Abstract
Histone methylation is crucial for regulating chromatin structure, gene transcription and the epigenetic state of the cell. LSD1 is a lysine-specific histone demethylase that represses transcription by demethylating histone H3 on lysine 4 (ref. 1). The LSD1 complex contains a number of proteins, all of which have been assigned roles in events upstream of LSD1-mediated demethylation2-4 apart from BHC80 (also known as PHF21A), a plant homeodomain (PHD) finger-containing protein. Here we report that, in contrast to the PHD fingers of the bromodomain PHD finger transcription factor (BPTF) and inhibitor of growth family 2 (ING2), which bind methylated H3K4 (H3K4me3)5,6, the PHD finger of BHC80 binds unmethylated H3K4 (H3K4me0), and this interaction is specifically abrogated by methylation of H3K4. The crystal structure of the PHD finger of BHC80 bound to an unmodified H3 peptide has revealed the structural basis of the recognition of H3K4me0. Knockdown of BHC80 by RNA inhibition results in the de-repression of LSD1 target genes, and this repression is restored by the reintroduction of wild-type BHC80 but not by a PHD-finger mutant that cannot bind H3. Chromatin immunoprecipitation showed that BHC80 and LSD1 depend reciprocally on one another to associate with chromatin. These findings couple the function of BHC80 to that of LSD1, and indicate that unmodified H3K4 is part of the ‘histone code’7. They further raise the possibility that the generation and recognition of the unmodified state on histone tails in general might be just as crucial as post-translational modifications of histone for chromatin and transcriptional regulation.
Recent studies have identified a subset of PHD fingers that bind methyl lysine5,6,8,9. To investigate the role of BHC80, a PHD finger-containing protein (Fig. 1a) of the LSD1 co-repressor complex, in transcriptional repression, we determined whether BHC80 also binds histone tails through its PHD finger. As shown in Fig. 1b, BHC80 binds the first 21 residues of histone H3 (lane 3), but not residues 21–44 or histones H4, H2A or H2B (lanes 4, 8–11). Unexpectedly, the BHC80—H3 interaction is disrupted by methylation of K4, but is insensitive to modifications at K9 or K14 (Fig. 1b, lanes 5–7 and Supplementary Fig. 1a). Native BHC80 in the LSD1 complex also binds H3K4me0, and this binding is similarly compromised by H3K4 dimethylation (Fig. 1c). BHC80 in a reconstituted, three-component complex (LSD1, the co-repressor CoREST and BHC80) also retains the ability to bind H3K4me0 (Supplementary Fig. 1b).
Figure 1. BHC80 binds histone H3 through the PHD zinc finger.
a, Diagram of BHC80 domain architecture. AT hook, a motif that binds AT-rich DNA. b, In vitro assays of binding of recombinant BHC80 to histone tail peptides. ac, acetyl group. c, Native BHC80 in purified LSD1 complex also preferentially binds unmodified H3 tail. WB, western blot, using antibodies against BHC80. d, PHD finger of BHC80 is necessary for H3 tail binding. e, The PHD finger is sufficient for H3 tail binding. f, Methylation of H3K4 inhibits PHD finger binding to the H3 tail. g, ITC measurement of binding of the BHC80 PHD finger to unmodified, mono-, or di-methylated H3K4 peptides (residues 1–10). KD values are means (±s.d.) of at least three experiments using varied peptide and protein concentrations.
Although full-length BHC80 binds H3K4me0, deletion of the PHD finger (BHC80ΔPHD) significantly impairs this interaction (Fig. 1d). A glutathione S-transferase (GST) fusion of the finger (BHC80 residues 486–543) binds the H3 tail and retains the histone-binding specificity of full-length BHC80 (Fig. 1e), indicating that the PHD finger alone is necessary and sufficient for binding the H3K4me0 peptide. Furthermore, all three methylation states of H3K4 inhibit binding, with mono-methylation having the least adverse effect (Fig. 1f). Isothermal titration calorimetric (ITC) analysis of the PHD finger determined a dissociation constant (KD) of ∼30 μM for the unmodified H3 peptide, ∼460 μM for H3K4me1, and no binding to H3K4me2 (Fig. 1g), consistent with the pulldown results.
The co-crystal structure of the BHC80 PHD domain (residues 486–543) bound to the unmodified H3 tail (residues 1–10) was solved at the resolution of 1.43 Å (Supplementary Table 1). All ten residues of the H3 peptide were observed (Supplementary Fig. 2) bound to one of the two molecules in the asymmetric unit (Supplementary Fig. 3a). Like all structurally characterized PHD (and RING) fingers, the BHC80 PHD finger adopts a ‘cross-braced’ topology of Zn2+-coordinating residues (Fig. 2a, b). The H3 peptide binds to the surface of the PHD finger as an anti-parallel β-sheet, with H3R2—H3R8 forming backbone hydrogen bonds with G498—M502 of BHC80 (Fig. 2b). The cognate PHD finger contacts the first 8 residues of the H3 peptide, but H3 residues 9 and 10 interact with a neighbouring PHD molecule (Supplementary Fig. 3b). The substrate specificity of the BHC80 PHD finger is determined primarily through the recognition of the H3 amino terminus, H3K4 and H3R8 (Fig. 2c). Three main chain carbonyl oxygen atoms (residues 523, 524 and 525) form a hydrogen bond ‘cage’ that recognizes the N terminus of H3. The side chain of H3A1 inserts into a shallow hydrophobic pocket formed by L512, W527 and P523 (Fig. 2c, inset). The H3 peptide-binding site is further defined by M502, which inserts between H3R2 and H3K4, and the side chain of D489, which inserts between the side chains of H3K4 and H3R8, forming an electrostatic bridge between the two (Fig. 2c). The importance of D489 and M502 to H3 binding was confirmed by mutagenesis; mutation of D to A and M to W, respectively, abolished PHD binding to the unmodified histone H3 tail (Fig. 2d).
Figure 2. Structure of BHC80 PHD with H3 1–10.
a, Cross-brace topology of the PHD domain. Residues involved in peptide binding are underlined. Bold residues are those involved in chelating Zn. b, The H3 peptide binds BHC80 as an anti-parallel β-strand (grey). c, Peptide binding is specified by the insertion of M502 between H3R2 and H3K4 and D489 between H3K4 and H3R8. D489 and E488 bind H3K4, and H487 (4.0 Å away) restricts methyl-lysine binding (Supplementary Fig. 4). The N-terminal amine of H3 is caged by carbonyl oxygens, and H3A1 is recognized by a shallow hydrophobic pocket. d, Mutation of D489 and M502 disrupt PHD binding to unmodified H3. WT, wild type.
As well as interacting with D489, the epsilon amino group of H3K4 forms a hydrogen bond with the main chain carbonyl oxygen of E488 (Fig. 2c, inset). A β-carbon from H487, 4.0 Å away, further restricts the lysine-binding site. Indeed, a modelled mono-methyl lysine is allowed only a 15° range of motion before clashing with other atoms (Supplementary Fig. 4). This might account for the > 15-fold reduction in binding of BHC80 to the H3K4me1 peptide when compared with the H3K4me0 peptide (Fig. 1g). A second or third methyl group would clash with the D489 side chain, E488 carbonyl, or H487 β-carbon, consistent with the observation that the BHC80 PHD finger does not bind H3K4me2 or H3K4me3 peptides (Fig. 1f, g). Therefore, molecular recognition of unmodified lysine is primarily through bonds to the unmodified epsilon amino group, and steric exclusion of methyl groups. This mode of binding is distinct from the caging of di- and tri-methyl lysine by aromatic residues, as identified in the polycomb and heterochromatin protein 1 (HP1) chromodomains10-13.
The aromatic cage, though absent from BHC80, is also present in structurally characterized methyl-lysine-binding PHD fingers, such as those of BPTF and ING2 (refs 14,15). The PHD fingers of BHC80, BPTF and ING2 adopt highly similar folds (Fig. 3a). All engage the H3 peptide as an anti-parallel β-sheet on the same face, with recognition of the H3 N-amine and the H3A1 side chain. H3R2 is buried in a pocket in BPTF and ING2, but is not contacted by BHC80. In BHC80, M502 occupies the space left open for R2 binding by a conserved glycine (G) in BPTF and ING2 (ref. 12). Only BHC80 contacts H3R8, whereas in BPTF and ING2, the H3 peptide meanders off the face of the PHD finger before R8. The BPTF PHD finger features a full aromatic cage (Fig. 3b) reminiscent of the binding of the HP1 or polycomb chromodomain to trimethyl lysine12,13, whereas ING2 has only half a hydrophobic cage, with a serine and methionine finishing the H3K4 binding pocket, similar to recognition of H3K4me3 by the chromo-ATPase/helicase-DNA-binding double chromodomain16,17. As a family, PHD fingers show flexibility in peptide binding (Fig. 3c), making it difficult to predict whether an individual PHD finger is a histone-binding module, and whether it binds lysine or methyl lysine, on the basis of its primary sequence. Indeed, the robustness of the PHD scaffold, and its plasticity as a binding module, have been noted18. Our data indicate that the array of PHD fingers in chromatin-interacting and -modifying proteins, many of which lack the consensus binding sequence for H3K4me3 that is found in the BPTF and ING PHD fingers, might have histone-binding activities of unknown specificity. In this regard, the DNA methyltransferase 3-like protein, Dnmt3L, a regulatory factor that is required for de novo DNA methylation of imprinting control regions in female germ cells and of retro-transposons in male germ cells19-21, was found to bind H3K4me0 through its Cys-rich PHD-like domain22. Structural comparison reveals that the N-terminal Cysrich domain of DNMT3L is similar to the PHD finger of BHC80 (Supplementary Fig. 5). The structural similarity as well as their common mode of interaction with histone H3K4me0 raises the question of whether de novo DNA methylation (by DNMT3) is linked to the action of H3K4 demethylases and their associated complex components.
Figure 3. Structural comparison of PHD fingers.
a, Superimposition of PHD domains of BHC80 (PDB 2PUY, green, H3 grey), BPTF (PDB 2F6J, red, H3 pink) and ING2 (PDB 2GQ6, blue, H3 purple). b, Recognition of H3K4 by BHC80 (left) and binding of H3K4me3 by BPTF (middle) and ING2 (right); colours as in a. c, Sequence alignment of the PHD fingers of BHC80, BPTF and ING2. Zinc-binding residues are bold, K4-binding residues blue and the R2-binding pocket, red. Residues that form the N-amine and A1 binding pockets are green. Residues that form the anti-parallel β-sheet with peptide are underlined. Stars mark invariant residues, and colons denote conservation.
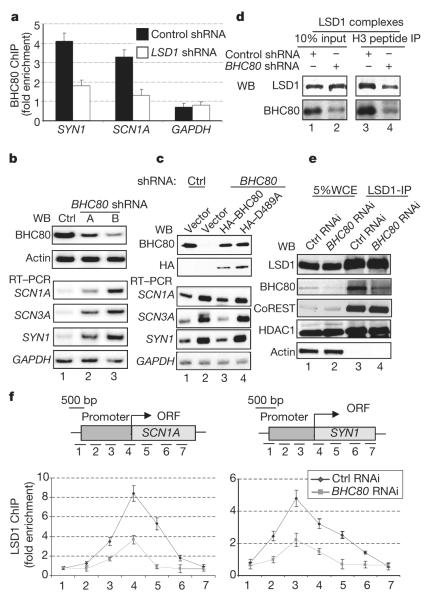
Having established the specificity of BHC80 for H3K4me0, we next investigated whether LSD1-mediated demethylation of H3K4me2 affects BHC80 binding at target promoters in vivo. Previous studies reported that depletion of LSD1 by RNA interference (RNAi) resulted in an increase in H3K4me2 at the synapsin 1 (SYN1) and voltage-gated sodium channel 1α (SCN1A) promoters1,4. Importantly, chromatin immunoprecipitation (ChIP) analysis showed that the binding of BHC80 to these promoters was reduced in the LSD1-depleted cells, although the global level of BHC80 was unaffected (Fig. 4a and data not shown). These data are consistent with a model in which LSD1-mediated demethylation of H3K4me2 is important for the association of BHC80 with chromatin.
Figure 4. BHC80 binding to the H3 tail is important for LSD1-mediated repression.
a, The occupancy of promoters by BHC80 is reduced in HeLa cells where LSD1 is knocked down by RNAi. Glyceraldehyde-3-phosphate dehydrogenase (GADPH) is used as a control. b, BHC80 was effectively depleted by BHC80 shRNAs and expression of SCN1A, SCN3A and SYN1 were de-repressed in the BHC80 RNAi cells. Ctrl, control. c, RNAi-resistant wild-type BHC80, but not the PHD point mutant D489A, restored target gene repression in BHC80 RNAi cells. d, Reduction of BHC80 in the LSD1 complex reduced the association of LSD1 with H3 peptide. IP, immunoprecipitate. e, Inhibition of BHC80 does not affect LSD1 complex formation in solution. f, RNAi of BHC80 results in decreased LSD1 occupancy at its target genes. Error bars in a and f represent s.e.m. calculated from three independent experiments.
We further investigated the function of BHC80 in LSD1-mediated transcriptional repression by depleting BHC80 using two independent short hairpin RNA (shRNA) constructs (Fig. 4b, top). The inhibition of BHC80 expression resulted in a consistent de-repression of a number of LSD1 target genes, including SCN1A, SCN3A and SYN1 (Fig. 4b). Introduction of the RNAi-resistant wild-type BHC80, but not the RNAi-resistant PHD finger mutant D489A, into the BHC80 knockdown cells restored the repression of LSD1 target genes, indicating that binding of H3K4me0 by BHC80 is important for LSD1-mediated gene repression. D489A was expressed at a similar level to that of the wild-type protein (Fig. 4C) and assembled into the LSD1 complex in the transfected cells (data not shown).
The binding of BHC80 to H3K4me0, which is a demethylation product of LSD1, indicates that BHC80 functions downstream of LSD1. How is a downstream effector of LSD1 required for efficient LSD1-mediated repression? Recent studies have indicated that the regulation of histone methylation is highly dynamic, and might involve the actions of both methylases and demethylases at the same target promoters23. BHC80 may be necessary to help to maintain LSD1 at the target promoters, thereby preventing the re-methylation of H3K4. Consistent with this idea, BHC80 has been shown to interact physically with LSD1 (ref. 24). To address this model experimentally, we compared the binding of LSD1 to H3 using LSD1 complexes prepared from either wild-type or BHC80-knockdown cells. A reduction of BHC80 in the LSD1 complex resulted in decreased binding of LSD1 to K4 unmethylated histone H3 peptides in vitro (Fig. 4d), indicating that BHC80 is important for the association of LSD1 with its reaction product H3K4me0. Consistent with this, recombinant BHC80 could significantly increase binding of LSD1 to the H3K4me0 peptide in the presence of CoREST in vitro (Supplementary Fig. 1b). These findings indicate that BHC80 is required for the association of LSD1 with histone H3 after demethylation.
We further investigated the effect of depletion of BHC80 on the LSD1 protein complex and its occupancy at target promoters. Knockdown of BHC80 had no effect on the assembly of the LSD1 complex in solution by co-immunoprecipitation analysis (Fig. 4e). In contrast, occupancy of LSD1 at its target gene loci was reduced as a result of BHC80 knockdown (Fig. 4e). These findings support a model in which BHC80 is required for stable association of the LSD1 complex with its target promoters. An alternative, but not mutually exclusive, model is that binding of BHC80 to the demethylated H3 might be important for LSD1 to mediate demethylation of the neighbouring nucleosome. This propagation mechanism is similar to the one that has been proposed for the H3K9 methyltranferase Suv39H and HP1, in which the Suv39H methylase and the H3K9me-binding HP1 function together to propagate the repressive signal at the target loci25-27. Many histone demethylases contain histone tail-binding modules in the form of PHD or Tudor domains. It will be interesting to determine whether some of these histone tail-binding modules function similarly to what has been reported here for LSD1 and BHC80, or facilitate cross-talk among modifications at different residues on histone tails9,28,29. Together, our findings indicate that each and every subunit of the LSD1 complex has a unique role in coordinating and setting up a chromatin environment that is important for repression.
In sum, we have identified a PHD finger with a new histone tail-binding specificity (H3K4me0), and provided structural insights into the specific recognition and functional importance of unmethylated lysine. We speculate the existence of more such binding modules in the human proteome that are dedicated to recognizing the unmodified state of amino-acid residues on histone tails that are otherwise modified by various post-translational modification events. We anticipate that the identification of these modules and insights into their functions will shed significant new light on dynamic chromatin regulation.
METHODS SUMMARY
BHC80 (BC015714)-derived recombinant proteins were purified from bacteria with either 6×His or GST tags. The LSD1 complex was purified from a stable HeLa cell line with Flag-HA—LSD1 expression4. We carried out in vitro binding assays using purified recombinant BHC80 proteins or LSD1 complex with a panel of biotinylated histone peptides with specific modifications at indicated lysine residues. We used tranylcypromine in the LSD1 complex binding assays to inhibit its demethylase activity. The bound fractions were visualized by either Coomassie blue staining or western blotting after immobilization by streptavidin beads. shRNA vectors were made as described30 and cotransfected with the puromycin resistance gene into HeLa cells. Chromatin samples and RNA samples were prepared from drug-selected cells. The semi-quantitative ChIP assay and PCR with reverse transcriptase were done by adding 32P-dCTP during PCR amplification and analysed by phosphor-image quantification. PCR reactions were optimized within the linear range. ChIP control oligonucleotides were designed to amplify the intron region of the RNA PolII gene, which in principle is not affected by either LSD1 or BHC80 knockdown. For crystallography and ITC, the BHC80 PHD finger was expressed as a 6×His—SUMO fusion, and cleaved, leaving HisMet fused to the recombinant protein. The structure of the BHC80 PHD domain in complex with histone peptide was solved by three-wavelength Zn anomalous diffraction data (Supplementary Table 1). ITC was performed on a MicroCal VP-ITC by injecting synthetic H3 peptide (residues 1–10, K4me0, 1 or 2) into BHC80 at various concentrations.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
In vitro binding assays
Histone peptides (0.5 μg) were incubated with 2–5 μg purified recombinant BHC80 or GST—PHD finger or 50 ng LSD1 complex for 2 h at 4 °C in binding buffer (20 mM Tris-HCl 7.5, 150 mM NaCl, 0.1% Triton X-100). LSD1 complexes were purified by Flag-immunoprecipitation from a stable cell line with Flag-HA—LSD1 followed by 2x elution using 3Flag peptide (Sigma)4. After 1 h incubation, Streptavidin beads (Upstate 16-126) were washed four times and subjected to Coomassie blue staining. Tranylcypromine (Sigma P8511) was used in Fig. 1c at a concentration of 50 μM to inhibit the demethylation reaction.
RNAi and PCR with reverse transcriptase (RT—PCR)
Two BHC80 shRNAs were designed to target the sequences (5′-GGAGGCTCTTAAAGTGGAAAT-3′ and 5′-GGGCAGAGGCTGTCCAAAT-3′), and subcloned into a pBS-U6 vector. LSD1 shRNA was done as described1. HeLa cells were cotransfected with shRNA and pBabe-puro vectors. After 12 h of transfection, cells were split and selected by 1.5 μg ml-1 puromycin for 60 h. RNA extraction and RT—PCR were performed as described1,4. The RT—PCR results in Fig. 4b, c were achieved by radioactive PCR.
Chromatin immunoprecipitation
LSD1 ChIP experiments were done using primers that have been described1. Chromatin samples from 107 cells were sonicated to 200–500 bp in ChIP lysis buffer (50 mM HEPES/KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate and protease inhibitors), and incubated with 3 μg LSD1 antibody (ab-17221) or BHC80 antibody10 for each chromatin IP experiment. The recovered DNA was amplified in radioactive PCR by the indicated primers and quantified by phosphor image, and an unrelated genomic region was used as a PCR internal control for normalization.
Crystallography
The BHC80 PHD finger (residues 486–543) was expressed in BL21-Gold (DE3) E. coli cells (Stratagene) as a 6×His—Smt3 (yeast SUMO) fusion construct (generated in-house: see ref. 31) harboured in a modified pET28b vector (Novagen). Protein expression was induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside for 3 h at 37 °C in Luria-Bertani broth supplemented with 50 μM ZnCl2. The fusion protein was isolated on nickel-charged HiTrap Chelating HP (GE Healthcare). Following imidazole elution, the fusion was cleaved off by Ulp1 protease31, leaving two amino acids (HisMet) fused to the BHC80 PHD finger. The protein was further purified by HiTrap-Q and Superdex-75 (GE HealthCare). For co-crystallization, the PHD finger (final concentration ∼40 mg ml-1 in 20 mM Tris, pH 7.2, 100 mM NaCl, 5 mM DTT, and 25 μM ZnCl2) was mixed in a 1:1.5 ratio with an H3 peptide (residues 1–10, dissolved in the protein buffer and neutralized with NaOH). Clusters of rod-shaped crystals were obtained using the sitting drop vapour-diffusion method at 16 °C, with mother liquor containing 100 mM sodium citrate, pH 5.6, 20% polyethylene glycol 4000, and 20% isopropanol. In follow-up screens, we obtained single rhombohedral crystals with the hanging-drop vapour diffusion method, in mother liquor containing MES 6.2–6.5, 5–10% polyethylene glycol 4000, and 20% isopropanol. Both crystal forms diffract in the C2 space group with similar cell dimensions, but the latter were used to determine the structure. Crystals were cyroprotected by soaking in mother liquor supplemented with 40% glycerol. To prevent the evaporation of isopropanol, drops containing crystals were submerged in paraffin oil during handling.
All data were collected at the SER-CAT 22-ID beamline at the Advanced Light Source at Argonne National Laboratory on a MAR300 CCD detector. Zinc anomalous diffraction data were collected from two crystals and processed using HKL2000. SOLVE32 found four zinc sites. We used SOLOMON solvent flipping33 for density modification, which gave a clear solvent boundary and traceable density. Model building with O34 and refinement with CNS35 continued by using a 1.43-Å data set (from a third crystal).
Isothermal titration calorimetry
A SUMO-tagged fusion of the BHC80 PHD was exchanged into 25 mM Tris-HCl buffer, 50 mM NaCl, 2 mM β-mercaptoethanol (pH 7.2) by gel-filtration chromatography. Extensively lyophilized H3 1–10 peptides were dissolved in the same buffer. ITC measurements were carried out from 100–500 μM protein concentration, with 3–7 mM peptide concentration, on a MicroCal VP-ITC instrument at 25 °C. For each peptide, a reference titration of peptide into SUMO alone was subtracted from experimental data to control for heat of dilution and non-specific binding. Binding constants were calculated by fitting the data using the ITC data analysis module of Origin 7.0 (OriginLab Corporation).
Supplementary Material
Acknowledgements
We thank S. Iwase and T. Baba for the gift of BHC80 antibody and Yujiang Shi for discussion. This work was supported by grants from the NIH to X.C. and Y.S..
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information The X-ray structure of the BHC80 PHD domain in complex with the H3 tail peptide has been deposited to PDB as 2PUY.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Hakimi MA, et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl Acad. Sci. USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 4.Shi YJ, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 5.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 7.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 8.Shi X, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwase S, et al. The X—linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 12.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 17.Sims RJ, III, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan AH, et al. Engineering a protein scaffold from a PHD finger. Structure. 2003;11:803–813. doi: 10.1016/s0969-2126(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 19.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 20.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 21.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 22.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. (this issue) doi: 10.1038/nature05987. doi:10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwase S, et al. Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem. Biophys. Res. Commun. 2004;322:601–608. doi: 10.1016/j.bbrc.2004.07.163. [DOI] [PubMed] [Google Scholar]
- 25.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 26.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama J-I, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of histone H3 lysine 9 methylatin in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Sui G, Shi Y. Gene silencing by a DNA vector-based RNAi technology. Methods Mol. Biol. 2005;309:205–218. doi: 10.1385/1-59259-935-4:205. [DOI] [PubMed] [Google Scholar]
- 31.Malakhov MP, et al. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 34.Jones TA, et al. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.