Abstract
The MRN complex, composed of MRE11, RAD50 and NBS, plays important roles in responding to DNA double-strand breaks (DSBs). In metazoans, functional studies of genes encoding these proteins have been challenging because complete loss-of-function mutations are lethal at the organismal level and because NBS has multiple functions in DNA damage responses. To study functions of Drosophila NBS in DNA damage responses, we used a separation-of-function mutation that causes loss of the forkhead-associated (FHA) domain. Loss of the FHA domain resulted in hypersensitivity to ionizing radiation and defects in gap repair by homologous recombination, but had only a small effect on the DNA damage checkpoint response and did not impair DSB repair by end joining. We also found that heterozygosity for an nbs null mutation caused reduced gap repair and loss of the checkpoint response to low-dose irradiation. These findings shed light on possible sources of the cancer predisposition found in human carriers of NBN mutations.
Keywords: double-strand break repair, NBS, checkpoints, haploinsufficiency
1. Introduction
Cells have evolved multiple ways to preserve genome integrity. Successful genome maintenance ensures high-fidelity transmission of genetic material to daughter cells. This maintenance can be interrupted by DNA damage, which can be incurred either exogenously by genotoxic agents or endogenously due to metabolic errors. DNA double-strand breaks (DSBs), because they involve both strands, are highly toxic if unrepaired or repaired inaccurately. Mechanisms for repairing DSBs fall into two categories. In homologous recombination (HR), information from a homologous template is used to repair the DSB accurately. In contrast, in non-homologous end joining (NHEJ) the broken ends are joined with little or no use of homology. Cells lacking proteins required for either of these repair types manifest genome instability; in mammals, this can lead to cancer [1–3].
Prior to activation of repair, the DNA damage response is initiated through a cascade of events. Sensor proteins recognize the damage and trigger transducers, including checkpoint proteins that arrest the cell cycle. This is thought to allow time for repair processes to act. A given DNA damage response protein may be involved in one or more aspects of the response. A good example is the MRN/MRX complex, composed of MRE11, RAD50, and NBS (Xrs2 in S. cerevisiae), which participates in damage sensing, checkpoint activation, and DNA repair [4,5].
Null mutations in genes encoding MRN components are lethal in metazoans [6–8]. While lethality illustrates the importance of these genes, it also poses challenges for in vivo studies of their molecular functions. Nonetheless, substantial progress has been made in characterization of the component proteins of the MRN complex and of functions of the complex as a whole. The MRE11 subunit is an exonuclease [9,10]. Studies in S. cerevisiae have demonstrated that MRE11 is essential during repair of meiotic DSBs, but MRE11 is partially redundant with other exonucleases during DSB repair in vegetative cells [11–14]. In humans, mutations in MRE11 cause ataxia telangiectasia-like syndrome (ATLD) [15], which is characterized at the cellular level by chromosome instability.
The RAD50 subunit has important enzymatic and structural functions, including ATPase activity and DNA bridging activity, both of which are required for DSB repair [16–19]. No genetic disorders have been associated with mutation in RAD50, but mouse Rad50 mutants exhibit cancer predisposition and hematopoetic failure [20].
NBS/Xrs2 is the major regulator of the complex [21,22]. This protein has a nuclear-localization signal (NLS), and in its absence the MRN complex remains cytoplasmic even in the presence of DNA damage [6,23,24]. For this reason, functional studies carried out in nbs mutants can be used to understand the nuclear function of the complex as a whole. Specific mutations in NBN (the gene that encodes human NBS) that allow expression of a partially functional protein cause Nijmegen Breakage Syndrome [25–27]. Clinical features manifested by these patients include microcephaly, immunodeficiency, and lymphorecticular malignancies, and cells from these patients are hypersensitive to IR and have defects in DNA damage responses. In addition, linkage analyses have shown that heterozygous carriers of NBN mutations, which may be as frequent as 1 in 150 to 1 in 190 in some populations, are predisposed to several types of cancer [28–32]. Cells from NBN carriers also show gross genome instability. This may be explained by a recent study that found that the checkpoint protein ATM is not activated normally in response to low-dose ionizing radiation in cells heterozygous for an NBN mutation [33].
The MRN/MRX complex has been proposed to function during early steps of DSB repair. Models for meiotic recombination and repair of DSBs by homologous recombination (HR) in mitotic cells require that the 5′ ends of DSBs are resected to produce intermediates with 3′-ended single-stranded overhangs, which are then bound by strand invasion proteins. Meiotic cells from S. cerevisiae mre11 mutants show an accumulation of unprocessed DSBs [13], leading to the proposal that the MRX complex participates in resection. In vitro studies demonstrated both exonuclease and endonuclease activities in the Mre11 protein [34]. It was suggested that coupling of these two activities might be required for resection, since the exonuclease activity had the opposite polarity of that required for resection. More recently, CtIP/Sae2, Exo1, and BLM/Sgs1 have been found to have important roles in resection, sometimes acting redundantly [35–37].
The role of the MRN/MRX complex in NHEJ remains unclear. Several studies have found a role for MRX in NHEJ in S. cerevisiae [38–41]. Experiments using HeLa cell extracts have implicated MRN in NHEJ [42], and studies of skin fibroblasts from NBS patients found a defect in microhomology-directed NHEJ [43]. In contrast, genetic experiments in the fission yeast Schizosaccharomyces pombe, studies with cell-free extracts from Xenopus laevis, and genetic analysis of chicken DT40 cells all failed to find a requirement for the MRN complex in NHEJ [44–46].
We sought to use Drosophila as another model in which to study roles of the MRN complex in DNA damage responses. We assayed end joining and homologous repair simultaneously in an in vivo assay in mutants with reduced or altered NBS function. We found that either reducing levels of NBS or removing the N-terminal forkhead-associated (FHA) domain caused a defect in gap repair by HR. Among products repaired by NHEJ, loss of the FHA domain was associated with decreased usage of short (1–5 bp) microhomologies and increased usage of a 10-bp microhomology. Reducing the level of NBS also resulted in a profound defect in the DNA damage-dependent cell cycle checkpoint. These finding provides further insights into in vivo NBS functions in metazoans.
2. Materials And Methods
2.1 Drosophila stocks and transgenic constructs
Drosophila stocks were maintained on standard medium at 25°. The null mutation used was nbs1 [6,47]. The hypomorphic allele used here was a P element insertion generated during the Berkeley Drosophila Gene disruption project and is available at the Bloomington stock center (# 21141) y1 w67c23; P{EPgy2}nbsEY15506. The P element is inserted in the 2nd exon of nbs, within coding sequences. A derivative of the P{EPgy2}nbsEY15506 hypomorphic allele was generated for use in assays that involve P transposase. Males carrying P{EPgy2}nbsEY15506 and P transposase were generated and crossed to w− females, and progeny that had lost the w+ marker from P{EPgy2} were screened for the absence of one end of the P element. The derivative we recovered, named nbsSM9, lacks the 3′ P element end and has approximately 3 kb of the original 10 kb P{EPgy2} remaining.
2.2 RNA analysis
For RNA blots, total RNA was isolated from wild type w1118, nbsP, nbsSM9 and nbs1 homozygous larvae by homogenizing 10 larvae in one ml Trizol reagent. This was followed by organic phase-separation and RNA precipitation using standard methods. Polyadenyleated RNA was selected using the Poly(A)Purist kit (Ambion). A probe from the 5′ end of nbs (Fig. 1b) was used to detect nbs transcripts. The structure of the transcripts of nbs mutants was determined with the 5′ RACE system for rapid amplification of cDNA ends, version 2.0 (Invitrogen).
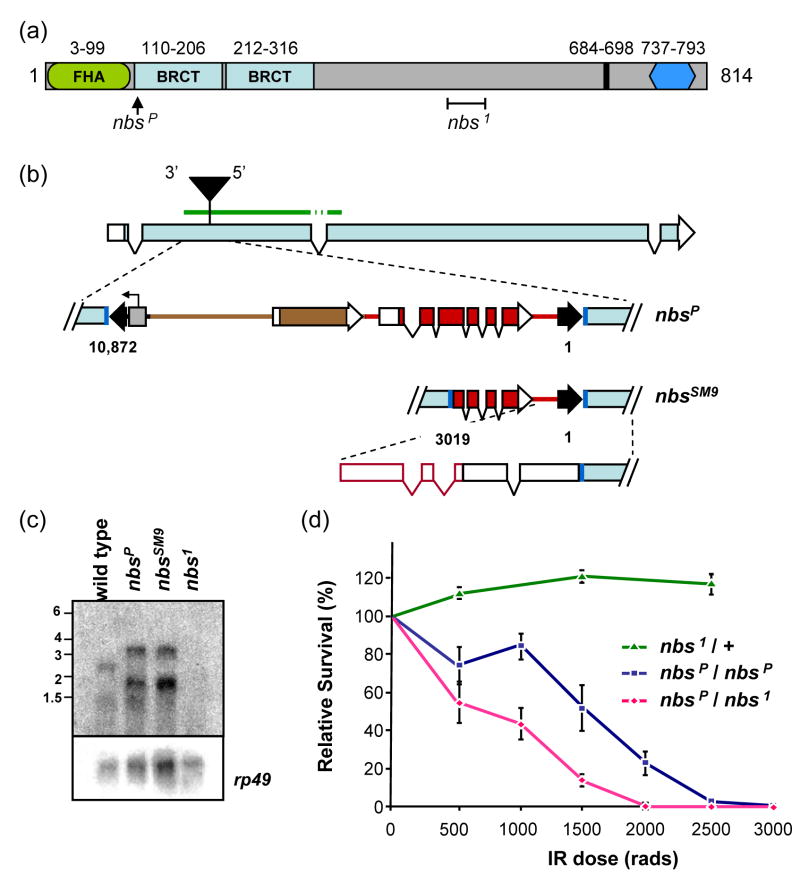
Figure 1. The Drosophila NBS protein and nbs mutations.
(a) The predicted Drosophila melanogaster NBS protein is 814 residues. Conserved sequences include an FHA domain (green oval), two BRCT repeats (light blue rectangles), a putative nuclear localization signal (black line), and the MRE11 binding site (dark blue hexagon). The positions of two mutations are indicated: nbsP (insertion of a P element) and nbs1 (238-bp deletion denoted by a bracket).
(b) Structure of nbsP and nbsSM9 mutations. The nbsP allele is caused by insertion of a 10-kb P element construct into coding sequences in the second exon of nbs. Dark blue lines represent the 8-bp insertion site sequence that is duplicated at each end of the P element. Black arrowheads indicate P element sequences, including inverted repeats at the ends that are required in cis for transposition. This construct carries a y+ body color gene and a w+ eye color gene (brown and red, respectively; rectangles indicate exons; protein-coding sequences filled), and a promoter under the control of an S. cerevisiae UAS sequence (gray box; arrow indicates start and direction of transcription). The P element is inserted such that the 5′ P element end is near the 3′ end of nbs. The nbsSM9 derivative has lost approximately 7 kb, including one inverted repeat, all of y+, and much of the w+ gene, retaining nucleotides 1-3019. The diagram at the bottom of this panel depicts the structure of the transcript determined by 5′ RACE. The green bar above the illustration of the intron/exon structure indicates the position of the probe used to detect transcripts (the probe was from cDNA, so intron and P element sequences were not included).
(c) Transcripts in nbs mutants. A blot of polyA-selected RNA from wild-type and nbs mutant L3 larvae was probed with a fragment from the 5′ end of nbs. Transcripts of about 1200 and 2600 nt are detected in wild-type larvae. In nbsP and nbsSM9 mutants, larger transcripts (1900 and 3300 nt) are seen; no transcripts are detected in the nbs1 mutant. The membrane was stripped and re-probed for rp49 as a loading control.
(d) Sensitivity of nbs mutants to ionizing radiation. An IR sensitivity assay of nbs mutants was carried out at the doses of gamma irradiation indicated on the X axis. Each point is the mean survival of mutants relative to survival of heterozygous siblings (for nbsP/nbsP and nbs1/nbsP) or wild-type siblings (for nbs1/+). Error bars indicate standard deviation (n = 6–14 bottles for each point).
2.3 IR sensitivity measurements
Males and females balanced with TM3, Sb were crossed and allowed to lay eggs overnight at 25° C on grape agar plates. Plates were then changed for a second collection. Second collection plates were incubated for two days, then exposed to gamma irradiation from a 137Cs source; the first collection was used as an un-irradiated control. After irradiation the grape agar with larvae was divided into four sectors and transferred to standard medium in bottles, then incubated at 25° C until adults eclosed. Adults were counted to determine the ratio of mutant to non-mutant. For homozygous and heteroallelic combinations, non-mutants were nbs/TM3, Sb flies, which we had determined were not hypersensitive to IR at the doses used (Fig. 1d). Relative survival of mutants was determined by normalizing the mutant to non-mutant ratio in irradiated bottles to the ratio in unirradiated bottles.
2.4 Double-strand break repair assays
The P{wa} assay has been described in detail previously [48–51]. We counted progeny from multiple vials (n=53 for +/+, n=67 for nbs1/+, and n=94 for nbs1/nbsSM9), each with a single male parent. Each vial was treated as a separate experiment. Statistical comparisons were done for each pair of genotypes, using a Mann-Whitney test done with InStat 3.05 (Graphpad Software, Inc.). Tract lengths were compared through Fisher’s exact test. Junction sequences from repair events were sequenced to understand the mechanism of joining. PCR was carried out with the forward primer 5′-CCCTGTCTGAAGTTCCGTAG-3′ and reverse primer 5′-CCCTCGCAGCGTACTATTGAT-3′, and products were sequenced with the forward primer.
We measured SSA with the assay of Rong and Golic [51]. In this assay, repair by SSA, which is the most common mechanism in wild-type flies, gives white-eyed progeny. We used a PCR assay to distinguish between SSA and deletion formation, which can also give rise to white-eyed progeny. To detect imprecise NHEJ among red-eyed progeny, PCR was conducted using the forward primer 5′-TGTGTGTTTGGCCGAAGTAT-3′ and the reverse primer 5′-CGCGATGTGTTCACTTTGCT-3′. Products were digested in vitro with I-SceI enzyme (New England Biolabs). Those that did not cut were sequenced using the forward primer. Statistical comparisons were done by Kurskal-Wallis non-parametric ANOVA.
2.5 Cell cycle checkpoint assay
We measured the G2/M DNA damage checkpoint using an assay described previously [52]. Wing imaginal discs were dissected out of L3 larvae one hour after exposure to 1000 or 4000 rads of gamma radiation from a 137Cs source. A rabbit polyclonal antibody to phospho-histone H3 (Upstate Cell Signaling Solutions, # 06-570) was used to mark mitotic cells. Mitotic cells were counted from at least seven discs for each genotype and dose. For statistical comparisons, counts were normalized to the mean for unirradiated, then compared in a Welch-corrected unpaired t test, using Instat 3.05 (Graphpad Software, Inc.).
3. Results
3.1 The Drosophila NBS protein and nbs mutations
The structure of the Drosophila NBS protein has been described previously [6,47]. The amino-terminal half of the protein contains a forkhead-associated (FHA) domain and two BRCT domains [53] (Fig. 1a). In other species, these domains mediate phospho-protein interactions and, in some assays, are required for checkpoint signaling [54,55]. There is an MRE11 binding site near the carboxy-terminus. Vertebrate NBS has a nuclear localization signal (NLS) that controls entry of the MRN complex into the nucleus [6,23,24]. Using ScanProsite on the ExPASy database [56], we detected a putative NLS at residues 684–698 of Drosophila NBS. Under similar stringency, no NLS motifs were found in MRE11 or RAD50.
Two genetically null mutations in Drosophila nbs have been characterized previously [6,47,57]. We used nbs1, which is a 238-bp deletion and one bp insertion beginning at codon 507, resulting in a frameshift and premature termination (Fig. 1a). We did not detect any transcripts in nbs1 mutants when we probed an RNA blot with a probe near the 5′ end of nbs (Fig. 1c), suggesting that the mutant mRNA is degraded by nonsense-mediated decay.
We obtained a stock with a 10 kb P element insertion into the second exon of nbs (Fig. 1a and 1b); we refer to this mutation as nbsP. Null mutations in nbs cause lethality at the pharate adult stage (i.e., apparently fully-developed adults that do not eclose from pupal cases). This lethality is believed to be related to chromosome instability resulting from telomere fusions that occur in the absence of MRN [47]. Flies homozygous for nbsP or heteroallelic for nbsP and nbs1 are viable as adults, and we did not detect elevated telomere fusions in neuroblasts of larvae from these genotypes (data not shown). These results indicate that nbsP is not a null allele, despite the fact that the insertion is within protein-coding sequences.
For use in an assay in which DSBs are generated by P transposase, we generated a derivative of nbsP that is stable in the presence of transposase (see Materials and Methods and Fig. 1b). To determine the effects of the insertions in nbsP and nbsSM9 we analyzed transcript structures. We first probed RNA blots with sequences from the 5′ half of nbs. Wild-type flies have a transcript of about 2.6 knt, which is the size predicted by the gene structure (Fig. 1c). A shorter transcript is also detected, but we have been unable to determine its structure through RT-PCR or analysis of the EST database sequences.
In both nbsP and nbsSM9 flies, nbs transcripts are about 700 nt longer than in wild-type flies (Fig. 1c). By 5′-RACE analysis, we determined that transcription in nbsP and nbsSM9 initiate within the P element, just downstream of the 3′ end of the w gene. No genes have been annotated for this region of the genome, and no ESTs have been found to include this sequence. Two introns with canonical splice donor and acceptor sites are removed from the transcript. Transcription continues through the 5′ P element end, in the opposite direction of the native P transposase promoter, and another intron is removed from the P element-derived sequences. Translation is predicted to begin at the last three nucleotides of P element sequence and continue in-frame into nbs sequences (Fig. 1b). In the predicted translation product, the first residue of the first BRCT domain is changed from leucine to methionine, but the domain is otherwise intact. Expression levels of nbsP and nbsSM9 transcripts both appear similar to wild-type. Importantly, the predicted protein produced by the nbsP and nbsSM9 alleles lacks the N-terminal FHA domain.
3.2 Hypersensitivity of nbs mutants to ionizing radiation
In other organisms, nbs mutation results in hypersensitivity to IR [58]. Oikemus et al. [47]found that irradiation resulted in elevated levels of chromosome breaks in Drosophila nbs null mutant larvae compared to wild-type larvae. We used the nbsP and nbsSM9 mutations to assay sensitivity to IR as determined by survival to adulthood (Fig. 1d). Both nbs1/nbsSM9 and nbs1/nbsP are similarly hypersensitive to IR (Fig. 1d and data not shown). However, larvae homozygous for nbsP are significantly less sensitive than nbs1/nbsP larvae at doses higher than 500 rads (two-tailed P <0.005 by unpaired t test, Welch corrected). This result agrees with our conclusion that nbsP and nbsSM9 are hypomorphic alleles. This could be due to reduced expression or activity or to a separation of function resulting from the loss of the FHA domain.
3.3 Synthesis-dependent strand annealing in nbs mutants
We used a P element excision assay [48] to assess the ability of nbs mutants to carry out gap repair by synthesis-dependent strand annealing (SDSA). The P{wa} element used in this assay carries the apricot allele of the white gene (wa), and is inserted into an intron of sd, an essential gene on the X chromosome (Fig. 2a). The wa allele is a copia retrotransposon inserted into an intron of w; the insertion decreases w expression to give apricot-colored eyes instead of red eyes. Excision of the P{wa} element was carried out in males by crossing in P transposase. Excision generates a 14-kb gap relative to the sister chromatid, which is the only template for HR repair. Gap repair in pre-meiotic germline cells occurs largely through SDSA [48]. If synthesis from each end extends through a copia long terminal repeat (LTR), the LTRs may anneal to one another to give a repair product that lacks the copia insertion except for a single LTR. This allows nearly normal expression of w, resulting in red eyes (Fig. 2a). We classify this repair type as ‘completed SDSA’. It is also possible for HR using the sister chromatid to restore the entire P{wa}, probably also through SDSA. Since this outcome is indistinguishable from failure to excise, which accounts for the majority of chromosomes recovered [48], we do not include it in our analysis. SDSA is sometimes aborted prior to synthesis or annealing of LTRs, and repair is completed by NHEJ. This process destroys expression of w, resulting in yellow eyes when the repaired chromosome is recovered in trans to an intact P{wa} (Fig. 2a). Yellow eyes can also result from repair by NHEJ without synthesis. This is rare in wild-type flies, but is common if strand invasion is prevented, as in mutants that lack the Rad51 ortholog of SPN-A [49].
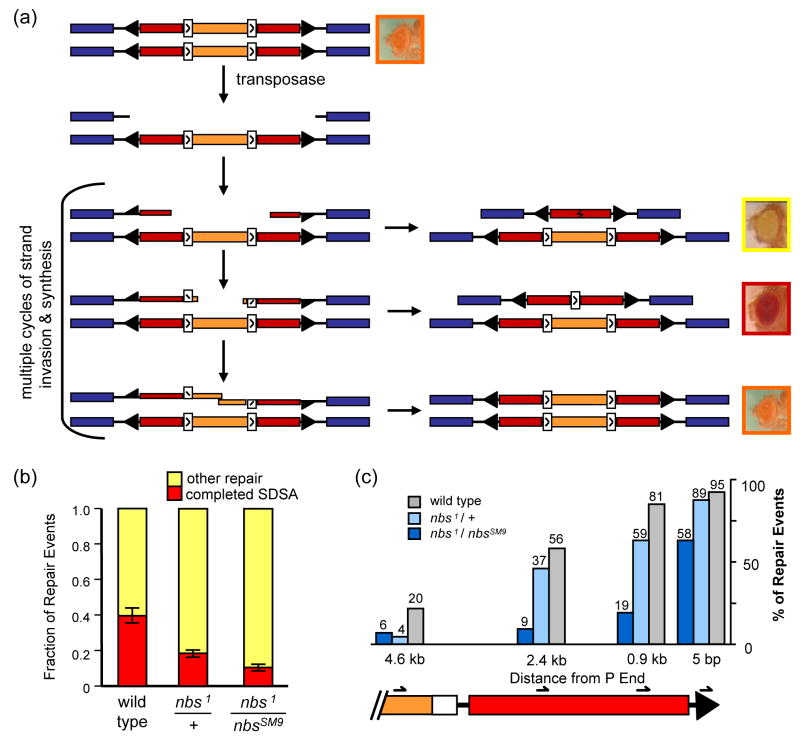
Figure 2. Repair of breaks generated by P excision.
(a) P-element based DSB repair assay. Each line represents a sister chromatid. Blue rectangles denote exons of sd, an essential gene on the X chromosome. A P{wa} element is inserted into a sd intron. Black arrowheads represent P element inverted repeat sequences. Red rectangles are halves of the w gene, which is interrupted by insertion into an intron of a copia retrotransposon (orange) with LTRs (white rectangles). This transgene confers apricot eye color to otherwise w− mutants. Transposase catalyzes excision of the element, leaving a 14-kb gap relative to the sister. This gap is usually repaired by SDSA, with synthesis occurring from both ends independently. In some cases, repair occurs by end joining, with or without synthesis; this leads to loss of w expression and yellow-eyed progeny. If synthesis from both ends extends past the LTRs, these sequences can anneal as in SDSA, leaving a single LTR in the w intron, which allows enough expression of w to give red eyes. It is also possible for the entire gap to be filled, restoring the P{wa} element and giving apricot-colored eyes.
(b) Results from P-element based DSB repair assay. Repair outcome is scored as completed SDSA (red eyes) or other repair (yellow eyes). Bars show the contribution that each class makes to the total of all repair events scored, and error bars indicate standard error of the mean (n=53 vials for wild type, 67 vial for nbs1/+, and 94 vials for nbs1/nbsSM9). Total number of progeny counted and mean percentages of red-eyed and yellow-eyed flies for each genotype are given in Table S1.
(c) Molecular analysis of repair synthesis tract lengths. Repair events from yellow-eyed progeny were analyzed to determine the extent of repair synthesis, if any. The right end of the P{wa} element is shown (coloring as in A). Four PCR reactions were carried out to measure synthesis; the position of the innermost primer used in each is indicated. Bars indicate percentage of events analyzed that had synthesis tracts of at least 5 bp, 0.9 kb, 2.4 kb, and 4.6 kb (n=38 for wild type, 59 for nbs1/+, and 81 for nbs1/nbsSM9). P values were determined by Fisher’s exact test. P<0.0001 for wild type vs. nbs1/nbsSM9; P<0.05 for wild type vs. nbs1/+, except at 5 bp synthesis (P=0.12). P<0.01 for nbs1/nbsSM9 vs. nbs1/+, except at 4.6 kb (P=1). See also Figure S1.
We conducted the P{wa} assay in wild-type males and in nbs1/nbsSM9 and nbs1/+ mutants. The total percentage of progeny representing repair (red-eyed and yellow-eyed progeny) was similar in all three cases (15.5%, 15.7%, and 16.1%, respectively; Table S1). However, the fraction of repair events attributed to completed SDSA (red-eyed progeny) was significantly lower in nbs1/+ mutants than in wild-type flies (Fig. 2b; P=0.0002), revealing haploinsufficiency for nbs in gap repair. SDSA was even less frequent in nbs1/nbsSM9 mutants (P=0.0002 compared to nbs1/+ and P<0.0001 compared to +/+). The difference between nbs1/+ and nbs1/nbsSM9 suggests that the FHA domain has a role in promoting the completion of SDSA during gap repair, although we can’t exclude the possibility that there is less NBS protein from the nbsSM9, thereby decreasing dosage even further in nbs1/nbsSM9 compared to nbs1/+.
To understand repair mechanisms in nbs mutant flies, we analyzed products recovered in yellow-eyed progeny (i.e., those not attributed to completed SDSA). We hypothesized that nbs might be required to process the ends of the break to allow strand invasion and synthesis, and that in nbs mutants there would be an increase in repair events with no detectable synthesis. In agreement with this hypothesis, the number of repair events that did not have evidence for synthesis was significantly increased in nbs1/nbsSM9 (P<0.0001 compared to either +/+ or nbs1/+). Synthesis was not entirely abolished in these flies, however. Six percent of the yellow-eyed class had tracts of at least 4.6 kb, and almost 10% of all repair events in this genotype (those recovered in red-eyed progeny) had extensive synthesis from both ends. The ability to do some synthesis could be due to perdurance of maternal NBS or residual function in the truncated protein produced by the nbsSM9 allele, or it may be that NBS facilitates processing of DSB ends for HR but is not essential.
We also detected decreased synthesis tract lengths in nbs mutants (Fig. 2c and S2). Tracts were significantly shorter in nbs1/nbsSM9 mutants than in wild-type flies for tracts less than 4.6 kb (P<0.0001 in for each length assayed), and in nbs1/+ mutants at the 0.9 kb and 4.6 kb measurements (P=0.026 and P=0.0125, respectively). This finding suggests that Drosophila NBS plays a role in gap repair after the initial end resection. Furthermore, nbs1/nbsSM9 mutants had significantly shorter tracts than nbs1/+ (P<0.0001 in for each length assayed), suggesting that the FHA domain is required for this function.
3.4 Single-strand annealing in nbs mutants
As a more stringent genetic test of the role of NBS in processing DSB ends, we conducted an assay for single-strand annealing (SSA). In this assay (Fig. 3a) a DSB is introduced by the I-SceI enzyme [51]. The I-SceI recognition sequence (I-site) is located between a truncated, 3.6 kb copy of the w gene and a functional, 4.6 kb copy. SSA therefore requires resection at least 3.6 kb to the left and 4.6 kb to the right, and gives a product that retains only a truncated, nonfunctional copy of w. Although SSA is the most frequent repair event from this assay in wild-type larvae [51], we also determined the frequency of deletions and imprecise NHEJ (see Materials and Methods).
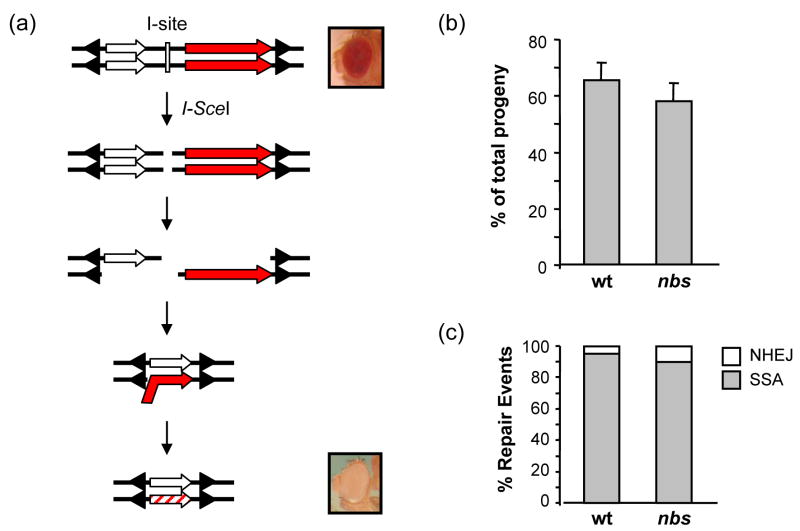
Figure 3. Repair of I-Sce1 induced breaks.
(a) Single-strand annealing assay. A schematic of the P{X97-Isite} construct [51], in double-stranded format. The construct carries an I-SceI recognition site (I-site, white rectangle) flanked by a partial w gene of 3.5 kb (white arrow) and a wild-type w gene of 4.5 kb (red arrow); arrowheads represent P element ends. Expression of I-SceI leads to cutting at the I-site to produce a DSB. Repair by SSA requires resection through both copies of the w gene, followed by annealing of complementary sequences, trimming, and ligation. The product has only a partial w gene, resulting in white eyes.
(b) SSA frequency. The percentage of progeny that inherited a chromosome on which repair occurred by SSA was calculated for each of 20 male parents. Bars show the mean percentages, with error bars showing SEM. There was no significant different between wild-type and nbs1/nbsSM9 larvae.
(c) Distribution of repair events recovered. SSA, deletion, and imprecise NHEJ frequencies were determined as described in Materials and Methods. Bars show the relative distribution of repair events from SSA and from imprecise NHEJ; no deletions were detected among repair events from these genotypes. There was no significant difference in SSA and imprecise NHEJ between wild-type and nbs1/nbsSM9 larvae.
We conducted this assay in the more severe nbs genotype, nbs1/nbsSM9. Wild-type and nbs1/nbsSM9 larvae were heat shocked simultaneously at 38° for one hour to express I-SceI. The frequencies of SSA, deletions, and imprecise NHEJ were not significantly different between nbs1/nbsSM9 and wild-type larvae (Fig. 3b and 3c). Thus, by this assay, we could not detect a function for NBS in SSA.
3.5 End joining in nbs mutants
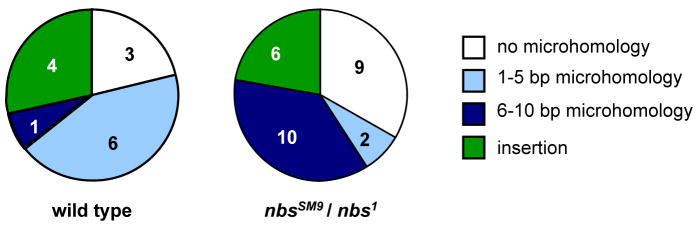
In the two assays described above, we recovered repair products produced by NHEJ. The number of events involving NHEJ was not reduced in nbs mutants, indicating that these mutants are competent to carry out at least some types of NHEJ. To determine whether Drosophila NBS or the FHA domain might be required for specific types of NHEJ, we sequenced repair junctions. In the P{wa} gap repair assay, repair events that do not achieve completed SDSA involve a non-canonical, DNA ligase IV-independent end-joining pathway [59]. In wild-type flies, we recognize four classes of junction structures: junctions without apparent microhomology, junctions with short (1–5 bp) microhomology, junctions with longer (6–10 bp) microhomology, and junctions with insertions [48]. All four junction classes were also seen in repair products from nbs1/nbsSM9 mutants (Fig. 4 and S1). The class with short microhomology was significantly reduced in nbs mutants relative to wild-type larvae (P = 0.012). In contrast, the class with longer microhomology was increased, though the difference was not statistically significant (P = 0.0636).
Figure 4. Comparison of junction types from end joining.
Pie charts indicate relative numbers of each type of junction from wild-type and nbs1/nbsSM9 larvae. Numbers in each slice are actual numbers of the corresponding class. The differences between wild-type and nbs mutant are significantly different only for the 1–5 bp microhomology class (two-tailed P=0.012 by Fisher’s exact test). Data for wild-type are from Adams et al. [48]. Junction sequences are listed in Supplemental Table S1.
3.6 Haploinsufficiency of nbs for the DNA damage-dependent checkpoint
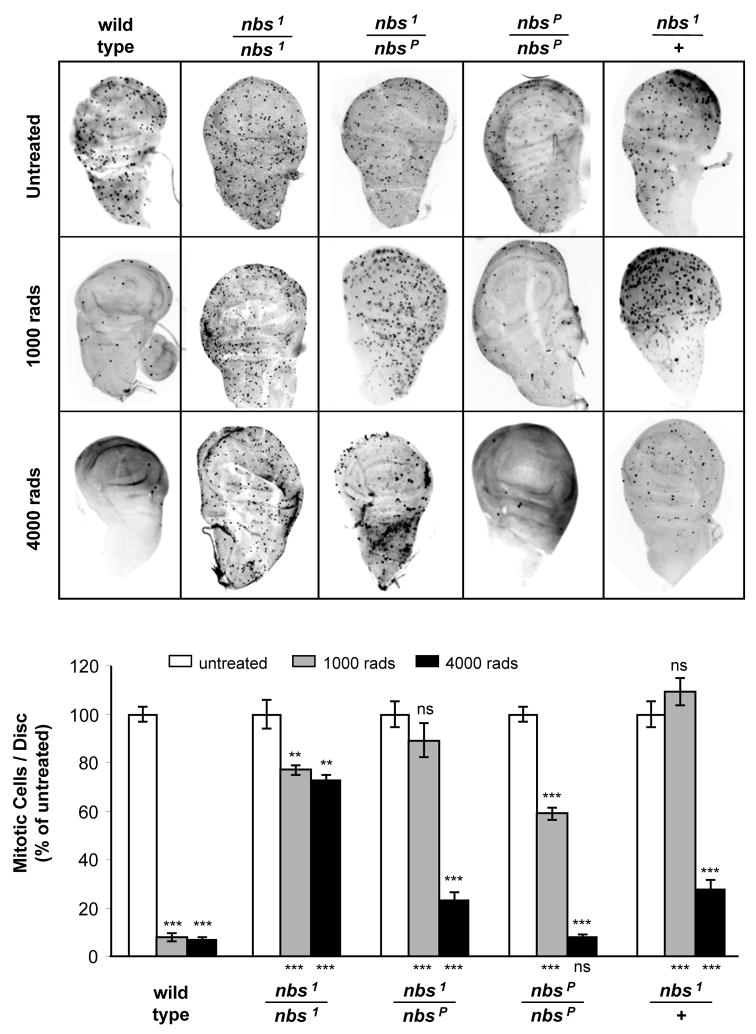
We observed haploinsufficiency of nbs in gap repair (Fig. 2b). We also examined another DNA damage response in which NBS has been implicated: the DNA damage-dependent cell cycle checkpoint. In Drosophila, the DNA damage checkpoint is most apparent at the G2/M boundary [52,60]. As described previously [6,47], the G2/M checkpoint induced by IR is nearly absent in null (nbs1/nbs1) mutants at both low-dose (1000 rads) and high-dose (4000 rads) irradiation (Fig. 5). Notably, heterozygosity for the null allele (nbs1/+) also caused loss of the checkpoint at low-dose irradiation and a significant weakening of the checkpoint at high-dose irradiation. Thus, Drosophila nbs is also haploinsufficient for the DNA damage-dependent checkpoint function.
Figure 5. The checkpoint response in nbs mutants.
(a) Mitotic cells in larval imaginal discs. Each panel shows a wing imaginal disc labeled with an antibody to phospho-histone H3 to mark mitotic cells. Discs were dissected out of wandering third instar larvae that were unirradiated or irradiated with 1000 and 4000 rads of gamma rays.
(b) Quantification of checkpoint defects. Mitotic cells were counted for at least seven wing discs from each genotype and treatment. Bars show number of mitotic cells per disc, normalized to the number in untreated discs of the same genotype. Error bars indicate SEM. Statistical significance relative to unirradiated of the same genotype is indicated above each bar. Significance relative to wild-type larvae at the same irradiation dose is shown below each bar. ** P<0.001; *** P<0.0001; ns P>0.05.
The nbsP allele, which is hypomorphic with respect to IR sensitivity, is also hypomorphic for the checkpoint response. Larvae homozygous for nbsP had a normal checkpoint at high-dose irradiation. At low-dose irradiation, the checkpoint was stronger than in null mutants, though significantly reduced relative to wild-type larvae (Fig. 5). In addition, the checkpoint response in nbs1/nbsP animals was not significantly different from that in nbs1/+. These findings suggest that the FHA domain of NBS is not essential for the G2/M checkpoint in Drosophila.
4. Discussion
4.1 The role of Drosophila NBS in homology-directed DSB repair
We used a P element excision assay to study the role of NBS in SDSA. The ability to complete repair by SDSA was significantly reduced in animals heterozygous for the null mutation nbs1. SDSA was further reduced in animals that were heteroallelic for nbs1 and nbsSM9, which removes the FHA domain. This suggests that the FHA domain is important for SDSA repair, but it is also possible that the protein produced by nbsSM9 is less abundant than that produced by a wild-type allele. If this is the case, then the increased severity of nbs1/nbsSM9 relative to nbs1/+ could be due to a further decrease in NBS levels.
In yeast and mice, the MRX/MRN complex is required during the early steps of DSB repair, where it is thought to participate in resection [61,62]. The results from our P element excision assay are consistent with a similar role for the Drosophila MRN complex. In this assay, 97% of repair events from wild-type males showed evidence for synthesis from the right side of the break (all of the red-eyed progeny, which accounted for 42% of all repair events, plus 95% of the yellow-eyed progeny; this is an underestimate because it does not include those repair events that regenerated a complete P{wa}, which could not be distinguished from non-excision). In contrast, only 62% of repair events from nbs1/nbsSM9 showed evidence of synthesis (P <0.0001). The decreased ability to initiate synthesis could be due to failure to process the 5′ ends to produce 3′ overhangs that are competent to participate in strand invasion. The defect was incomplete, since more than half of the repair products we analyzed from these nbs mutants showed evidence for synthesis. This could be due to partial function retained by the nbsSM9 mutation or to maternally-deposited NBS still present at the time of excision. Alternatively, other proteins may have overlapping or partially redundant roles in resection, as seems to be the case in S. cerevisiae [11–14].
As noted above, SDSA was reduced in nbs1/+ heterozygous animals; however, the decrease in synthesis initiation in this genotype was not significantly different from wild-type. We suggest that the reduced ability of nbs mutants to complete SDSA is due to a defect in reinitiating repair after the first round of synthesis. Among repair events that had synthesis, tracts were significantly shorter in nbs mutants than in wild-type flies. For example, among the yellow-eyed progeny of wild-type, 82% had repair products with at least 900 bp of synthesis (Fig. 2c). In contrast, tracts of at least 900 bp were found in only 59% of yellow-eyed flies from nbs1/+ and 19% from nbs1/nbsSM9, both of which are significantly different from wild-type (P<0.0001 in both cases). If the sole defect in nbs mutants is in resection, then we would expect that among those events in which it was possible to initiate synthesis, tracts should be similar in length to those of wild-type flies. Since we did not obtain this result, we conclude that Drosophila MRN has some role in addition to initial resection.
Previous studies led to the suggestion that repair of large gaps requires multiple cycles of strand invasion, synthesis, and displacement of the nascent strand [49]. Based on this model for repair, we envision three possible downstream requirements for MRN. First, it is possible that the newly synthesized strand is made double-stranded either during repair synthesis or when annealing fails. Additional resection would then be required before re-invasion can occur. Second, some end processing may be a prerequisite for loading strand invasion proteins onto single-stranded DNA prior to each cycle of invasion and synthesis. Reduced availability of NBS in mutants would limit the number of synthesis cycles, resulting in a tendency for short repair tracts. Third, NBS may be required as a sensor for recognition of an intermediate generated during each round of strand invasion and synthesis. Detection of such intermediates may be required to generate a signal that allows strand invasion proteins to re-load onto the single-stranded region for re-invasion and extension of the synthesis tract. Reduced ability to recognize these intermediates in nbs mutants might result in increased use of end joining to complete repair. This function would have to be independent of MEI-41, the Drosophila ortholog of ATR, because mei-41 mutants do not have a defect in synthesis tract lengths [63]. In any of these cases, the function for NBS after initial synthesis that we uncovered could be the same as the initial function; it is possible that such a function is only detectable in gap repair assays, in which multiple cycles of processing, strand invasion, synthesis, and dissociation are required for accurate repair.
Although reduced initiation of synthesis in the P element excision assay is consistent with a role for NBS in resection, we did not detect any requirement for NBS in the SSA assay. We employed this assay because SSA in this case requires >3.5 kb of resection to one side and almost 5 kb to the other. The finding that SSA was as frequent in nbs1/nbsSM9 mutants as in wild-type larvae suggests that either NBS is not essential for resection, or that the residual activity of NBSSM9 is sufficient for this function. Alternatively, it is possible that maternal contribution of NBS was sufficient to complete any end processing necessary for SSA in early larval development.
4.2 Drosophila NBS in end joining
The breaks generated by P excision or I-SceI cleavage can be repaired by several types of NHEJ. In the P excision assay, events in which SDSA is initiated but not completed are finished through a DNA ligase IV-independent end-joining pathway [59]. This type of repair was not reduced in nbs1/nbsSM9 or nbs1/+ mutants; indeed, there was an increase in this class compared to wild-type flies, corresponding to the decreased repair by SDSA (Fig. 2b, yellow-eyed class; Table S1). Similarly, in the SSA assay, which involves repair of a DSB generated by I-SceI cleavage, the fraction of events in which repair occurred by imprecise end joining was not significantly different between wild-type and nbs mutants (Fig. 3c). Thus, the ability to repair DSBs by end joining did not appear to be defective in the assays we used.
In S. cerevisiae, MRX is required for a class of NHEJ called microhomology-mediated end joining (MMEJ), a process characterized by use of microhomologies longer than five base pairs [reviewed in 64]. In the P{wa} assay, there was a significant (P=0.012) decrease in junctions with microhomologies up to five base pairs in nbs1/nbsSM9 mutants compared to wild-type flies (Fig. 4). Longer microhomologies were significantly more frequent than short microhomologies in nbs1/nbsSM9 mutants (two-tailed P=0.0194 by Fisher’s exact test), but not in wild-type flies (P=0.0768). Thus, MMEJ does appear to occur more frequently in nbs1/nbsSM9 mutants than in wild-type flies. This suggests that either MRN or the FHA domain of NBS is not required for MMEJ in Drosophila. Indeed, MRN or the FHA domain may be required to avoid MMEJ, possibly by promoting a type of end joining that relies on short microhomologies.
4.3 The FHA domain of Drosophila NBS
The FHA domain is absent in both nbsP and nbsSM9 mutants. Our data suggest that the FHA domain plays a role in completion of SDSA during gap repair, but is not essential for this process. Further, we found that among gap repair events that did not complete SDSA, the FHA domain is important for promoting longer synthesis tracts. These different readouts are likely to represent the same function: nbsSM9 mutants have a reduced ability to initiate and produce long synthesis tracts, which results in fewer cases of completed SDSA.
Loss of the FHA domain did not significantly affect the rate of SSA repair, nor the rate of imprecise NHEJ in the SSA assay. This suggests that the FHA domain is not involved in the resection required in this assay. We also found that the FHA domain does not have a significant role in the G2/M checkpoint, consistent with a previous study in human cells that showed the first BRCT domain, but not the FHA domain, is involved in cell survival after exposure to IR [65].
A caveat to these interpretations is that we were not able to measure the level of NBS protein present in nbs1/nbsSM9 mutants. In humans, the most common allele that causes Nijmegen breakage syndrome is a small deletion that causes expression of the FHA and first BRCT repeat on one polypeptide, and the rest of the protein on another [26]. This mutation does not completely eliminate NBS function, suggesting that there is stable expression of the C-terminal segment that, like NBSSM9, lacks the FHA domain [26]. Our data show that nbs transcript levels are not grossly reduced in nbsSM9 mutants (Fig. 1c); however, these blots were made from mRNA from whole larvae, whereas the SDSA and SSA assays probed germline function only. To our knowledge, the cryptic promoter downstream of w that is used to generate these transcripts has not been previously reported, and it is unknown whether it displays any tissue specificity.
4.4 Haploinsufficiency of nbs
Linkage analysis in human populations has shown that heterozygous carriers of nbs mutation are predisposed to various types of cancers [28,29], and cells heterozygous for an NBN mutation have been found to be defective in activation of the checkpoint protein ATM after low-dose ionizing radiation [33]. In Drosophila mutants heterozygous for the null mutation nbs1, we found that the checkpoint was absent after low-dose irradiation and significantly reduced after high-dose irradiation (Fig. 5). Previous studies have shown that Drosophila ATM and MRE11 are essential for this checkpoint after 500 rads, but not after 4000 rads [66]. It was suggested that ATM is required to amplify the signal due to a low degree of damage. Similarly, in the case of lesser damage, the cell may need a normal dose of NBS to amplify the signal and establish the checkpoint. At high doses of IR, the lower level of NBS present in nbs1/+ larvae may be sufficient.
We did not detect hypersensitivity to IR in nbs1/+ heterozygotes. Likewise, cells from human carriers of NBN mutations are not hypersensitive to IR [67]. However, spontaneous chromosomal translocations are elevated in human carriers, and in mice heterozygous for an Nbn mutation, suggesting defects in DSB repair [28,30,67]. Consistent with this interpretation, we found that nbs1/+ heterozygous flies had reduced SDSA repair of gaps generated by P element excision. This is the first report of defects in HR in nbs/NBN heterozygotes in a model organism. Our results may have important implications for understanding the cancer predisposition of human carriers of NBN mutations.
Supplementary Material
Acknowledgments
We thank Paul Brewer-Jensen and Lillie Searles for assistance with RNA blots, Yikang Rong for the P{X97} insertion, and members of the Sekelsky and Duronio laboratories for helpful discussions. M.C.L. was supported by the Cell and Molecular Biology Program. This work was supported by a grant from the National Institutes of Health (GM61252) to J.S.
Footnotes
Conflict of Interest statement: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chu G. Double strand break repair. J Biol Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 2.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967–8980. doi: 10.1038/sj.onc.1206136. [DOI] [PubMed] [Google Scholar]
- 5.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Reports. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciapponi L, Cenci G, Gatti M. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics. 2006;173:1447–1454. doi: 10.1534/genetics.106.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang J, Bronson RT, Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 2002;21:1447–1455. doi: 10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demuth I, Frappart PO, Hildebrand G, Melchers A, Lobitz S, Stockl L, Varon R, Herceg Z, Sperling K, Wang ZQ, Digweed M. An inducible null mutant murine model of Nijmegen breakage syndrome proves the essential function of NBS1 in chromosomal stability and cell viability. Hum Mol Genet. 2004;13:2385–2397. doi: 10.1093/hmg/ddh278. [DOI] [PubMed] [Google Scholar]
- 9.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 11.Lewis LK, Storici F, Van Komen S, Calero S, Sung P, Resnick MA. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166:1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Usher M, Hu Y, Kmiec EB. Nuclease activity of Saccharomyces cerevisiae Mre11 functions in targeted nucleotide alteration. Appl Environ Microbiol. 2003;69:6216–6224. doi: 10.1128/AEM.69.10.6216-6224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakada D, Hirano Y, Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol Cell Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NGJ, Raams A, Byrd PJ, Petrini JHJ, Taylor AMR. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 16.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 17.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 20.Bender CF, Sikes ML, Sullivan R, Huye LE, Le Beau MM, Roth DB, Mirzoeva OK, Oltz EM, Petrini JH. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu K, Antoccia A, Sakamoto S, Kobayashi J, Matsuura S, Tauchi H. NBS1 and MRE11 associate for responses to DNA double-strand breaks. International Congress Series. 2007;1299:158. [Google Scholar]
- 22.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 Is Required for Proper Nuclear Localization and Multiple Functions of NBS1. J Biol Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- 25.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 26.Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 27.Varon R, Dutrannoy V, Weikert G, Tanzarella C, Antoccia A, Stockl L, Spadoni E, Kruger L-A, Masi Ad, Sperling K, Digweed M, Maraschio P. Mild Nijmegen breakage syndrome phenotype due to alternative splicing. Hum Mol Genet. 2006;15:679–689. doi: 10.1093/hmg/ddi482. [DOI] [PubMed] [Google Scholar]
- 28.Dumon-Jones V, Frappart PO, Tong WM, Sajithlal G, Hulla W, Schmid G, Herceg Z, Digweed M, Wang ZQ. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263–7269. [PubMed] [Google Scholar]
- 29.Seemanova E. An increased risk for malignant neoplasms in heterozygotes for a syndrome of microcephaly, normal intelligence, growth retardation, remarkable facies, immunodeficiency and chromosomal instability. Mutation Res. 1990;238:321–324. doi: 10.1016/0165-1110(90)90024-6. [DOI] [PubMed] [Google Scholar]
- 30.Tanzanella C, Antoccia A, Spadoni E, di Masi A, Pecile V, Demori E, Varon R, Marseglia GL, Tiepolo L, Maraschio P. Chromosome instability and nibrin protein variants in NBS heterozygotes. Eur J Hum Genet. 2003;11:297–303. doi: 10.1038/sj.ejhg.5200962. [DOI] [PubMed] [Google Scholar]
- 31.Varon R, Seemanova E, Chrzanowska K, Hnateyko O, Piekutowska-Abramczuk D, Krajewska-Walasek M, Sykut-Cegielska J, Sperling K, Reis A. Clinical ascertainment of Nijmegen breakage syndrome (NBS) and prevalence of the major mutation, 657del5, in three Slav populations. Eur J Hum Genet. 2000;8:900–902. doi: 10.1038/sj.ejhg.5200554. [DOI] [PubMed] [Google Scholar]
- 32.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebi H, Matsuo K, Sugito N, Suzuki M, Osada H, Tajima K, Ueda R, Takahashi T. Novel NBS1 heterozygous germ line mutation causing MRE11-binding domain loss predisposes to common types of cancer. Cancer Res. 2007;67:11158–11165. doi: 10.1158/0008-5472.CAN-07-1749. [DOI] [PubMed] [Google Scholar]
- 34.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 35.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 40.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmbos PL, Daley JM, Wilson TE. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol. 2005;25:10782–10790. doi: 10.1128/MCB.25.24.10782-10790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howlett NG, Scuric Z, D’Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair (Amst) 2006;5:251–257. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Di Virgilio M, Gautier J. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J Cell Biol. 2005;171:765–771. doi: 10.1083/jcb.200506029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, Takata M, Takeda S, Matsuura S, Komatsu K. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 47.Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, Brodsky MH. Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet. 2006;2:e71. doi: 10.1371/journal.pgen.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 49.McVey M, Adams M, Staeva-Vieira E, Sekelsky J. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–1992. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rong YS, Golic KG. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 2003;165:1831–1842. doi: 10.1093/genetics/165.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, Rubin GM. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000;14:666–678. [PMC free article] [PubMed] [Google Scholar]
- 53.Becker E, Meyer V, Madaoui H, Guerois R. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics. 2006;22:1289–1292. doi: 10.1093/bioinformatics/btl075. [DOI] [PubMed] [Google Scholar]
- 54.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 55.Yu X, Chini CCS, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 56.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leicht BG, Bonner JJ. Genetic analysis of chromosomal region 67A–D of Drosophila melanogaster. Genetics. 1988;119:579–593. doi: 10.1093/genetics/119.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McVey M, Radut D, Sekelsky J. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics. 2004;168:2067–2076. doi: 10.1534/genetics.104.033902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 61.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LaRocque JR, Jaklevic B, Su TT, Sekelsky J. Drosophila ATR in double-strand break repair. Genetics. 2007;175:1023–1033. doi: 10.1534/genetics.106.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao S, Renthal W, Lee EY. Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 2002;30:4815–4822. doi: 10.1093/nar/gkf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bi X, Gong M, Srikanta D, Rong YS. Drosophila ATM and Mre11 are essential for the G2/M checkpoint induced by low-dose irradiation. Genetics. 2005;171:845–847. doi: 10.1534/genetics.105.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neubauer S, Arutyunyan R, Stumm M, Dork T, Bendix R, Bremer M, Varon R, Sauer R, Gebhart E. Radiosensitivity of ataxia telangiectasia and Nijmegen breakage syndrome homozygotes and heterozygotes as determined by three-color FISH chromosome painting. Radiat Res. 2002;157:312–321. doi: 10.1667/0033-7587(2002)157[0312:roatan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.