Abstract
We investigate the relationship between landscape heterogeneity and the spatial distribution of small mammals in two areas of Western Sichuan, China. Given a large diversity of species trapped within a large number of habitats, we first classified small mammal assemblages and then modelled the habitat of each in the space of quantitative environmental descriptors. Our original two step “classify then model” procedure is appropriate for the frequently encountered study scenario: trapping data collected in remote areas with sampling guided by expert field knowledge.
In the classification step, we defined assemblages by grouping sites of similar species composition and relative densities using an expert-class-merging procedure which reduced redundancy in the habitat factor used within a multinomial logistic regression predicting species trapping probabilities. Assemblages were thus defined as mixtures of small mammal frequency distributions in discrete groups of sampled sites.
In the modelling step, assemblages’ habitats and environments of the two sampled areas were discriminated in the space of remotely sensed environmental descriptors. First, we compared the discrimination of assemblage/study areas by linear and non-linear forms of Discriminant Analysis (Linear Discriminant Analysis versus Mixture Discriminant Analysis) and of Multiple Regression (Generalized Linear Models versus Multiple Adaptive Regression Splines). The “best” predictive modelling technique was then used to quantify the contribution of each environmental variable in discriminations of assemblages and areas.
Mixtures of Gaussians provided a more efficient model of assemblage coverage in environmental space than a single Gaussian cluster model. However, non-linearity in assemblage response to environmental gradients was consistently predicted with lower deviance and misclassification error by Multiple Adaptive Regression Splines. The two study areas were mainly discriminated along vegetation indices. However, although the Normalized Difference Vegetation Index (NDVI) could discriminate forested from non-forested habitats, its power to discriminate assemblages in Maerkang, where a greater diversity of forest habitat was observed, was seen to be limited, and in this case NDVI was outperformed by the Enhanced Vegetation Index (EVI). Our analyses highlight previously unobserved differences between the environments and small mammal communities of two fringe areas of the Tibetan plateau and suggests that a biogeograph-ical approach is required to elucidate ecological processes in small mammal communities and to reduce extrapolation uncertainty in distribution mapping.
Keywords: Small mammal assemblages, Habitat distribution modelling, Mixture Discriminant Analysis, Multiple Adaptive Regression Spline, Environmental gradients
1 Introduction
1.1 Modelling distributions of assemblages
Modelling spatial distributions of species is a developed and promising research field which can both explain and predict the effects of environmental descriptors on species presence/absence in space. This relies on defining species’ habitats (Guisan and Zimmerman, 2000) following Hutchinson’s concept of ecological niche (1957). Recent reviews are found in Pulliam (2000) and Hirzel et al. (2002).
In situations where a large number of species co-occur in a large number of sampled sites, habitat definition for individual species can be complicated by species interactions. Defining habitat for rare or “shy” species can be impossible when presence is difficult to detect. By contrast, the full extent of habitats for dominant species can be elusive when the species is found in a large proportion of sampled sites. In these situations community level modelling constitutes a useful tool that provides a synthesis of such data sets by reducing their complexity to a much smaller set of higher-level entities. One such higher level entity can be that of an assemblage, i.e. a group of taxonomically related species which share the same habitat (Ferrier and Guisan, 2006). The habitat of an assemblage can therefore be defined in reference to a group of sites in which similar species composition and densities are observed.
Community-level spatial modelling involves three main steps (Ferrier and Guisan, 2006): i) classification of species and/or sites into groups, ii) statistical formulation to assess relationships between groups and environmental descriptors, and iii) predictive mapping of groups. Classification (i) is often realized through ordination methods without incorporating environmental information, e.g. the TWINSPAN algorithm (Hill, 1979; Legendre and Legendre, 1998). Grouping species into discrete assemblages involves making assumptions on species distributions (Olden, 2003): first, grouped species are treated in a similar way regarding their responses to environmental descriptors; then, species are assumed to belong to discrete mutually exclusive groups. In regards to Hutchinson’s concepts of ecological niche and associated species’ niche specificity and marginality, such assumptions seem at odds with species distributions in nature (Doledec et al., 2000; Hirzel et al., 2002). Moreover, clustering procedure can be biased by the subjective choice of similarity criterion and decision threshold (Anderson and Clements, 2000). In the modelling step (ii) groups are typically modelled against environmental variables, individually via GLM or GAM, or simultaneously using polychotomous regression or discriminant analysis (Lehmann et al., 2002; Gibson et al., 2004; Ferrier and Guisan, 2006). Modelling techniques have been deeply investigated in a “model then classify” approach by which species responses are grouped into assemblages after prediction of their occurrences (Lehmann et al., 2002; Gibson et al., 2004; Ferrier and Guisan, 2006). Joint modelling of multi-species responses has been recently developed to account for species interactions. This can help elucidate those variables that have strong effects on the whole community but might not be identified relevant in species level analyses (Leathwick et al., 2006). This has been successfully achieved through non-linear models such as Multiple Adaptive Regression Splines (MARS) (Friedman, 1991; Moisen and Frescino, 2002; Leathwick et al., 2006) and Artificial Neural Networks (Olden, 2003; Olden et al., 2006). However, these methods consider the model response to be presence of the individual species in each sampled site, defined using a threshold probability (usually 0.5) on observation frequencies, which can be viewed as a constraint for rarely observed species or sparse data sets.
1.2 Modelling landscape disturbance effects on small mammal distributions
Effects of landscape disturbances on small mammal communities have been investigated by defining assemblages using several classification methods on the basis of species trapping data. For example, Butet et al. (2006) used ordination methods and species/sites proximity in co-inertia axes while Giraudoux et al. (1998) defined assemblages by subjective criteria. Clustering algorithms have also been used to classify habitats into groups according to species composition dissimilarity (Krasnov et al., 1996).
On the basis of an a priori expert defined and qualitative habitat nomenclature, Raoul et al. (2008) produced an objective and reproducible classification of assemblages. The set of sampled habitat classes was reduced into a smaller set using information theory and assuming a multinomial model for the small mammal trapping data. Each a posteriori habitat class plus associated estimates of species trapping frequencies defined an assemblage. This approach presents several advantages over currently popular classification methods in the “Classify then model” approach: first, classification of assemblages was performed via a reclassification of sampled habitat classes a priori identified in the field and thus incorporates information gained from an expert oriented sampling strategy (Pearce et al., 2001); secondly, by considering each assemblage as a localised picture of species composition and relative densities, classification was performed at the habitat level instead of the species level, thus the crude assumptions of species responses mentioned above were avoided; finally, in the reclassification step, models were compared and selected using Akaike Information Criterion (AIC) which can be viewed as an objective method (Burnham and Anderson, 1998). However, this classification method limits the definition of assemblages’ habitats to qualitative and a priori habitat classes limited to the sampling design. Consequently it suffers from predictive power limitations, i.e. assemblage definitions cannot be spatially extrapolated beyond the sampled sites.
1.3 Context, hypothesis and objectives
In China, the spatial distribution of small mammal species has been shown to be modified by landscape disturbances such as overgrazing and fencing practices on the Tibetan plateau (Wang et al., 2004; Raoul et al., 2006), deforestation in Gansu (Giraudoux et al., 1998) and afforestation in Ningxia (Raoul et al., 2008). We aimed to investigate the relationship between landscape heterogeneity and the spatial distribution of small mammal assemblages in two forested areas located in the fringes of the Tibetan plateau (Sichuan, China). There, forest management leads to landscape heterogeneity and likely drives changes in the spatial distributions of small mammals.
Our main methodological challenge was to develop a predictive model for trapping data sets of diverse taxon, realized in remote areas and thus constrained by a limited sampling effort, a large number of rare species trapped with low frequency and an expert oriented sampling design. There is currently a need to adapt habitat modelling methodologies in order to fit and extract the maximum information possible from such data sets which is common in conservation studies. Here, we developed an original two step “Classify then model” procedure to address these issues.
In the classification step, we applied the Raoul et al. (2008) expert-class-merging procedure to summarise our trapping data by defining assemblages. Then, our major technical contribution was to incorporate such assemblage definitions into a predictive modelling framework. In the modelling step, the predictive limitations of the initial expert-class-merging assemblage definitions was overcame by extending the definition of the habitat associated with each assemblage using a set of quantitative variables extracted from remote sensors. By doing so, we addressed some currently debated methodological issues in the field of species distribution modelling. The modelling step aimed to answer two questions:
which modelling technique and associated response-factor relationships predict assemblage occurrence with lowest prediction error?
what are the contributions of each environmental variable in assemblage discrimination and prediction?
Among the theoretical hypothesis supporting current species modelling methodological frameworks, the shape of response curves has often been neglected and there is a need to consider it at the interface between statistical methods and ecological realism (Austin, 2002, 2007; Guisan et al., 2006). The richness of existing statistical models offers the opportunity to compare different ecological theories underlying observed patterns of species distribution in environmental space. While previous studies have shown non-linear models provide better fits and predictions of multiple species responses than linear models (Doledec et al., 2000; Olden, 2003; Leathwick et al., 2006), we tested if this observed pattern could hold when species responses are considered as a whole, i.e when assemblages and not species are used as the response variable. Several methods exist to discriminate known groups by continuous variables. We compared the discrimination ability of two widely used classifiers, multiple logistic regression and discriminant analysis, and tested several forms of assemblage response curves: linear versus non-linear and single Gaussian versus Gaussian mixture respectively.
The selection of a relevant set of predictors for building predictive models also remains an active current issue which is complicated by the nature of environmental predictors (direct/indirect) and their interactions (Austin, 2007). Here, instead of selecting a single “best” model, we investigated the overall contribution of each environmental variable within a set of models.
Because trapping data were collected in two distinct areas, the same questions were answered at the regional spatial extent to investigate discrimination of the two study areas in the environmental space.
2 Material and methods
2.1 Small mammal species data sets
2.1.1 Study areas
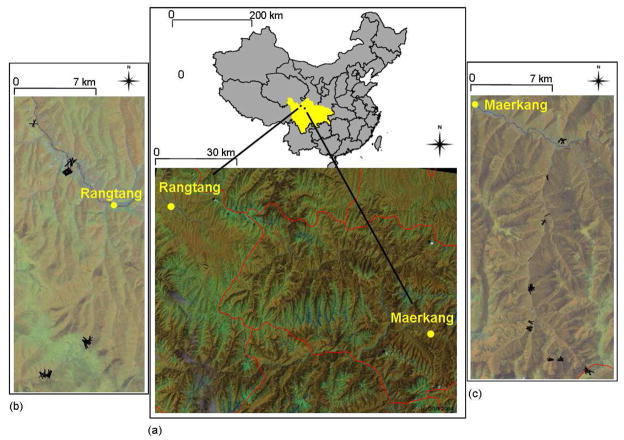
Two study sites were investigated in western Sichuan province (central China) in the vicinities of Rangtang and Maerkang cities, (approximatively 100 kilometres apart) (figure 1), in June 2004 and July/September 2005 respectively. The sampling area in Maerkang ranged from 2950 to 4100 meters altitude. The 2005 mean annual temperature and yearly average rainfall were 8.9 °C and 811.5 mm respectively (data source: Maerkang Center for Disease Control). Forested areas were either coniferous or birch and oak. Afforestation measures were in place and a large number of young plantation forests were found. Rhododendron forests and pastures were observed at higher elevations. Villages were often surrounded by ploughed fields and were situated close to rivers with abandoned terraced fields at higher elevation. In Rangtang, elevation of sampled area ranged from 3350 to 3900 meters and in 2005 yearly average temperature and yearly average rainfall were 5.4 °C and 854.3 mm respectively. Landscape was mainly composed of grassland and shrub. Forest was less abundant than in Maerkang and was more frequently coniferous than broad-leaf.
Figure 1.
a) Location of Maerkang and Rangtang study areas in Sichuan province, China. Lines delineate province and county boundaries. (b) and (c) represent locations of traplines (lines) around Rangtang and Maerkang cities respectively. Locations are plotted on a false colour composite satellite image in which red, green and blue correspond to Landsat ETM bands 4, 5 and 3.
2.1.2 Sampling protocol
Sampling was undertaken in a priori defined habitats identified in the field i.e. habitats classified on the basis of apparent dissimilarities in vegetation structure and dominant genus composition. In Maerkang and Rangtang, 18 and 12 habitats were sampled respectively (table 2). Four habitats were found to be similar in the two locations: “Forest Rhododendron/coniferous”, “Forest coniferous willow bushes understory”, “Stream bushes” and “Slope bushes” (table 2).
Table 2.
Maerkang (M1 to M4) and Rangtang (R1 to R4) assemblages obtained from expert-class-merging procedure. The number of traplines (Nb lines), habitat composition, number of trap-nights per sampled habitat (Nb trap-nights), species trapped (Dominant and Other) and Simpson diversity index are reported for each assemblage.
| Assemblage | Nb lines | Habitat composition | Nb Trap-nights | Dominant species | Others species | Simpson diversity |
|---|---|---|---|---|---|---|
| M1 | 20 | Bushes North Slope | 75 | Niviventer Confucianus | Chodsigoa hypsibia | 2.60 |
| Bushes former Terrace | 744 | Apodemus latronum | ||||
| Bushes Birch Culture | 300 | |||||
| Grassland | 447 | |||||
| Forest Oak near Village | 301 | |||||
| Set a side Field | 749 | |||||
| Stream near Culture | 300 | |||||
|
| ||||||
| M2 | 7 | Culture | 1050 | Micromys minutus | Eozapus setchuanus | 2.08 |
| Eo spalax fontanieri | ||||||
| Apodemus latronum | ||||||
|
| ||||||
| M3 | 34 | Forest Regeneration & Plantation | 719 | Qchotona cansus | Apodemus peninsulae | 7.86 |
| Stream Bushes & Trees | 325 | Apodemus draco | ||||
| Forest Coniferous & Willow Bushes | 600 | Apodemus latronum | ||||
| Forest Oak | 444 | Chodsigoa hypsibia | ||||
| Stream Willow Bushes | 294 | |||||
| Stream Bushes | 444 | Sorex thibetantus | ||||
| Forest Rhododendron | 746 | Sorex cylindricauda | ||||
| Forest Rhododendron & Coniferous | 747 | Uropsilus soricipes | ||||
| Sicista concolor | ||||||
| Microtus irene | ||||||
|
| ||||||
| M4 | 5 | Forest white Birch wet | 306 | Apodemus draco | Apodemus latronum | 1.97 |
| Forest red Birch Understorey | 438 | Niviventer confucianus | ||||
| Uropsilus soricipes | ||||||
|
| ||||||
| R1 | 23 | Field bank | 717 | Apodemus peninsulae | Microtus limnophilus | 1.07 |
| Slope grass & Sparse bushes | 300 | Eozapus setchuanus | ||||
| Field bank | 369 | |||||
| Culture border | 671 | |||||
|
| ||||||
| R2 | 18 | Bottom valley grassland & bushes | 600 | Apodemus peninsulae | Microtus limnophilus | 1.25 |
| Bottom valley grassland & bushes (Namuda) | 577 | |||||
| Village garden | 599 | |||||
| Forest coniferous & willow bushes | 898 | |||||
|
| ||||||
| R3 | 2 | Fenced grassland | 48 | Microtus limnophilus | Apodemus peninsulae | 1.25 |
|
| ||||||
| R4 | 14 | Forest rhododendron & coniferous | 175 | Qchotona cansus | Microtus irene | 2.48 |
| Slope bushes | 746 | Apodemus peninsulae | ||||
| Stream bushes | 1041 | Microtus limnophilus | ||||
Extensive standard trapping (Giraudoux et al., 1998, Raoul et al, 2006) was undertaken in each habitat. Each standardized trapline consisted of 25 traps spaced three meters apart. Two types of traps were used: small break back traps (SBBT), for animals not heavier than 100g and big break back traps (BBBT) for larger individuals. Each trap was set for three nights (unless non-controlled factors dictated otherwise, e.g. trap theft), checked every morning and re-set/re-baited as necessary. We use the term trap-night to refer to a single trap set for one night. A total of 8095 trap-nights (4603 in Maerkang; 3492 in Rangtang) in 122 traplines (66 in Maerkang; 56 in Rangtang) were set; differential trapping effort by habitat is reported in table 2.
Species were identified at the Centre de Biologie et Gestion des Populations (J.P. Quéré) using the following references: Corbet (1978); Fen and Zheng (1985); Gromov and Polyakov (1978); Gromov and Erbajeva (1995); Smith and Xie (2008). Apodemus penninsulae, Apodemus draco and Apodemus latronum identifications were confirmed using cytochrome b sequencing. Nomenclature followed Wilson and Reeder (2005).
2.2 Assemblage definition
2.2.1 Statistical model
The response variable, the category (i.e species or empty trap) observed in a given trap-night, was assumed to follow a multinomial distribution (Raoul et al., 2008). Relationships between habitat classes and species distribution were modelled using log linear multinomial regression with a priori selected habitat classes as explanatory variables (McCullag and Nelder, 1989). The three nights per trap and trap type were included as factors in the regression. Within each study site, a preliminary model comparison via AIC indicated no evidence for a night effect, so the factor was removed for further analyses.
2.2.2 Expert-class-merging procedure
Following Raoul et al. (2008), pairs of habitat classes (Hi and Hj) were “merged” by imposing an equality constraint on their regression coefficients (i.e. β Hi = βHj). Iterative application of this constraint to all habitat class pairs lead to K new models which were compared via the difference in AICc (ΔAICci j) between each new model and the original (unconstrained) one. The model/merge providing the largest ΔAICci j identified the most redundant habitat class distinction and the two corresponding classes were fused into a single super-class. The process was iterated until no further evidence of class redundancy was observed i.e. once ΔAICci j < 2 (Burnham and Anderson, 1998). Finally, we computed the Simpson diversity index of each resulting assemblage. All statistical algorithms were implemented in R (R Development Core Team, 2008). Multinomial models were fitted using the multinom function of the nnet package.
2.3 Assemblage habitat modelling
2.3.1 Environmental descriptors
The environment at trapline locations was described using remotely sensed data layers corresponding to: spectral responses of Landsat Enhanced Thematic Mapper (ETM) bands; elevation; slope; and sun exposure. A (July 2005) multi-spectral ETM image from Landsat 7 was obtained from the U.S. Geological Survey ( landsat.usgs.gov). ETM bands 3, 4 and 7, corresponding to red, near-infrared and middle infrared respectively, each with a 30 meters resolution, were used for the analyses. Other ETM bands were omitted from further analysis due to strong correlations with the selected bands. Using the ETM image, the Normalized Difference Vegetation Index (NDVI) and Enhanced Vegetation Index (EVI) were computed (Huette et al., 1999). NDVI quantifies the difference between photosynthetic activity related absorption in the visible range and reflectance in the near-infrared which is related to electro-magnetic emission by plants. NDVI is thus correlated to vegetation biomass. However, the NDVI is known to suffer from saturation when vegetative biomass is high and to be sensitive to canopy background in open forested areas (Huette et al., 2002). Therefore, we computed EVI, defined as
where L is the canopy background and snow correction, C1 and C2 are coefficients of aerosol “resistance” terms and G the gain factor with L = 1, C1 = 6, C2 = 7.7 and G = 2.5.
A digital elevation model (DEM) was obtained from the SRTM program ( http://srtm.usgs.gov/). Slope, aspect and elevation were derived from the DEM in GRASS GIS ( http://grass.itc.it/). The sun index (SI), which provides a proxy for net incident solar radiation, was estimated as (Gibson et al., 2004):
In general traplines were not confined within the bounds of single pixels. In order to construct the data matrix the mean of each variable was estimated from 10 points regularly spaced along each trapline. This was performed using spgrass6, the R/GRASS interface available at http://r-spatial.sourceforge.net/xtra/xtra.RHnw.html.
2.3.2 Discriminant models
We explored if the set of environmental descriptors could: explain small mammal assemblage distributions locally, i.e. within each study area; and discriminate between the two study areas using data from trapline locations, a regional level analysis. Essentially this entailed discrimination of previously identified assemblages/traplines in the environmental space and thus presents a supervised classification problem. Model performances of two currently popular methods were compared: Discriminant Analysis (DA) and Logistic Multiple Regression (LMR). Both techniques were tested: in linear form, Linear Discriminant Analysis (LDA) (Fisher, 1936) and multinomial Generalized Linear Model (GLM); and in non-linear form, Mixture Discriminant Analysis (MDA) and Multiple Adaptive Regression Splines (MARS).
MARS
Multiple Adaptive Regression Splines (Friedman, 1991) enable modelling multiple categorical responses with weak assumptions on the shape of the response. A set of pairwise basis functions provide a set of nonlinear transformations on covariates enabling regression modelling in an enlarged space defined by the set of basis functions and their products and thus enabling identification of complex non-linear responses (Hastie et al., 2001, 1994). In the context of species distribution modelling, MARS incorporated within GLMs has been shown to outperform other non-parametric and non-linear modelling techniques on real (Leathwick et al., 2006) and simulated (Moisen and Frescino, 2002) data sets. Here we test performance of MARS incorporated into a multinomial GLM.
MDA
Non-linear decision boundaries in classification problems can also be modelled indirectly, i.e. without specifying and parameterising the form of the non-linear decision boundary. Mixture Discriminant Analysis is such an approach, generalising the linear decision boundaries of LDA by incorporating mixtures of multivariate Gaussians to discriminate a single class (Hastie and Tibshirani, 1993). MDA divides a given class into multiple subclasses each following a multivariate normal distribution with unique mean vector and a common covariance matrix. As far as we know, this method has been rarely tested in the context of niche spatial modelling except by Ter Braak et al. (2003). Here, maximum likelihood based parameter estimation was achieved using the expectation maximisation (EM) algorithm (cf. Appendix 1). Starting values for the EM algorithm were obtained by the Learning Vector Quantisation clustering algorithm (Hastie and Tibshirani, 1993).
All computations were performed in R (R Development Core Team, 2008). LDA was computed using the lda function of the “MASS” package. MARS and MDA were computed using the mars and mda functions of the mda package.
2.3.3 Evaluation and comparison of modelling techniques’ performances
In order to compare modelling techniques on the basis of their statistical properties, we only compared models of similar structure in their explanatory components, i.e. including all available explanatory variables (Segurado and Araujo, 2004; Potts and Elith, 2006).
Evaluation criteria
Following Pearce and Ferrier (2000), models were evaluated on their ability to i) provide reliable predictions (as soft classifiers) and ii) discriminate between occupied and unoccupied sites (as hard classifiers). Prediction reliability was quantified using residual deviance (RD) (Pearce and Ferrier, 2000; Leathwick et al., 2006) equivalent to minus twice the log likelihood of the fitted probabilities. i.e. for N observations
Discriminative performance (ii) is composed of omission (true positive fraction) and commission (false positive fraction) error (Pearce and Ferrier, 2000; Anderson et al., 2003). Here, predicted assemblages were mutually exclusive, i.e the commission error of one assemblage corresponded to the omission error of another. Consequently, discriminative performance was assessed by omission error only using the true presence misclassification error rate (Segurado and Araujo, 2004).
Model validation
Predictive ability was assessed using the above mentioned criteria on independently re-sampled testing data sets using the bootstrap 632+ (Efron and Tibshirani, 1995) which provides the least biased and variant model re-sampling evaluation method (Hastie et al., 2001). Bootstrap 632+ was developed to reduce optimistic error estimation that arises with the original bootstrap due to commonality between the observations of training and testing data sets. Originally used to estimate error rates of per-subject prediction rule (e.g. misclassification error rate) it has recently and successfully been applied to evaluate predictive models of species or disease distribution (Steyerberg et al., 2001; Leathwick et al., 2006; Potts and Elith, 2006). We computed the bootstrap 632+ in R (R Development Core Team, 2008), using the bootpred function of the boostrap package and the errorest function of the ipred package. For each criteria the mean and standard deviation across 1000 bootstrap samples was calculated.
2.3.4 Effects of environmental descriptors on assemblage distributions
To simplify the assessment of effect size we selected the classification methods that provided the lowest bootstraped misclassification error rate and residual deviance.
Effect Size
Multicollinearity between factors is known to inflate the variance of regression coefficients, alter model predictive performance and complicates identification of real effects of factors on data variability (Legendre and Legendre, 1998). Hierarchical partitioning (MacNally, 2000, 2002), widely used in explanatory modelling (Gibson et al., 2004; Greaves et al., 2006; Olivier et al., 2000), permits to partition the variance explained by a model in order to isolate the independent versus joint contributions of each variable. We used this method to estimate the independent effects of each variable on the log-likelihood. Significance of these independent effects was assessed using a permutation test. The independent effect was re-estimated using 100 random permutations of the covariates, the mean and standard deviation of the resulting distribution permitting a z-test of the original independent effect.
The effect size of each environmental descriptor on the probability of observing each assemblage were assessed. All possible additive combinations of variables were fitted. For each combination we calculated the mean predicted probability of each assemblage at sites where that assemblage was observed. The difference in predicted probabilities
were computed, where μj1 and μj0 represent the mean predicted probabilities across models which included and excluded the jth variable respectively. Means and 95% quantile intervals were estimated over 200 cross-validation iterations. Predictive effects were considered for further analysis if the lower 95% quantile was greater than zero.
Response shape along environmental gradients
The shape of assemblage responses were visualised by plotting predicted probabilities with respect to each relevant environmental variable (Elith et al., 2005). Predictions of assemblage occurrence probabilities were made across the range of the variable of interest whilst all other variables were fixed to their mean value among sites corresponding to the assemblage in question.
All computations were made in R (R Development Core Team, 2008). Independent effects were estimated using the combos and partition functions of the hier.part package. The z.test was adapted from the rand.hp function of the same package.
3 Results
3.1 Trapping success: small mammal data
A total of 173 small mammals were trapped in Rangtang (90) and Maerkang (83) including 10 rodent, 1 lagomorph and 4 insectivore species (table 1). Four species were trapped in both areas: Ochotona cansus, Eozapus setchuanus, Apodemus peninsulae, and Microtus irene.
Table 1.
Sampling success for each species (number of trapped individuals) obtained in each study area. The order of each species is indicated: Rodent (R), Insectivore (I) or Lagomorph (L).
| Location |
|||
|---|---|---|---|
| Order | Maerkang | Rangtang | |
| Empty traps | 4516 | 3396 | |
| Apodemus draco (Apda) | R | 17 | 0 |
| Apodemus latronum (Apla) | R | 14 | 0 |
| Apodemus peninsulae (Appe) | R | 3 | 68 |
| Eospalax fontanirerf (Eofo) | R | 1 | 0 |
| Eozapus setchuanus (Eose) | R | 1 | 1 |
| Microtus irene (Miir) | R | 1 | 1 |
| Microtus limnophilus (Mili) | R | 0 | 13 |
| Micromys minutus (Mimi) | R | 6 | 0 |
| Niviventer confucianus (Nico) | R | 19 | 0 |
| Sicista concolor (Sico) | R | 1 | 0 |
| Chodsigoa hypsibia (Chhy) | I | 10 | 0 |
| Sorex cylindricauda (Socy) | I | 1 | 0 |
| Sorex thibetanus (Soth) | I | 2 | 0 |
| Uropsilus soricipes (Urso) | I | 2 | 0 |
| Ochotona cansus (Occa) | L | 5 | 7 |
3.2 Assemblage definition
Maerkang
Four assemblages were identified (M1 to M4) (table 2, Appendix 3. a). Assemblage M1, which included seven habitats subjected to human influences, was dominated by the rat Niviventer confucianus. M2, the Culture habitat, was not merged with other classes and was dominated by Micromys minutus which was specific to that habitat. M3 included forest and bush was the richest (n = 11) and most diversified (7.86) assemblage. Trapping probabilities were an order of magnitude lower in M3 than in other assemblages and nothing was trapped in Forest Oak and Stream bushes. M4 grouped white and red birch forests and had the lowest diversity. Trapping frequencies of Apodemus draco in M4 were greater than for any other species in any other assemblage.
Rangtang
Four assemblages were identified (R1 to R4) (table 2, Appendix 3. b). R1, which included habitats in close vicinity to culture, was dominated by Apodemus peninsulae and provided the lowest diversity among all assemblages. In R2, which included valley bottom bushes, Forest coniferous/willow bushes and Village garden, Apodemus peninsulae was dominant but trapped with lower probability than in R1. R3 corresponded to the Fenced grassland class which was not merged with other classes. Only two species were found: Microtus limnophilus which was trapped at highest probability in R3, and Apodemus peninsulae. R4 grouped Forest rhododendron/coniferous with Slope and Stream bushes. Diversity was highest in this assemblage and Ochotona cansus was specific to it.
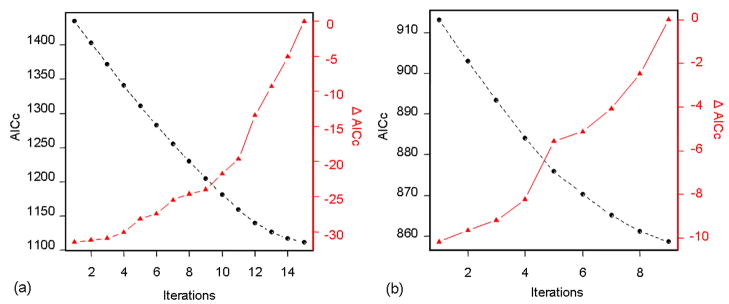
Details of AICc and Δ AICc evolution according to the number of the expert-class-merging procedure iterations, for Maerkang and Rangtang study areas, are available in the Appendix 2.
3.3 Assemblage habitat modelling
Modelling techniques were applied to discriminate all assemblages previously defined except assemblage R3 since it included only 2 traplines (table 2).
3.3.1 Modelling techniques performances (table 3)
Table 3.
Residual deviance (RD) and misclassification error rates (Error) obtained for each modelling procedure (Linear Discriminant Analysis (LDA), Mixture Discriminant Analysis (MDA), Multinomial logistic regression (MN) and Multiple Adaptive Regression Spline (MARS)), for each study case, on training (Train) and testing (Boot) data sets. For the test data set, means (Boot mean) and standard deviances (Boot sd), estimated over 1000 bootstrap 632+ iterations, are indicated.
| LDA | MDA | MN | MARS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RD | Error | RD | Error | RD | Error | RD | Error | ||
| Maerkang | |||||||||
|
| |||||||||
| Train | 86.645 | 0.212 | 55.251 | 0.121 | 70.886 | 0.197 | 0.001 | 0.000 | |
| Boot mean | 111.322 | 0.270 | 105.491 | 0.258 | 107.589 | 0.313 | 53.585 | 0.221 | |
| Boot sd | 0.092 | 0.001 | 0.109 | 0.001 | 0.084 | 0.001 | 0.049 | 0.000 | |
|
| |||||||||
| Rangtang | |||||||||
|
| |||||||||
| Train | 46.815 | 0.182 | 71.922 | 0.073 | 13.060 | 0.055 | 0.000 | 0.000 | |
| Boot mean | 67.082 | 0.308 | 65.095 | 0.265 | 44.391 | 0.253 | 41.144 | 0.232 | |
| Boot sd | 0.092 | 0.001 | 0.069 | 0.001 | 0.129 | 0.001 | 0.046 | 0.001 | |
|
| |||||||||
| Study sites | |||||||||
|
| |||||||||
| Train | 51.693 | 0.106 | 17.506 | 0.033 | 41.230 | 0.081 | 0.000 | 0.000 | |
| Boot mean | 56.292 | 0.111 | 46.620 | 0.073 | 57.401 | 0.101 | 16.927 | 0.029 | |
| Boot sd | 0.001 | 0.000 | 0.012 | 0.000 | 0.027 | 0.000 | 16.927 | 0.029 | |
Discriminant Analysis
For discriminant analysis of Maerkang assemblages and study sites, bootstrapped RD and error rate of MDA were approximately twice the RD and error estimated on training data. MDA was a more discriminant and reliable predictor than LDA in terms of lower bootstrapped error rates (Δ Error(Maerkang)LDA−MDA = 0.012; Δ Error(Region)LDA−MDA = 0.038) and RD (Δ RD(Maerkang)LDA−MDA = 5.831; Δ RD(Region)LDA−MDA = 9.672). In Rangtang, training data based estimates indicated MDA gave a more deviant fit than LDA although again bootstrapped RD (Δ RD(Rangtang)LDA−MDA = 1.99) and error (Δ Error(Rangtang)LDA−MDA = 0.043) were lower than for LDA.
Mixture Discriminant Analysis was also a more discriminatory and reliable model than the multinomial GLM in the Maerkang (Δ Error(Maerkang)MN−MDA = 0.055; Δ RD(Maerkang)MN−MDA = 2.089) and regional level (Δ Error(Region)MN−MDA = 0.028; Δ RD(Region)MN−MDA = 10.781) analysis. However, in Rangtang, MN outperformed MDA (Δ Error(Rangtang)MDA−MN = 0.012; Δ RD(Rangtang)MDA−MN = 20.704).
Finally, in each of the three analysis, MARS perfectly fitted the training data (RDMARS,train = 0.00; ErrorMARS,train = 0.00). Despite this overfitting, MARS was the most reliable and discriminatory model in the prediction test, providing bootstrap predictor errors 0.037, 0.021 and 0.044 lower than the next most discriminant predictor of Maerkang assemblages, Rangtang assemblages and study sites respectively.
3.3.2 Effects of environmental descriptors on assemblage distributions
Maerkang
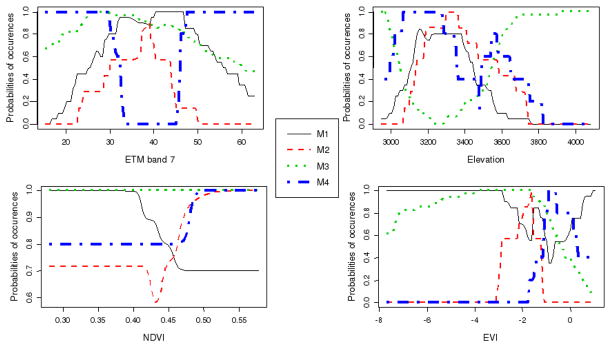
Significant independent contributions to model goodness-of-fit were observed for elevation and ETM band 7 (table 4). Among all variables, elevation provided the largest independent contribution (I%= 39.7). It increased predicted probabilities for all assemblages bar M2 on training and bootstrapped data sets (table 5). A threshold was observed at about 3600 meters altitude above which M1 was seldom observed, the probability of M2 and M4 dropped rapidly and M3 became predominant (figure 2). Enhanced Thematic Mapper band 7 consistently discriminated all assemblages. The probability of M2 (cultures) reached a distinct peak at intermediate values (40) where the probability of M4 approached zero.
Table 4.
Percentage of the independent contribution over all independent effects (I %) and total contributions (Total) in model goodness of fit estimated for each variable and for each study case. The statistical significance (sig.) of the Z.score based on upper 0.95 confidence limit is indicated by (*)
| ETM7 | Elevation | Slope | SI | NDVI | EVI | |
|---|---|---|---|---|---|---|
| Maerkang | ||||||
| I% | 20.912 | 39.196 | 11.461 | 2.208 | 11.068 | 15.156 |
| Z.score | 2.13 | 9.36 | 0.17 | −0.15 | −0.03 | 0.52 |
| sig. | * | * | ns | ns | ns | ns |
| Total | 23.096 | 39.691 | 15.099 | 3.601 | 11.091 | 16.627 |
|
| ||||||
| Rangtang | ||||||
| I % | 16.410 | 23.313 | 21.160 | 2.263 | 29.058 | 7.790 |
| Z.score | 1.9 | 5.88 | 4.13 | −1.1 | 2.92 | −0.27 |
| sig. | * | * | * | ns | * | ns |
| Total | 17.251 | 18.888 | 9.694 | 2.369 | 26.061 | 5.812 |
|
| ||||||
| Region | ||||||
| I % | 26.842 | 37.132 | 8.529 | 0.316 | 21.024 | 6.156 |
| Z.score | 12.68 | 16.4 | 3.79 | 0.19 | 7.35 | 2.74 |
| sig. | * | * | * | ns | * | * |
| Total | 31.888 | 58.427 | 16.210 | −0.549 | 39.418 | 13.850 |
Table 5.
Contributions of each variable to change in the mean predicted probabilities of each assemblage at sites where the given assemblage was observed. Estimates based on training data (Mean Train) and cross validation means (Mean CV P1-P0) and confidence intervals (qt0.05 CV P1-P0) are shown. Grey cells correspond to variable effects which lower 95% quantile was greater than zero.
| Maerkang | Rangtang | |||||||
|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | R1 | R2 | R4 | ||
| ETM 7 | Mean Train | 0.158 | 0.254 | 0.096 | 0.255 | 0.101 | 0.113 | 0.098 |
|
| ||||||||
| Mean CV P1-P0 | 0.107 | 0.143 | 0.049 | 0.212 | 0.030 | 0.003 | 0.015 | |
| qt0.05 CV P1-P0 | 0.079 | 0.068 | 0.021 | 0.133 | 0.008 | −0.017 | 0.000 | |
| qt0.95 CV P1-P0 | 0140 | 0.234 | 0.073 | 0.293 | 0.053 | 0.018 | 0.029 | |
|
| ||||||||
| elevation | Mean Train | 0.363 | 0.133 | 0.278 | 0.433 | 0.161 | 0.128 | 0.123 |
|
| ||||||||
| Mean CV P1-P0 | 0.250 | 0.060 | 0.329 | 0.283 | 0.207 | 0.073 | 0.127 | |
| qt0.05 CV P1-P0 | 0.212 | −0.007 | 0.290 | 0.160 | 0.178 | 0.038 | 0.078 | |
| qt0.95 CV P1-P0 | 0.278 | 0.121 | 0.357 | 0.364 | 0.237 | 0.102 | 0.163 | |
|
| ||||||||
| slope | Mean Train | 0.076 | 0.092 | 0.038 | 0.197 | 0.150 | 0.228 | 0.071 |
|
| ||||||||
| Mean CV P1-P0 | 0.018 | 0.028 | −0.021 | 0.036 | 0.190 | 0.307 | 0.052 | |
| qt0.05 CV P1-P0 | −0.012 | −0.050 | −0.044 | −0.046 | 0.159 | 0.271 | 0.028 | |
| qt0.95 CV P1-P0 | 0.049 | 0.075 | 0.003 | 0.096 | 0.222 | 0.342 | 0.079 | |
|
| ||||||||
| SI | Mean Train | 0.005 | −0.003 | 0.003 | −0.012 | −0.002 | 0.007 | 0.008 |
|
| ||||||||
| Mean CV P1-P0 | −0.016 | −0.042 | −0.009 | −0.030 | −0.009 | −0.006 | −0.011 | |
| qt0.05 CV P1-P0 | −0.032 | −0.195 | −0.019 | −0.058 | −0.011 | −0.009 | −0.017 | |
| qt0.95 CV P1-P0 | −0.002 | 0.017 | −0.001 | −0.001 | −0.007 | −0.004 | −0.008 | |
|
| ||||||||
| NDVI | Mean Train | 0.087 | 0.106 | 0.045 | 0.140 | 0.084 | 0.195 | 0.300 |
|
| ||||||||
| Mean CV P1-P0 | 0.034 | 0.011 | −0.003 | 0.043 | 0.041 | 0.084 | 0.218 | |
| qt0.05 CVP1-P0 | 0.008 | −0.030 | −0.019 | −0.006 | 0.014 | 0.057 | 0.186 | |
| qt0.95 CV P1-P0 | 0.063 | 0.066 | 0.014 | 0.092 | 0.069 | 0.111 | 0.245 | |
|
| ||||||||
| EVI | Mean Train | 0.104 | 0.237 | 0.055 | 0.180 | 0.043 | 0.042 | 0.058 |
|
| ||||||||
| Mean CV P1-P0 | 0.029 | 0.104 | 0.000 | 0.093 | −0.014 | −0.014 | −0.010 | |
| qt0.05 CV P1-P0 | −0.008 | 0.037 | −0.019 | 0.035 | −0.024 | −0.021 | −0.016 | |
| qt0.95 CV P1-P0 | 0.063 | 0.179 | 0.022 | 0.169 | −0.005 | −0.006 | −0.004 | |
Figure 2.
Responses (predicted probabilities) of Maerkang assemblages along those continuous gradients for which predictive power for at least one assemblage was identified (table 5).
Despite the non-significance of their independent contributions, vegetation indices influenced mean predicted probabilities (table 4, table 5). On average, inclusion of NDVI increased predicted probabilities of M1 by 0.087. A change was observed at NDVI = 0.45 with M1 being more probable than M2 or M4 below and less probably than other assemblages above this point respectively (figure 2). Inclusion of EVI significantly increased the probability to predict M2 and M4 whose bootstrapped means increased by 0.104 and 0.093 respectively. The range of EVI corresponding to M2 occurrence being lower than that of M4 (birch forests). Finally, slope and SI had no predictive power.
Rangtang
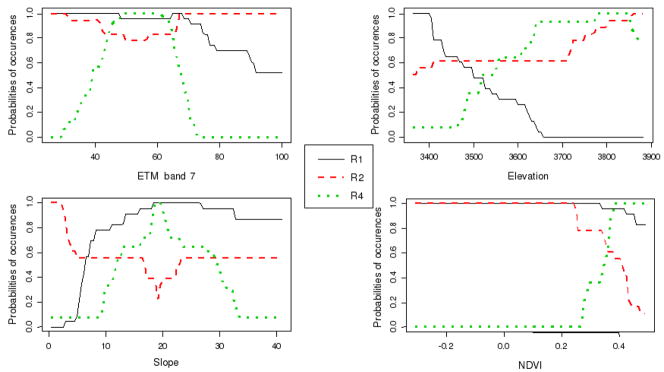
Independent effects of NDVI, elevation, ETM band 7 and slope were all significant (table 4). NDVI provided the highest independent contribution (I%=29.058) and significantly improved mean predicted probabilities for all assemblages, this improvement being greatest for R4 (Forest rhododendron/coniferous and Slope bushes) (0.30) which was not observed when NDVI < 0.25 (table 5, figure 3). By contrast, above this threshold, the probability of R2, and to a lesser extent R1, dropped considerably.
Figure 3.
Responses (predicted probabilities) of Rangtang assemblages along those continuous gradients for which predictive power for at least one assemblage was identified (table 5).
Elevation increased predicted probabilities for all assemblages, the size of the effect being greatest for R1 (fields and bushes) which was predominant at lower altitudes and did not occur above 3650 meters. Above this level, the probability of R4 reached a plateau and the probability of R2 also increased. Inclusion of ETM band 7 significantly increased prediction accuracy for R1 and R4. Band 7 values greater than 70 corresponded to predominant and subordinant predicted probabilities of R2 and R4 respectively, the latter being optimally distributed in the range 50–65. Inclusion of slope significantly increased predictive probabilities at R2, R1 and R4 sites (ordered by decreasing effect size). R2 was the predominant assemblage in flat areas and R1 became predominant when slopes became steeper than 10 °. Finally, SI and EVI had no predictive effects.
Between study sites
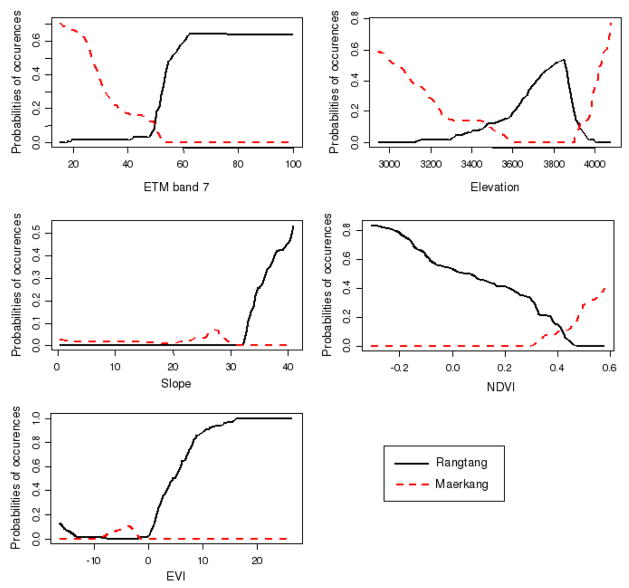
All variables had significant and independent contributions in model goodness-of-fit except SI which had a negative contribution in the model likelihood (table 4). Rangtang was associated with ETM band 7 > 50, steep slopes, EVI > 0, NDVI < 0.4 and elevations in the range 3600–3900 meters. Maerkang was associated with ETM band 7 < 50, NDVI > 0.4 and elevations below and above 3500m and 3900m respectively (figure 4).
Figure 4.
Responses (predicted probabilities) of the two study areas along those continuous gradients for which predictive power for at least one assemblage was identified.
4 Discussion
4.1 Assemblages as mixtures of species distributions
The ability of the expert-class-merging procedure to cluster observations from a large number of habitats into a small number of super-classes highlights that apparently different habitats in fact provide approximately equivalent habitat quality for many small mammals species. However, the influence of a species in the assemblage definition is directly related to trapping frequency, the definition being dominated by those species trapped in largest number (e.g. Apodemus peninsulae), whilst rare or shy species trapped in low numbers (e.g. Eozapus setchuanus) play a relatively small role in the assemblage definition. It is well known that spatial distributions of small mammals can be driven by population dynamics (Giraudoux et al., 1997, 2007). The assemblages defined in the present study constituted a spatio-temporal snapshot of a process of complex interactions between multi-species metapopulations (Guisan and Thuiller, 2005). The current data set, with its lack of a temporal component, thus limits analyses to the description of “potential habitats” for the identified small mammal assemblages (Guisan and Thuiller, 2005; Araujo and Guisan, 2006; Guisan et al., 2006).
4.2 Assemblage habitat distribution in environmental space
Linear or non-linear Discriminant Analysis for small sample size problems?
Linear Discriminant Analysis is said to perform well when sample size is small because of the simple boundaries it provides between classes (Hastie et al., 1997). However, our analyses showed prediction reliability and class discrimination were consistently lower for MDA than for LDA. MDA’s discrimination of assemblages in several sub-classes, permitting identification of non-linear boundaries, clearly helped capture important extra within class variability. This result re-enforces the need to use non-linear models in community modelling even when sample size is small (Munoz and Felicisimo, 2004).
MDA allows multi-modal representation of an assemblage in environmental space. By contrast, MN assumes assemblage responses can be represented as simple linear combinations of variables rescaled to the 0–1 range via a link function. Despite a larger number of parameters, MDA was more discriminant and less deviant than MN for discrimination of Maerkang assemblages and site environments. This result highlights the importance of modelling sub-class clusters in those case studies. By contrast, Rangtang assemblages were better discriminated by MN than by MDA suggesting a more homogeneous distribution of those assemblages in environmental space. MN was also a more performant predictor of Rangtang data than LDA despite the two models sharing the same logit regression form (Hastie et al., 2001). Regardless of linearity/non-linearity assumptions, discriminant analysis was not the most appropriate method for modelling the distribution of Rangtang assemblages for which sample sizes were smallest.
Parametric or non-parametric non-linear modelling of assemblage distributions?
Multiple Adaptive Regression Splines advantageously extends the logistic response of multinomial GLMs to include multi-modal variation of the linear predictor with respect to a given variable. MARS has previously been shown to be a more appropriate mapping technique for Fagus species than Logistic Multiple Regression (Munoz and Felicisimo, 2004) although more generally there has been difficulty to demonstrate better performance of MARS over linear models on real data sets (Moisen and Frescino, 2002). Here, MARS consistently provided more discriminatory and reliable predictions than LDA, MDA and GLM in all three of our cases studies. The Non-linear structures in class distributions were more accurately captured by a model which didn’t rely on parametric assumptions regarding assemblage distribution in environmental space. These promising prediction results of MARS were obtained despite strong evidence of overfitting (i.e. selection of an excessive number of basis functions) the training data. This observation is encouraging since the superior predictive performance of MARS could clearly be improved via the increased parsimony obtainable with model selection procedures such as step-wise pruning (Friedman, 1991).
Assuming each a priori habitat was representable as a single cluster in environmental space, a mixture representation of an assemblage (i.e. super-class), including the diversity of species responses, and thus the requirement of non-linear decision boundaries for assemblage discrimination, becomes natural. This complexity arises in part from the fact that most of the variables in question were, at best, indirectly related to small mammal dynamic processes. Guisan and Zimmerman (2000) suggested that the use of direct environmental variables provides a physiologically mechanistic character to a model that is not apparent when indirectly related variables are used. We further suggest that the use of indirect variables in fact complicates response curves. Multi-modal responses in the space of indirect variables might correspond to relatively homogeneous responses among variables directly related to processes.
4.3 Explanatory and predictive power of environmental descriptors
Within versus between study area discrimination
The observed between site differences in small mammal fauna, environmental conditions and pertinence of environmental variables provided strong evidence that two distinct biogeographical zones had been identified. The degree of abruptness/smoothness in the transition between these zones was not identifiable given the scale of the sampling design and clearly further work is required before the process of small mammal communities on the fringes of the Tibetan plateau are fully elucidated.
The discrimination between Maerkang and Rangtang at high and low NDVI values respectively suggests greater productivity in terms of vegetative biomass in Maerkang. This corresponds with our a priori land cover classification since forested habitats were more widespread and diversified in Maerkang than in Rangtang. By contrast, within study sites, NDVI and EVI were useful discriminators of different assemblages. In Maerkang, NDVI significantly improved predictions of M1 (bushes, grassland an oak forests near villages), while EVI helped discriminate birch forests (M4) and culture (M2). In Rangtang, NDVI discriminated forest (R4), valley-bottom vegetation (R2) and non-forested bush (R1), whereas EVI displayed no discriminative ability. The fact that NDVI was less useful in Maerkang, where nine in eighteen habitats were forested, than in Rangtang, where only two in twelve habitats were forested habitats, probably reflects the saturation problem, i.e. NDVI’s limited ability to discriminate among forest types (Huette et al., 1999).
Variables such as elevation, ETM band 7 and slope had descriptive and predictive power but their indirect relation to small mammal resources renders interpretation of their effects difficult (regardless of the mixture problem outlined above). Moreover, these variables are subjected to the law of relative site constancy (Guisan and Zimmerman, 2000; Randin et al., 2006) which complicates the comparison of assemblage distributions between the two study areas along these gradients.
The discriminatory power of environmental variables differed according to the study area (Maerkang versus Rangtang) and to the spatial extent of the training area (local versus regional analysis). For predictive mapping purposes, one should be aware of such variation in order to select a set of environmental variables appropriate for the required extent, resolution and location of the map (Guisan and Thuiller, 2005).
Improving habitat definition
Numerous improvements to the current habitat descriptions can be envisaged. Firstly, numerous species have been shown to respond to landscape level effects whereby densities respond to composition or structure of landscape in a surrounding neighbourhood. Examples include Tetrao urogallus (Graf et al., 2005), Echinococcus multilocularis (Giraudoux et al., 2003), Arvicola terrestris (Fichet-Calvet et al., 2000; Giraudoux et al., 2007; Morilhat et al., 2007) and Microtus arvalis (Delattre et al., 1999, 2006; Duhamel et al., 2000), the latter two being small mammal species of the sub-family Arvicolinae. Here effects of composition and structure in buffers surrounding traplines were not considered, largely due to identifiability issues associated with the required increased level of data mining and the small sample size.
Secondly, trapline locations were often observed to lie within mixed pixels where numerous habitat types contribute to the observed spectral response. Improved predictive performance might therefore be achievable by addressing the mixed pixel problem for which fuzzy set approaches (Foody, 2000) and super-resolution techniques (Tatem et al., 2002) exist.
Thirdly, descriptors of structure and composition of the first vegetative strata, being more directly related to small mammal resources would be pertinent (Catling and Coops, 1999; Pearce et al., 2001; Gibson et al., 2004). This move to direct variables would be of particular relevance in a predictive modelling setting since indirect variables limit model transferability across large areas (Guisan and Thuiller, 2005; Randin et al., 2006). Understory modelling with very high resolution satellite remotely sensed data has recently been applied in sparse forests (Jianxi et al., 2007). Further, helicopter-borne laser scanner data has been used to provide high resolution DEMs from which understory structural and textural characteristics are easily extracted (Hirata et al., 2003). Further research is needed before remotely sensed indices of understory structure can be incorporated into small mammal species distribution models.
Toward building a process based models
As outlined above, statistical models can help resolve some theoretical hypotheses on species/environment relationships (e.g. shape of the response curves, selection of influent environmental factors) and thus provide basis for the development of process based models. Inversely, process based models can serve statistical modelling and an iterative procedure incorporating both approaches had been advocated (Austin, 2007). Here, prediction precision might be increased once a deeper understanding of the ecological processes driving small mammal distributions on the fringes of the Tibetan plateau is achieved. It would be helpful to investigate landscape composition effects on small mammal communities in time and space. For this purpose, there is need to develop process based landscape metrics of direct relevance to small mammal resources. Ultimately, such relationships could be incorporated into a Structural Equation Modelling framework (Guisan et al., 2006; Austin, 2007).
4.4 Conclusion and perspectives
Our results showed mixtures of Gaussians provided better descriptions of the environmental space occupied be small mammal assemblages than single cluster model. However, the Gaussian mixture model was in turn outperformed by MARS with its flexible ability to detect non-linearity. ETM band 7, vegetation indices, elevation and slope all helped discriminate assemblages. Their predictive effects varied according to the location and spatial scale of the training area. However, predictive performance might be improved given further investigation of methodological issues and a deeper understanding of underlying ecological processes. i.e. given: a deeper knowledge of the responses of multiple-species to environmental variation; a closer match between spectrally derived variables and small-mammal resources; or, an improved data set characteristics (sampling protocol). In our study, MARS was prone to over-fitting, thus its transferability and superior performance over other techniques should be assessed cautiously prior to extrapolation beyond the sampling area (Randin et al., 2006). Moreover, because it is a classifier (i.e assumes the pre-defined groups exist), used here to model assemblage responses jointly, prediction of new (i.e. untrained) assemblages in space and time was not possible. Two distinct biogeographical areas regarding small mammal assemblages and environmental conditions were identified here, but the current sampling design does not permit elucidation of the transition between the two areas. Mapping small mammal assemblages in the area between Rangtang and Maerkang would require a sampling design spanning the ranges of each species within both the geographical and environmental space separating the two areas (Murphy, 2007).
Supplementary Material
Acknowledgments
This work was supported by grants number RFATW-00-002 and RO1 TW001565 from the Fogarty International Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Center or the National Institutes of Health. We address many thanks to our Chinese colleagues working for the Sichuan Center for Diseases Control of Chengdu for their logistic support. We express many thanks to Kurt Galbreath, department of Ecology and Evolutionary Biology, Cornell University, Ithaca (New York), for his help in identification of Ochotona species. We address a special thanks to Stéphane Chrétien of the Department of Mathematics, UMR CNRS 6623, for his constructive critiques and corrections of the statistical component of this study. We also address many thanks to Alexandre Hirzel for his interest and attention in a pre-submission review of this paper.
4.5.1 Appendix 1: MDA algorithm
The algorithm computes the mixture density for each class j divided in Rj subclasses each defined by their own mean μ(j,r) and a common covariance matrix Σ, such as:
where D(x,y) is the Mahalanobis distance between x and the y and the mixing probability πjr are unknown model parameters. Then, the conditional log-likelihood for the data lmix is computed and maximised by the EM (Expectation Maximisation) algorithm with:
Expectation Maximisation (EM) algorithm is an iterative two-step procedure: first, it estimates the mixing probabilities for each subclass and class and then, it conditionally optimises the mean and covariance for each subclasses.
4.5.2 Appendix 2: Expert-class-merging procedure
Appendix 2.
Redundancy reduction of the expert-class-merging procedure for (a) Maerkang and (b) Rangtang data sets illustrated by decreasing AICc and increasing Δ AICc w.r.t. to the number of iterations of the procedure.
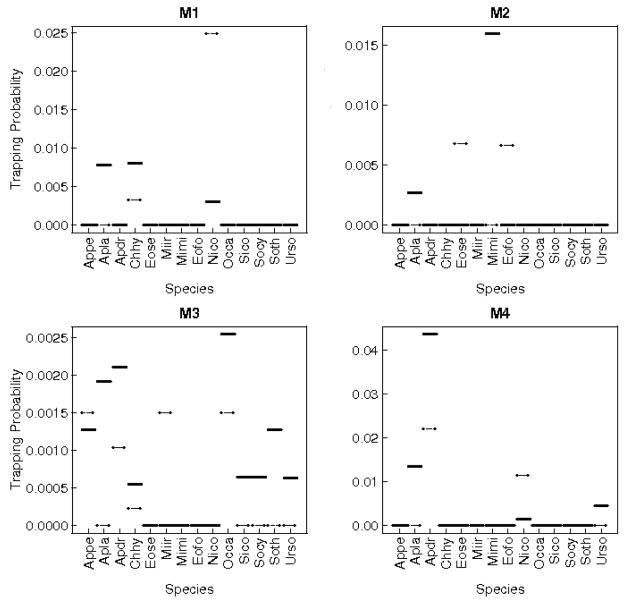
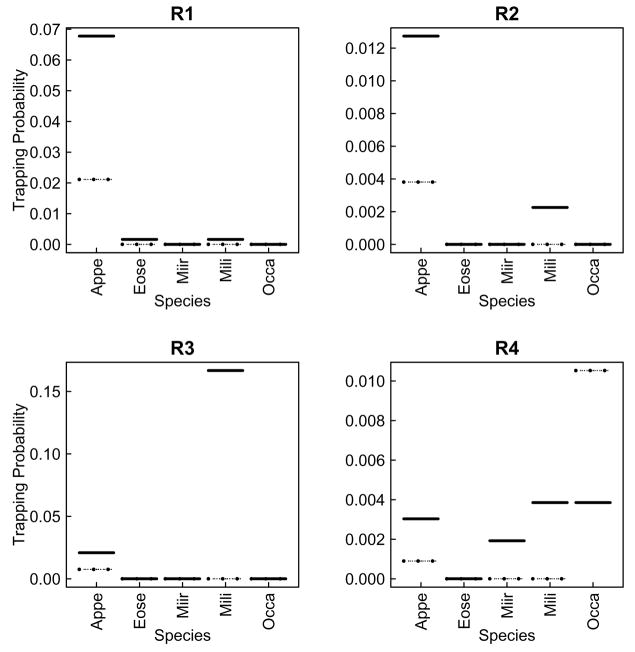
4.5.3 Appendix 3: Species predicted probability distributions in Maerkang and Rangtang assemblages
Appendix 3.a.
Species trapping joint probabilities predicted by the multinomial model for each assemblage (M1 to M4), in Maerkang. Dashes and full lines correspond to small and big traps respectively. Correspondence between species names and their abbreviations is available in table 2.
Appendix 3.b.
Species trapping joint probabilities predicted by the multinomial model for each assemblage (R1 to R4), Rangtang. Dashes and full lines correspond to small and big traps respectively. Correspondence between species names and their abbreviations is available in table 2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson M, Clements A. Resolving environmental disputes: a statistical method for choosing among competing cluster models. Ecol Appl. 2000;10(5):1341–1355. [Google Scholar]
- Anderson M, Lew D, Peterson A. Evaluating predictive models of species’ distributions: criteria for selecting optimal models. Ecol Model. 2003;162:211–232. [Google Scholar]
- Araujo M, Guisan A. Five (or so) challenges for species distribution modelling. J Biogeogr. 2006;33:1677–1688. [Google Scholar]
- Austin M. Spatial prediction of species distribution: an interface between ecological theory and statistical modelling. Ecol Model. 2002;157(18):101–118. [Google Scholar]
- Austin M. Species distribution models and ecological theory: A critical assessment and some possible new appraoches. Ecol Model. 2007;200:1–19. [Google Scholar]
- Burnham K, Anderson D. Model selection and multimodel inference. 2. Springer-verlag; New York: 1998. [Google Scholar]
- Butet A, Paillat G, Delettre Y. Factors driving small rodents assemblages from field boundaries in agricultural landscapes of western france. Landsc Ecol. 2006;21:449–461. [Google Scholar]
- Catling PC, Coops NC. Prediction of the distribution and abundance of small mammals in the eucalypt forests of south-eastern australia from airborne videography. Wildl Res. 1999;26:641–650. [Google Scholar]
- Corbet G. British Museum (Natural History) Cornell University Press; Londres: 1978. The mammals of the Palearctic Region: a taxonomic review. [Google Scholar]
- Delattre P, Clarac R, Melis J, Pleydell D, Giraudoux P. How moles contribute to colonization success of water voles in grassland:implications for control. J Appl Ecol. 2006;43(2):353–359. [Google Scholar]
- Delattre P, De Sousa B, Fichet-Calvet E, Quéré J, Giraudoux P. Vole outbreaks in a landscape context: evidence from a six year study of microtus arvalis. Landsc Ecol. 1999;14(4):401–412. [Google Scholar]
- Doledec S, Chessel D, Gimaret-carpentier C. Niche separation in community analysis: a new method. Ecology. 2000;81:2914–2927. [Google Scholar]
- Duhamel R, Quéré J, Delattre P, Giraudoux P. Landscape effects on the population dynamics of the fossorial form of the water vole (Arvicola terrestris sherman) Landsc Ecol. 2000;15(2):89–98. [Google Scholar]
- Efron B, Tibshirani R. Technical report (tr-477) Dept. of Statistics; Stanford University: 1995. Cross-validation and the bootstrap: Estimating the error rate of a prediction rule. [Google Scholar]
- Elith J, Ferrier S, Huettmann F, Leathwick J. The evaluation strip: a new robust method for plotting predicted responses from species distribution models. Ecol Model. 2005;186:280–289. [Google Scholar]
- Fen Z, Zheng C. Studies on the pikas (genus Ochotona) of China. taxonomic and distribution. Acta Theriol Sin. 1985;5:269–289. [Google Scholar]
- Ferrier S, Guisan A. Spatial modelling of biodiversity at the community level. J Appl Ecol. 2006;43(3):393–404. [Google Scholar]
- Fichet-Calvet E, Pradier B, Quéré J, Giraudoux P, Delattre P. Landscape composition and vole outbreaks: evidence from an eight year study of Arvicola terrestris. Ecography. 2000;23(6):659–668. [Google Scholar]
- Fisher RA. The use of multiple measurments in taxonomic problems. Ann Eug. 1936;7:179–188. [Google Scholar]
- Foody G. Estimation of sub-pixel land cover composition in the presence of untrained classes. Comput Geosci. 2000;26:469–478. [Google Scholar]
- Friedman JH. Multivariate adaptive regression splines. Annals of Statistics. 1991;19:1–67. [Google Scholar]
- Gibson L, Wilson B, Aberton J. Landscape characteristics associated with species richness and occurence of small native mammals inhabiting a coastal heathland: a spatial modelling appraoch. Biol Conserv. 2004;120:75–89. [Google Scholar]
- Giraudoux P, Craig P, Delattre P, Bao G, Bartholomot B, Harraga S, Quéré J, Raoul F, Wang Y, Shi D, Vuitton D. Interactions between landscape changes and host communities can regulate Echinococcus multilocularis transmission. Parasitology. 2003;127:121–131. [PubMed] [Google Scholar]
- Giraudoux P, Delattre P, Habert M, Quéré J, Deblay S, Defaut R, Duhamel R, Moissenet M, Salvi D, Truchetet D. Population dynamics of fossorial water vole (Arvicola terrestris scherman): a land use and landscape perspective. Agric Ecosyst Environ. 1997;66:47–60. [Google Scholar]
- Giraudoux P, Pleydell D, Raoul F, Vaniscotte A, Ito A, Craig PS. Echinococcus multilocularis: why are multidisciplinary and multiscale approaches essential in infectious disease ecology? Tropical Medicine and Health. 2007;35(4):293–299. [Google Scholar]
- Giraudoux P, Quéré J, Delattre P, Bao G, Wang X, Shi D, Vuitton D, Craig P. Distribution of small mammals along a deforestation gradient in Southern Gansu, central China. Acta Theriol. 1998;43(4):349–362. [Google Scholar]
- Graf R, Bollmann K, Suter W, Bugmann H. The importance of spatial scale in habitat models: Capercaillie in the swiss alps. Landsc Ecol. 2005;20(6):703–717. [Google Scholar]
- Greaves R, Sanderson R, Rushton S. Predicting species occurence using information-theoretic appraoches and significance testing: An example of dormouse distribution in Cumbria, UK. Biol Conserv. 2006;130:239–250. [Google Scholar]
- Gromov IM, Erbajeva MA. The mammals of Russia. Russian Academy of Sciences; St Petersburg: 1995. [Google Scholar]
- Gromov IM, Polyakov IY. Voles (Microtinae) In: Brill EJ, editor. Fauna of the USSR, Mammals. Vol. 3. Publishing Company; 1978. [Google Scholar]
- Guisan A, Lehmann A, Ferrier S, Austin M, Overton JCRATH. Making better biogeographical predictions of species’ distributions. J Appl Ecol. 2006;43(3):386–392. [Google Scholar]
- Guisan A, Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecology Lett. 2005;8(9):993–1009. doi: 10.1111/j.1461-0248.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- Guisan A, Zimmerman N. Predictive habitat distribution models in ecology. Ecol Model. 2000;135:147–186. [Google Scholar]
- Hastie T, Tibshirani R. Discriminant analysis by gaussian mixtures. J R Stat Soc Series B (Methodological) 1993;58(1):155–176. [Google Scholar]
- Hastie T, Tibshirani R, Buja A. Flexible discriminant analysis by optimal scoring. J Ame Stat Ass. 1994;89(428):1255–1270. [Google Scholar]
- Hastie T, Tibshirani R, Buja A. Flexible discriminant and mixture models. In: Kay J, Titterington D, editors. Neural Networks and Statistics. Oxford University Press; 1997. [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning. Springer-Verlag; Aug, 2001. [Google Scholar]
- Hill MO. Ecology and Systematics. Ithaca, N.Y: Cornell University; 1979. DECORANA - A FORTRAN program for detrended correspondence analysis and reciprocal averaging. [Google Scholar]
- Hirata Y, Sato K, Sakai A, Kuramoto S, Akiyama Y. The extraction of canopy-understory vegetation-topography structure using helicopter-borne lidar measurement between a plantation and a broad-leaved forest. Geoscience and Remote Sensing Symposium, 2003 IGARSS apos;03. Proceedings 2003 IEEE International. 2003;5:3222–3224. [Google Scholar]
- Hirzel A, Hausser J, Chessel D, Perrin N. Ecological niche factor analysis: How to compute habitat-suitability maps without absence data? Ecology. 2002;83(7):2027–2036. [Google Scholar]
- Huette A, Didan K, Miura T, Rodriguez E, Gao X, Ferreira L. Overview of the radiometric and biophysical performance of the modis vegetation indices. Remote Sens Environ. 2002;83:195–213. [Google Scholar]
- Huette A, Justice C, van Leeuwen W. Modis vegetation index (mod 13) algorithm theoretical basis document. version 3 1999 [Google Scholar]
- Jianxi H, Feng M, Wenbo X. Retrieval of vegetation understory information fusing hyperion and panchromatic quickbird data in the method of neural network. Geoscience and Remote Sensing Symposium, 2007. IGARSS 2007. IEEE International Volume (23–28) 2007:4315–4318. [Google Scholar]
- Krasnov B, Shenbrot G, Khokhlova I, Ivanitskaya E. Spatial patterns of rodent communities in the Ramon erosion cirque, Negev Highlands, Israel. J Arid Environ. 1996;32(3):319–327. [Google Scholar]
- Leathwick J, Elith J, Hastie T. Comparative performance of generalized additive models and multivariate adaptative regression splines for statistical modelling of species distributions. Ecol Model. 2006;199:188–196. [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology. 2. Elsevier Science B.V; Amsterdam: 1998. [Google Scholar]
- Lehmann A, Overton JM, Leathwick J. GRASP: generalized regression analysis and spatial prediction. Ecol Model. 2002;157:189–207. [Google Scholar]
- MacNally R. Regression and model building in conservation biology, biogeography and ecology: the distinction between and reconciliation of “predictive” and “explanatory” models. Biodivers Conserv. 2000;9:655–671. [Google Scholar]
- MacNally R. Multiple regression and inference in conservation biology and ecology: further comments on identifying important predictor variables. Biodivers Conserv. 2002;11:1397–1401. [Google Scholar]
- McCullag P, Nelder J. Generalized Linear Models. 2. London: Chapman and Hall; 1989. [Google Scholar]
- Moisen GG, Frescino TS. Comparing five modelling techniques for predicting forest characteristics. Ecol Model. 2002;157(2–3):209–225. [Google Scholar]
- Morilhat C, Bernard N, Bournais C, Meyer C, Lamboley C, Giraudoux P. Response of Arvicola terrestris scherman populations to agricultural practices and Talpa europaea abundance in Eastern France. Agric Ecosyst Environ. 2007;122:392–398. [Google Scholar]
- Munoz J, Felicisimo AM. Comparison of statistical methods commonly used in predictive modelling. J Veg Sci. 2004;15(2):285–292. [Google Scholar]
- Murphy HT. Accounting for regional niche variation in habitat suitability models. Oikos. 2007;116(12):99–110. [Google Scholar]
- Olden J. Species-specific approach to modelling biological communities and its potential for conservation. Conserv Biol. 2003;17:854–863. [Google Scholar]
- Olden J, Joy M, Death R. Rediscovering the species in community-wide predictive modeling. Ecol Appl. 2006;16(4):1449–1460. doi: 10.1890/1051-0761(2006)016[1449:rtsicp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Olivier I, Mac Nally R, York A. Identifying performance indicators of the effects of forest management on ground-active arthropod biodiversity using hierarchical partitioning and partial canonical correspondence analysis. For Ecol Manag. 2000;139:21–40. [Google Scholar]
- Pearce J, Cherry K, Drielsma M, Ferrier S, Whish G. Incorporating expert opinion and fine-scale vegetation mapping into statistical models of faunal distribution. J Appl Ecol. 2001;38:412–424. [Google Scholar]
- Pearce J, Ferrier S. Evaluating the predictive performance of habitat models developed using logistic regression. Ecol Model. 2000;133:225–245. [Google Scholar]
- Potts JM, Elith J. Comparing species abundance models. Ecol Model. 2006;199(2):153–163. [Google Scholar]
- Pulliam H. On the relationship betwen niche and distribution. Ecology Lett. 2000;3:349–361. [Google Scholar]
- R Development Core Team, 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0. URL http://www.R-project.org
- Randin CF, Dirnbock T, Dullinger S, Zimmermann NE, Zappa M, Guisan A. Are niche-based species distribution models transferable in space? J Biogeogr. 2006;33(15):1689–1703. [Google Scholar]
- Raoul F, Pleydell D, Quéré J, Vaniscotte A, Rieffel D, Takahashi K, Bernard N, Wang J, Dobigny T, Galbreath K, Giraudoux P. Small mammals assemblage response to deforestation and afforestation in Central China: a multinomial based modelling approach. Mammalia. 2008;72:320–332. [Google Scholar]
- Raoul F, Quéré J, Rieffel D, Bernard N, Takahashi K, Scheifler R, Ito A, Wang Q, Qiu J, Yang W, Craig P, Giraudoux P. Distribution of small mammals in a pastoral landscape of the Tibetan plateau (Western Sichuan, China) and relationship with grazing practices. Mammalia. 2006;70(3–4):214–225. [Google Scholar]
- Segurado P, Araujo MB. An evaluation of methods for modelling species distributions. J Biogeogr. 2004;31(10):1555–1568. [Google Scholar]
- Smith A, Xie Y. A guide to the mammals of China. Princeton University Press; Princeton: 2008. [Google Scholar]
- Steyerberg E, Harell JF, Borsboom G, Eijkemans M, Vergouwe Y, Habbema J. Internal validation of predictive models: Efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54(8):774–781. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- Tatem A, Lewis H, Atkinson P, Nixon M. Super-resolution land cover pattern prediction using a hopfield neural network. Remote Sens Environ. 2002;79:1–14. [Google Scholar]
- Ter Braak C, Hoijtink H, Akkermans W, Verdonschot P. Bayesian model-based cluster analysis for predicting macrofaunal communities. Ecol Model. 2003;160(3):235–248. [Google Scholar]
- Wang Q, Vuitton D, Qiu J, Giraudoux P, Xiao Y, Schantz P, Raoul F, Li T, Wen Y, Craig P. Partial fencing as a possible risk factor for human alveolar echinococcosis in pastoral herdsmen communities of Sichuan, China. Acta Trop. 2004;90:285–293. doi: 10.1016/j.actatropica.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Wilson DE, Reeder DM. Mammals species of the World : a taxonomic and geographic reference. Smithsonian Institution Press; Washington, Londres: 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.