond cleavages per dimer [5,6]. Along with biochemical experiments (reviewed in [7]) this established RNase III enzymes containing single RNase III domains function as homodimers. As Drosha and Dicer have two RNase III domains each, this suggested a number of mechanisms based on inter- or intramolecular dimerization with two active sites per molecule (reviewed in [4]).
Zhang et al. [8] have demonstrated a single reaction center, capable of simultaneously cutting both strands of dsRNA, is generated by the intramolecular dimerization of the two RNase III domains of Dicer [8]. They suggest, contrary to the prediction of two cleavage sites per monomer from the Aquifex RNase III crystal structure [6], that only one active site resides in each monomer. Notably, mutagenesis of human Dicer [8], and E. coli RNase III [8,9] demonstrate the residues of one of the two putative catalytic sites seen in the crystal structure are not essential for activity. The compound active site forms by bringing two catalytic sites together, each of which cleaves one strand. Thereby, both strands of RNA are cut, and by offsetting the two scission sites by 2-nt, an end with a 3′ 2-nt overhang is generated (this is the typical product of RNase IIIs). Since only one double-stranded cut is made, something other than the spacing of two double-stranded cut sites must govern product size. Two accessory domains, considered in the following sections, may solve this dilemma. Zhang et al. propose a PAZ–RNase III combination could act as the molecular ruler to guarantee production of appropriate size siRNAs by Dicer [8]. Similarly, Drosha may use a special dsRBD (perhaps its own, or that of its obligate binding partner, Pasha) to recognize and bind stem-loops, presenting pri-miRNAs to the compound active site such that a typical pre-miRNA (a stem-loop containing dsRNA of about 70-nt) is measured out and produced (see Fig. 1).
3. Twists on the ‘traditional’ dsRNA binding domain: implications for pri-miRNA processing
The dsRBD family is a conserved dsRNA-specific binding domain, of which a number of crystal and NMR structures have been solved (reviewed in [10,11]). They are presented briefly here to contrast their mode of dsRNA binding with that of the more novel modes of RNA binding in the RNAi pathway, and to discuss a recent structure of a hairpin binding dsRBD.
dsRBDs are ∼70 amino acid domains characterized by a αβββα fold, where the α-helices pack against one side of the antiparallel β sheet. In a typical dsRBD, Xenopus dsRNA binding protein A [12] (Fig. 2A), the protein–RNA interactions span two minor grooves and one major groove. The N-terminal helix contacts the 2′-hydroxyls of the sugars and functional moieties of the RNA bases in the minor groove both directly and through water. The β sheets themselves make no contacts with the RNA, but a loop interacts extensively with 2′-OH and RNA bases in the second minor groove. The C-terminal helix interacts primarily with the phosphodiester backbone of the intervening major groove. Most of the contacts with RNA bases are mediated either through water, or through moieties that are equivalent in all four bases, and therefore are sequence independent [12].
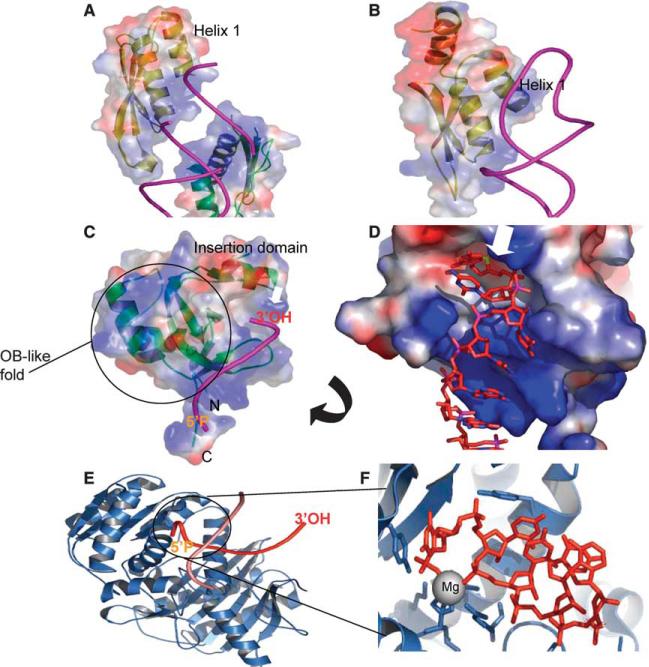
Fig. 2.
Novel modes of RNA recognition. (A) A typical dsRBD: Xenopus binding protein A (1DI2). A RNA helix is modeled pink, and the protein is rendered in transparent electrostatic contours (blue is basic, red acidic). Note the interaction of helices along the major groove, and the position of helix 1. A second dsRBD protein is visible, in the lower right. (B) A dsRBD, Saccharomyces Rnt1P (1T4L), recognizes hairpin loops. A novel third helix (top) pushes helix one into the loop of a hairpin RNA. (C) 3′-OH recognition by PAZ. Human Eif2c1 (1SI3) bound to RNA (pink) is shown. PAZ is green, with transparent electrostatic surface plot. The OB-fold (nucleotide binding fold) and the insertion domain are labeled. Note the glove-and-thumb like cleft they form, that the 3′-OH is inserted into. A basic groove (blue) the RNA binds along outside the cleft is visible. (D) A close-up view of PAZ, as in C (surface not-transparent, slightly rotated). See white arrows for orientation, and location of 3′-OH binding site. RNA is shown red in sticks. The terminal –OH is barely visible, buried in a cleft. It and the carbon it bonds have been colored yellow for clarity. (E) The PIWI domain (2BGG). Note the insertion of the 5′P red (labeled) into the binding site. Its complimentary strand (pink) is not annealed to it, and the 3′ overhang and first complimentary bases sit on the protein surface. (F) An enlarged view of (E), with protein in slate and RNA modeled as red sticks. The coordinated magnesium is a grey sphere, which is coordinated by the terminal carboxylate of the protein, protein side chains, and RNA phosphate oxygens. The 5′ base stacks against a conserved Tyr. Several other sidechain contacts are shown.
The most recent dsRBD structure reveals a unique additional C-terminal helix, which allows Rnt1p RNase III to bind hairpin structures [13]. This helix does not contact the RNA directly, but instead packs against the first helix of the dsRBD, forcing it into a conformation that favors binding short hairpins (Fig. 2B, compare the position of helix-1 with that of Fig. 2A). Drosha, or the associated double-dsRBD protein Pasha/DGCR8 (reviewed in [14]) that resides in complex with it, could employ a similar mechanism to recognize longer loops. This could account for Drosha's substrate specificity (requiring a hairpin dsRNA with a loop of greater than 10-nt versus other dsRNA [15]), and product specificity of generating ∼70-nt hairpin containing pre-miRNAs with 3′ 2-nt overhangs. In this model, Pasha/DGCR8 recognizes and binds the >10-nt loop of pri-miRNAs, orienting the RNA and contacting the RNase III domain such that the active site is positioned to cleave ∼30 base pairs from the 10-nt loop, yielding the 70-nt product (see Fig. 1A). The dsRBD of Drosha could bind dsRNA emerging out the other side of the RNase III cut site. Alternatively, the role of the two could be inverted.
This model is consistent with experimental evidence of Drosha/Pasha function. Drosha itself shows weak, non-specific RNase III activity, but full and specific activity in vitro requires only addition of Pasha [16]. Pasha is thought to bind the RNase III domains of Drosha [17], placing it in position to present the pri-mRNA to the catalytic site. Current biochemical data suggest Pasha/Drosha complexes not only recognize and measure from the pre-miRNA loop, but also require a dsRNA extension beyond the cleavage site, suggesting the dsRBD of Drosha needs to bind a dsRNA ‘tail’ extending from the RNase III domains for maximum activity [15]. Lastly, a recent study suggests that the Arg-rich domain of Pasha contacts ssRNA outside the hairpin, and that this contact is required for full activity. This may further select for primary miRNA transcripts, by requiring recognition of a single-stranded region, and potentially allows Pasha to scan along ssRNA transcripts for long hairpin structures [18]. Testing of this model awaits structural solutions of Pasha/Drosha complexes, in particular, with pri-mRNA bound.
4. The PAZ domain: discriminatory binding of 3′ ssRNA overhangs
PAZ, a 110 amino-acid domain found in Dicer and Argonaute proteins, is most notable for its ability to selectively bind 3′ 2-nt overhangs (first proposed by [19,20]). Zhang et al. have suggested that Dicer's PAZ and RNase III domains act as a molecular ruler to precisely measure out ∼21-nt siRNAs for cleavage (see Fig. 7 in [8]). Although this model awaits Dicer structures, multiple structures of the PAZ domain from Argonaute proteins explain why it is an attractive domain to assign this function. Contributing to our structural understanding of the PAZ domain are the solution structure of Ago1 PAZ [20], and the NMR [21] and crystal structures [19] of Ago2 PAZ. A crystal structure of human Eif2c1 PAZ in complex with a 9-nt dsRNA with 3′ 2-nt overhang [22], and the solution structure of Ago2 PAZ with single-stranded 5-nt RNA and DNA oligonucleotides [23] have revealed the physical basis for single-stranded 3′ end recognition by PAZ.
The central domain of PAZ is a left-handed twisting β-barrel of 5 or 6 β-stands, capped on one end by two α-helices (Fig. 2C). A conserved subdomain composed of a β-hairpin followed by a short helix inserts into a loop between two strands of the β-barrel. Interestingly, the PAZ domain ends where it begins, with the N- and C-termini forming an anti-parallel β-sheet. The central domain resembles the OB-fold [24] found in nucleic acid binding proteins, pointing to the function of the PAZ domain. In this case, the nucleic acid binding face shared with the OB-fold faces across a cleft towards the β-hairpin/α-helix subdomain not seen in OB-fold proteins. This portion of the molecule contains most of the conserved residues of the PAZ domain, and the cleft is lined with aromatic and basic residues (see Fig. 2C and D for electrostatic surface plots). Using NMR titration and binding assays along with mutation of conserved residues, it was confirmed the cleft region is responsible for nucleic acid binding [19–21]. In particular, mutation of PAZ cleft Arg, Lys, Phe, and Tyr residues substantially reduces RNA binding [20,22].
Solution structures of Ago2 PAZ in complex with 5-nt single-stranded DNA and RNA [23], and a crystal structure of human Ago Eif2c1 PAZ in complex with 9-mer RNA with 3′ 2-nt overhangs [22] detail the molecular basis for PAZ recognition of single-stranded 3′ RNA ends. The PAZ domain showed no significant structural rearrangement upon nucleic acid binding. Interestingly, the nucleic acids adopt different conformations in each structure. The 3′ end of each nucleic acid binds deep into a conserved hydrophobic pocket between the β-barrel central domain and the β-hairpin/α-helix insertion subdomain. Recognition of the 3′ end is primarily by steric exclusion, as a paired base, or terminal phosphate group would not be accommodated. (Fig. 2D), consistent with findings that the PAZ domain binds ssRNA and duplex siRNAs with 2-nt 3′ overhangs [19–21] with micromolar affinity, but binds blunt or 5′ overhang dsRNAs quite poorly [19,21]. In the human Ago Eif2c1 PAZ/siRNA mimic structure, the terminal nucleotide is essentially saturated with contacts. In particular, its base stacks against a conserved Phe with one face, and makes a contact with a hydrophobic side chain with the other face. Mutation of either residue strongly reduces RNA binding. Both the phosphodiester backbone preceding the terminal nucleotide and the terminal sugar ring are extensively hydrogen bonded [22]. A few contacts in this pocket may explain the preference of PAZ domains for RNA versus DNA [21], as is demonstrated by the different conformations of the RNA versus DNA oligonucleotides induced when a backbone amine designated to hydrogen bond with the 2′-OH of the terminal ribose instead bonds with a DNA base [23]. The different locations of this and subsequent nucleotides in the ssRNA, ssDNA, and siRNA mimic may reflect variability in nucleic acid binding in the PAZ domain, or a potential mechanism for discrimination of ssRNA from dsRNA. Following the 2-nt binding pocket, the nucleic acids take a greater than 90° turn and follow the PAZ domain cleft. Binding along the cleft is primarily by electrostatic interactions with the phosphate backbone of the strand whose 3′ end is buried. Contacts with the complimentary strand are notably absent.
In considering the role of PAZ in Dicer function, it should be noted that Dicer PAZ domains contain a 30-residue Arg and Lys rich loop (7 conserved between human and Drosophila Dicer) insertion in a loop near the nucleic acid binding cleft (see sequence alignment in [19]). This loop could contribute to binding the complimentary strand that is not engaged in the Ago PAZ cleft. A model of PAZ function in Dicer is shown in Fig. 1A (after Fig. 7 of [8]). In this model, a long dsRNA, or pre-miRNA binds PAZ at one end. PAZ recognizes the 3′ 2-nt overhang generated by Drosha, or the last cut made by Dicer. PAZ and RNase III are spaced such that when the 3′ overhang is bound, they act as a ruler to position the cut site exactly 22-nt away from the ends, activity seen in “3′ end-counting rule” elucidated in biochemical assays [25]. The dsRBD may aid in non-specifically binding incoming substrates, and potentially allow Dicer to cleave long dsRNA substrates processively. This model attractively fits several key observations. Dicer requires free ends, thereby cleaving from the ends of dsRNA and pre-miRNAs, shortening the substrate consecutively by siRNA-sized segments, and not cutting internally, generating multiple larger fragments [3,8,25,26]. This activity also suggests an enzyme with a single processing center. Additionally, both the PAZ and dsRBD domains are required for full Dicer activity. Intact dicer can slowly process blunt-ended dsRNA, however deletion of the dsRBD makes Dicer more dependent on substrates with 3′ overhangs [8]. This suggests the dsRBD contributes to non-specific binding of substrates, perhaps allowing a first ‘proper end’ to be generated on blunt dsRNA substrates. From this initial cut (which in pre-miRNAs is preexisting due to Drosha activity), subsequent ends are recognized by PAZ and rapidly cleaved. This proposal explains the observation of Lee et al. [3], who observed that the combination of Drosha and Dicer in processing of pri-miRNAs yielded a more heterogeneous population and greater yield of mature miRNA than dicer processing alone. This model does not explain the role of the helicase domain in Dicer. Based on other RNase III enzymes, and biochemical assays, it is unlikely ATP is required for catalysis itself. It has been suggested the helicase contributes to unwinding complex secondary structures in dsRNAs, processivity along dsRNA, or product release (see discussion in [26] for a summary of relevant data). Again, it should be noted the structural insights we have on the PAZ domain are in the context of Argonaute proteins, as no Dicer structures have, at this time, been solved.
5. Crystal structure of a full-length archaeal Argonaute
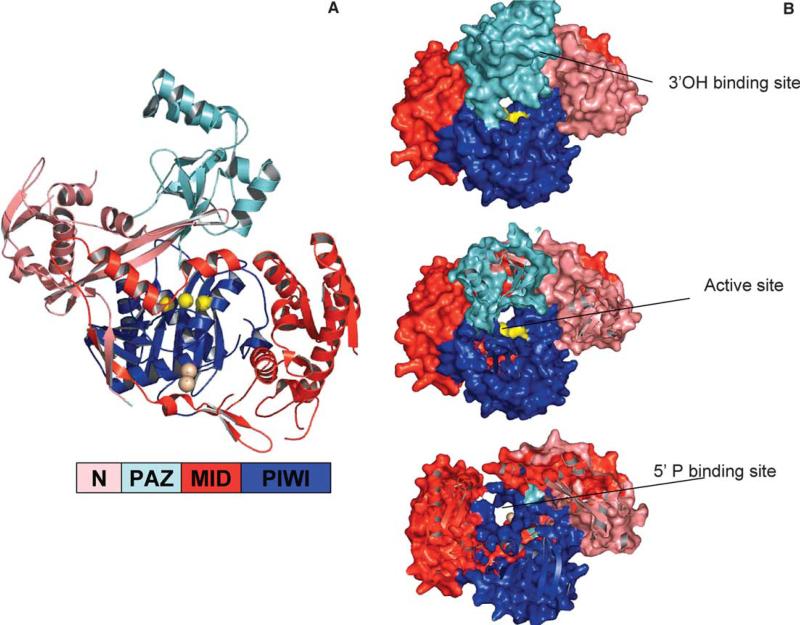
Guided by the structure of Pyrococcus furiosus Argonaute (PfAgo) [27], it has been demonstrated that human Argonaute2 possesses the Slicer activity of RNAi [28,29]. Further, this activity can be generated in vitro using only two components: recombinant Argonaute and an siRNA [29]. Argonaute is composed of four domains: a N-terminal domain, the PAZ domain, the “middle” Lac-Z like domain, and the PIWI domain (Fig. 3A). The N, Middle and PIWI domains form a bowl-like structure, over which the PAZ domain sits. The ∼100 amino acid N-domain consists of an α/β structure of one sheet and several helices, followed by a three-strand anti-parallel β sheet, upon which the PAZ domain sits. The PAZ domain is similar to other PAZ structures, except the insertion domain in the OB-fold is composed of two helices rather than a hairpin followed by a helix. Despite this difference, and large differences at the primary sequence level, residues of importance are conserved at the structural level, occupying similar spatial positions. The PAZ RNA binding cleft runs towards the bowl created by the other domains. The PIWI domain, described in detail in the next section, sits centrally in the bowl structure, flanked by the N and middle domains. Similar in fold to RNase H [27], PIWI harbors Slicer activity in a centrally located active site that faces across a cleft toward the PAZ domain (Fig. 3A). Putative guide-strand 5′ phosphate binding sites and RNA binding grooves are also located in the N−/Mid/PIWI bowl.
Fig. 3.
Argonaute/RISC. (A) P. furiosus Argonaute (PDB 1Z26). A color-guided key to the domains is presented. PAZ sits over the PIWI/N/MID bowl and active site. The liganding atoms for the catalytic metal are depicted as yellow balls for clarity. The tungstate binding site (5′P surrogate) is shown as tan spheres. (B) A guide strand channel. Looking down from the PAZ domain towards the active site, Z-sections are clipped off. Colors of domains are as in the key in (A). Wrapping down along a basic cleft from the PAZ 3′OH binding site (approximate position labeled), a RNA binding groove passes the active site (yellow), and runs down to the 5′P binding site (tan balls). A second cleft running perpendicular to this one at its entry may accommodate target strand RNA. For more detail, and models of siRNA placed into the grooves, see [27,29].
6. The PIWI domain: 5′ guide strand end recognition and RNase H-like catalytic site
In addition to the P. furiosus Argonaute (PfAgo) structure, three crystal structures of an archaeal PIWI protein (AfPIWI) have been solved: one unliganded [30], one with a 16-nt dsRNA with 2-nt 3′ overhang [31], and one with a 21-nt siRNA-like molecule [32]. In common, AfPIWI and PfAgo share the PIWI and ‘middle’ domains, but AfPIWI lacks a PAZ domain. AfPIWI's ‘middle’-homologous domain consists of β/α repeats, yielding a parallel β-sheet bracketed with two parallel α-helices on each side, reminiscent of the structure of Lac repressor (Fig. 2E). Like RNase H, PIWI consists of a conserved core of a 5-stranded β sheet surrounded by three helices [27]. In AfPIWI and PfAgo, an insertion in this domain links it to the rest of the molecule. The classic RNase H activity is cleavage of the 3′ O-P of the RNA strand in RNA–DNA hybrids, utilizing a divalent metal chelated by acidic residues [33]. There is still some controversy as to the number and function of metals in catalysis, but it seems that normal catalysis requires one metal (see [33] for a potential resolution of the dispute). Two invariant Asp residues of the PIWI domain are spatially conserved with those of RNase H, suggesting the location of the active site. In PfAgo, a metal was observed at the putative active site following soaking of Mn+ into the crystals. The metal is liganded by the two conserved Asp, and the third ligand of the catalytic metal was identified a His [29], in contrast to the Glu used by a typical RNase H. The metal-coordinating residues are not conserved in the catalytically inactive Ago1 or Ago4 (for a thorough review of non-catalytic RISCs, see [2]). Mutation of the catalytic-site His or of either Asp [29] yields inactive Argonaute2.
In the AfPIWI structures, a potential 5′ phosphate binding site and RNA binding channel lined with basic and polar residues was observed. It was confirmed AfPIWI binds siRNA, and two AfPIWI structures with RNA bound reveal molecular details of protein–RNA interactions [31,32]. In general, the 5′ phosphate of the guide strand is buried in a conserved pocket (Fig. 2E and F) generated by the Mid and PIWI domains. The 3′ overhang and the base that should pair with the 5′ end of the guide strand are not accommodated in this pocket. Rather, the 3′end has melted off, and sits out on the protein surface without being specifically contacted (Fig. 2E, light pink strand). The rest of the RNA molecule adopts a helical conformation, following a basic channel between the Mid and PIWI domains. Interestingly, like in the PAZ domain structures, the cognate strand makes few contacts with the protein. The position of the 5′ phosphate places the guide strand in an acceptable position relative to the putative active site in AfPIWI.
In more detail, the 5′ phosphate binding site formed by the coordination of a metal by the carboxylate of the C-terminus, along with three other protein-derived oxygens, a water, and oxygens from the phosphates of the first and third residues of the RNA (Fig. 2F). C-terminal residues of the PIWI domain are invariably hydrophobic, allowing this pocket to form by burial of C-terminal side chains in the face of the protein. Addition of a single Gly to the C-terminus reduces siRNA binding [30]. At least two Lys, a Tyr, and Gln, all invariant residues, also coordinate a network of hydrogen bonds with the 5′ phosphate. The 5′ base stacks with the aromatic ring of a conserved Tyr (Fig. 2F). Electrostatic and hydrogen bonds contact the phosphodiester backbone along the cleft. No sequence-specific contacts (or even contacts with 2′-OH groups) are made.
The PIWI domain of PfAgo also specifically binds 5′ phosphates of siRNA guide strands. To identify a 5′ end-binding site in PfAgo, tungstate was soaked into the molecule. A site consisting of several lysines was identified [29]. The putative 5′ phosphate binding site (tan spheres in Fig. 3A) identified is not the same as that found in AfPIWI structures. It sits further back along a basic groove, in the “PIWI box”, which, incidentally, has been shown to bind Dicer [34]. Whether the difference in 5′ end binding reflects a bona-fide difference between the AfPIWI and PfAgo proteins, an intermediate site used in RISC assembly, or something else awaits the structures of Argonaute with various RNAs bound.
7. Modeling in the RNA: guide strand placement and target strand recognition
RNA binding grooves are apparent in the PfAgo structure. One that could accommodate the guide strand follows along the PAZ domain as in the PAZ structures, and then down into the cleft between the PAZ and the bowl, and across the active site. A dsRNA modeled into this groove, starting with its 3′ end bound in the PAZ domain, positions the center of the guide strand over the active site, in a position that would ensure proper cleavage of the target strand-about 11-nt away from the 3′ end (see electrostatic plots and siRNA models in [27]). Additionally, when a dsRNA is modeled with into a RNA binding groove leading from the 5′ phosphate binding site to the active site, the position of the guide RNA to the active site is appropriate (about 10-nt) (see [29]). Taken together, the distance is ∼21nt. A readily visible path is represented in the Z-section of Fig. 3B, where looking down from the 3′ end-binding PAZ domain, a channel is apparent that crosses the active site (yellow) and ends at the PIWI 5′ end-binding site (tungstate marked as brown spheres). Other potential binding clefts (running across the bowl) may accommodate incoming target RNAs (see [30]).
Of particular interest in the structures to come will be the conformations of the protein and RNA in a Ago-ssRNA guide strand complex compared to Ago-dsRNA complexes with extensively paired and poorly paired target RNAs. In Slicer-competent RISCs, only extensive base pairing brings about strand cleavage, while less complementarity brings about a translational block [2,35]. Slicer activity is only licensed when the guide strand pairs with the target strand for at least a complete turn of the RNA helix (nt 2−12 from the 3′ end of the guide strand) [35]. Two interesting (and not mutually exclusive) scenarios can be considered. Argonaute could restrict access to the active site, such the guide/target strand hybrid must adopt a tight RNA helix to be cut, while bulgy mismatched dsRNA would be mispositioned relative to, or excluded from the catalytic site. Alternatively, the strong anchoring of the 5′ and 3′ end of the guide strand may allow the guide strand itself to serve as the sensor for extensive dsRNA helix formation. dsRNA helices are quite rigid [36], and the pull of the RNA on the PAZ and PIWI domain upon helix induction could license Slicer activity by a conformational change in Argonaute. Interestingly, despite the lack of contacts with the non-guide strand, and even the melting off of the complimentary strand end in a PIWI structure, both PAZ and PIWI bind dsRNA. In fact, PAZ binds dsRNA with 50-times higher affinity than ssRNA [22], perhaps owing to placement of backbone phosphates in a better conformation to bind electrostatic and polar grooves. Resolution of this question may shine light on double-stranded intermediates of RISC loading, and licensing of RISC slicer activity.
8. Defeating RNA silencing: viruses strike back
The anti-viral role of RNA silencing in plants was appreciated at the discovery of siRNA [37], and demonstrated in invertebrates shortly thereafter [38]. Recently, it has been demonstrated that several mammalian viruses encode proteins that block siRNA, including LaCrosse virus (encephalitis/meningitis) [39], poxviruses and influenza [40], among others [41,42]. When RNAi is blocked in human cells by expression of a plant-virus protein p19 (VP19), the growth of a primate virus is significantly enhanced, demonstrating a role for RNA silencing in antiviral defense in vertebrates [42].
With the view that RNA silencing is a conserved and ancient eukaryotic defense mechanism against viruses, transposons, and other molecular parasites, it is not surprising that viruses have developed multiple mechanisms to suppress it. For a comprehensive analysis of viral suppressors of RNA silencing, see [43], and the review by Morris and Qu in this issue. Here, the simplest mechanism, the direct sequestration of dsRNA and siRNA, is considered.
VP19 is not only necessary for tombus virus virulence in plants (Silhavy, 2002) but is a universal suppressor of RNAi: its overexpression in Drosophila [44] or human [42] cells blocks RNA silencing. VP19 specifically binds duplex siRNAs with quite high affinity (Kd = ∼0.17 nM) [45], preventing their unwinding and incorportation into RISC [44,46]. Two crystal structures of VP19, one from tomato bushy stunt virus (TBSV) [47], the other from Carnation Italian ringspot virus (CIRV) VP19 [45] have been solved, both in complex with 21-nt siRNA. VP19 binds siRNA as a homodimer (Fig. 4A), contacting the siRNA centrally with an 8-strand (4 from each monomer) concave β sheet and bracketing the end of the siRNA with two α helices (one from each monomer). In each monomer, three helices flank the non-RNA binding face of the β sheet, forming the core of the protein. Between this central core and the ‘bracketing’ helix of each monomer are flexible loops, but the helix is docked onto the core by side-chain electrostatic interactions. The dimer is formed by the C-terminal α helix of each monomer through salt bridges and hydrophobic contacts, and by anti-parallel hydrogen bonding of the preceding β sheet. In total, an otherwise solvent-accessible face of 1300Å2 is buried in the dimer interface [47].
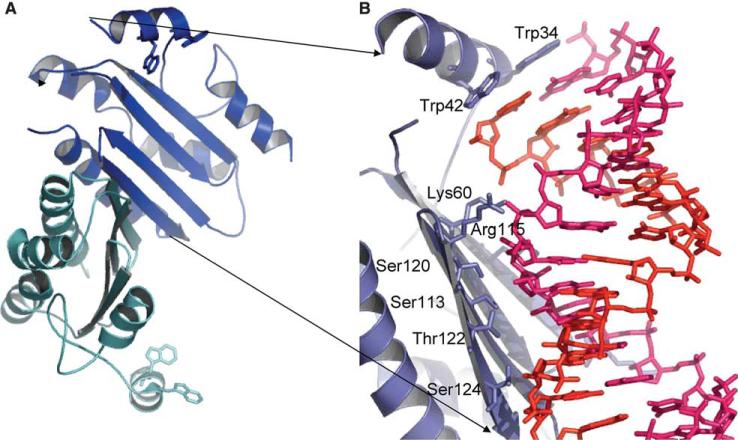
Fig. 4.
VP19 sequestration of siRNA. (A) CIRV VP19 (1RPU, RNA removed). Two monomers (blue and cyan) form an 8 strand, concave β-sheet with bracketing helices at the ends. (B) Tombusviral VP19 bound to siRNA (1 monomer shown). RNA strands are modeled as sticks, with one strand pink and one red. The bracketing helix places two tryptophans in position to stack over the terminal RNA bases. On the β-sheet surface, and Arg and a Lys interact with the phosphate backbone, and at the center of the RNA binding surface, a number of Ser and Thr mediate an extensive hydrogen bond network. Both the Trp brackets and RNA binding by an extended β-sheet are unique.
No sequence-specific interactions are seen in the structure. The central β sheet makes at direct contacts with phosphate groups via basic or polar residues (Fig. 4B). Extensive contacts with 2′-OH groups account for VP19 specificity for RNA versus DNA [46]. A number of serine and threonine residues in the central portion of the β-sheet coordinate a network of direct and water-mediated hydrogen bonds.
The ‘bracketing’ helix from each monomer contains a pair of tryptophans that stack with the 5′ and 3′ bases of each strand of the siRNA (Fig. 4B), serving as a molecular ruler, sizing out shorter (the Kd for a 19-nt is 320X weaker) or longer (the Kd for a 26-mer is 75X weaker) dsRNAs [45]. This gives VP19 a high specificity for 20−22 nt siRNA, though the 3′ 2-nt overhang is not essential for binding [45,47]. The tryptophan bracket is a unique sizing mechanism for dsRNA binding, distinct from typical dsRNA binding domains, which bind as monomers, generally with helical and loop regions interacting with the sugar-phosphate backbone. This perhaps owes to the existence of two classes of plant siRNAs with distinct function [48]. Sequestering short (∼21 nt) siRNAs that would have an antiviral function while sparing longer (∼26 nt) siRNAs required for epigenetic maintenance of the host genome may be beneficial to certain plant viruses. However, the mechanism of VP19 may be very specific; indeed, the closest characterized relative of VP19, VP14, an Aureusvirus RNA silencing suppressor, binds dsRNA without size selectivity. Indeed, viruses reply to vertebrate defenses with length-independent dsRNA binding proteins that dually sequester siRNAs and longer dsRNAs. The E3L protein of poxviruses (utilizing a typical dsRBD) and NS1 of influenza (through a novel RNA binding domain [49,50]) act as interferon antagonists and suppressors of RNA silencing. This dual role has been demonstrated to be dependent on their dsRBD(s) [40]. Recently, other viral virulence factors have been shown to block RNAi by binding both long dsRNA and genuine siRNAs [41]. The dsRNA ‘coating’ function seems to allow mammalian viruses to simultaneously block RNAi and to avoid triggering the interferon system of innate immunity [51] by sequestering cellular dsRNA. Although the structure of VP19 is known, and the E3L protein from poxvirae contains a canonical dsRDB, many of the viral suppressors of RNAi may constitute novel RNA binding folds, as neither their primary sequence, nor secondary sequence prediction correlates with known dsRBD folds.
9. Conclusions
Structural studies have shed light on the conserved core of the RNA silencing pathway. Models that have been formulated based on structures from related proteins await verification. Drosha/Dicer structures will be critical in defining the actual modes of substrate recognition that allow for such precise processing of RNAs. Structures of Argonaute in complex with guide and target strands bound will define the mechanism that allows Slicer activity to be dependent on extensive target strand complementarity. In the longer term, biochemical and structural analyses will define the actions of the RISC-loading complex, key accessory proteins to the pathway, and the diverse functions of RISC complexes.
Acknowledgments
Our apologies to all studies not included, particularly the wealth of dsRBD structures that were not discussed. Structural figures were generated with Pymol [52]. R.E.C was supported by a training Grant T32 GM008367 and is currently supported by U.S. Public Health Services Grant GM068680 (to X.C.).
References
- 1.Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. doi: 10.1016/j.tcb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 5.Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure (Cambridge) 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 6.Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure (Cambridge) 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 9.Sun W, Li G, Nicholson AW. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry. 2004;43:13054–13062. doi: 10.1021/bi049258i. [DOI] [PubMed] [Google Scholar]
- 10.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 11.Carlson CB, Stephens OM, Beal PA. Recognition of double-stranded RNA by proteins and small molecules. Biopolymers. 2003;70:86–102. doi: 10.1002/bip.10413. [DOI] [PubMed] [Google Scholar]
- 12.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leulliot N, Quevillon-Cheruel S, Graille M, van Tilbeurgh H, Leeper TC, Godin KS, Edwards TE, Sigurdsson ST, Rozenkrants N, Nagel RJ, Ares M, Varani G. A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J. 2004;23:2468–2477. doi: 10.1038/sj.emboj.7600260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can't cut it without a partner. Curr. Biol. 2005;15:R61–R64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking non-structured RNA sequences. J. Biol. Chem. 2005 doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 19.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 20.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 21.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 22.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 24.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 29.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 30.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keck JL, Goedken ER, Marqusee S. Activation/attenuation model for RNase H. A one-metal mechanism with second-metal inhibition. J. Biol. Chem. 1998;273:34128–34133. doi: 10.1074/jbc.273.51.34128. [DOI] [PubMed] [Google Scholar]
- 34.Kolb FA, Zhang H, Jaronczyk K, Tahbaz N, Hobman TC, Filipowicz W. Human dicer: purification, properties, and interaction with PAZ PIWI domain proteins. Meth. Enzymol. 2005;392:316–336. doi: 10.1016/S0076-6879(04)92019-8. [DOI] [PubMed] [Google Scholar]
- 35.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 36.Perez A, Noy A, Lankas F, Luque FJ, Orozco M. The relative flexibility of B-DNA and A-RNA duplexes: database analysis. Nucleic Acids Res. 2004;32:6144–6151. doi: 10.1093/nar/gkh954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 39.Soldan SS, Plassmeyer ML, Matukonis MK, Gonzalez-Scarano F. La Crosse virus nonstructural protein NSs counteracts the effects of short interfering. RNA. J Virol. 2005;79:234–244. doi: 10.1128/JVI.79.1.234-244.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in Mammalian cells. J. Virol. 2005;79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 43.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 44.Lakatos L, Szittya G, Silhavy D, Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 46.Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21-to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chien C-Y, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat. Struct. Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Lynch PA, Chien C-Y, Montelione GT, Krug RM, Berman HM. Crystal Structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 51.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. Available from: http://www.pymol.org. [Google Scholar]