Abstract
Elongator is a six subunit protein complex, conserved from yeast to humans. Mutations in the human Elongator homologue, hELP1, are associated with the neurological disease familial dysautonomia. However, how Elongator functions in metazoans, and how the human mutations affect neural functions is incompletely understood. Here we show that in Caenorhabditis elegans, ELPC-1 and ELPC-3, components of the Elongator complex, are required for the formation of the 5-carbamoylmethyl and 5-methylcarboxymethyl side chains of wobble uridines in tRNA. The lack of these modifications leads to defects in translation in C. elegans. ELPC-1::GFP and ELPC-3::GFP reporters are strongly expressed in a subset of chemosensory neurons required for salt chemotaxis learning. elpc-1 or elpc-3 gene inactivation causes a defect in this process, associated with a posttranscriptional reduction of neuropeptide and a decreased accumulation of acetylcholine in the synaptic cleft. elpc-1 and elpc-3 mutations are synthetic lethal together with those in tuc-1, which is required for thiolation of tRNAs having the 5′methylcarboxymethyl side chain. elpc-1; tuc-1 and elpc-3; tuc-1 double mutants display developmental defects. Our results suggest that, by its effect on tRNA modification, Elongator promotes both neural function and development.
Author Summary
The efficiency of protein synthesis can be modulated by alterations of various components of the translation machinery. In translation, transfer RNAs act as adapter molecules that decode mRNA into protein and thereby play a central role in gene expression. In the tRNA maturation process, a subset of the normal nucleosides undergoes modifications. Modified nucleosides in the tRNA anticodon region are important for efficient translation. We found that, in the worm C. elegans, components of the Elongator complex are required for the formation of a certain set of tRNA modifications in the anticodon region. We observed a reduced efficiency of translation as well as a lower production of neurotransmitters in Elongator mutant worms. Elongator is conserved in eukaryotes, and mutations in a subunit of human Elongator cause a severe neurodegenerative disease, familial dysautonomia (FD). It is unclear in humans whether Elongator acts on the translational level through tRNA modification to regulate neuronal processes. Our observations in C. elegans, together with the role of yeast Elongator in translation, show that the function of Elongator in tRNA modification is conserved. Inactivation of Elongator may cause neuronal defects by affecting translation.
Introduction
Regulation at the level of translation is one important way in which gene activity is controlled in metazoans. Several different mechanisms have previously been identified by which translation can be regulated during development or memory formation [reviewed in 1]. During anterior-posterior patterning of the Drosophila embryo, the translation of hunchback mRNA in the posterior region of the embryo is inhibited by binding of a protein complex to the Nanos response element in the hunchback 3′UTR [2]. In Caenorhabditis elegans, developmental timing is controlled by the small temporal RNAs, lin-4 and let-7, which act by forming heteroduplexes with their target mRNAs and, at least in some cases, suppressing their translation [3]. Translation efficiency is also regulated by phosphorylation of translational components at the initiation and elongation steps [4],[5]. For example, during memory formation in mice, translation of ATF4 mRNA is regulated by phosphorylation of initiation factor eIF2α [6].
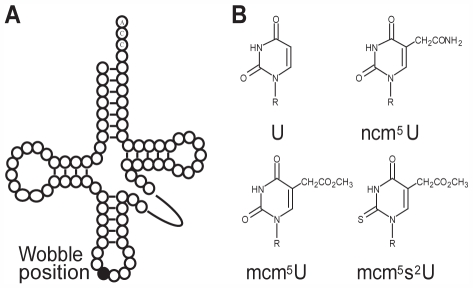
Another way in which the efficiency of translation can be modulated is by covalent modification of nucleosides in the anticodons of tRNAs. In the decoding of mRNA, modified nucleosides in the anticodon region, especially position 34 (wobble position) and position 37, have been suggested to be important for restriction or improvement of codon-anticodon interactions [7]–[10]. In S. cerevisiae, 25% of the tRNA species are covalently modified by the addition of either carbamoylmethyl (ncm) or methoxycarbonylmethyl (mcm) side chains to the 5′carbon of U34 [11]–[14]. A subset of these tRNAs contains a further modification on wobble uridines, addition of a thio group at the 2′position (Figure 1) [11],[13],[14]. In vivo, presence of an 5-carbamoylmethyluridine (ncm5U), an 5-methoxycarbonylmethyluridine (mcm5U) or an 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) improves reading of both A- and G-ending codons [14]–[16].
Figure 1. Schematic drawing of the tRNA secondary structure and modified wobble uridines.
(A) Secondary structure of tRNA with wobble position shown (•). (B) Wobble uridines can be modified to 5-carbamoylmethyluridine (ncm5U), 5-methoxycarbonylmethyl (mcm5U) or 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U).
In S. cerevisiae, formation of ncm and mcm side chains present at 5′position of wobble uridines requires the Elongator complex [12], which is composed of six subunits Elp1p – Elp6p [17],[18]. Yeast cells lacking Elongator activity are viable but display multiple defects including those in PolII transcription and exocytosis [16], [18]–[22]. However, these defects all appear to result from a primary defect in tRNA modification [16]. Elongator complex is conserved in eukaryotes and has also been purified from humans [23]. Inactivation of Elongator subunits in multicellular organisms causes multiple defects including those in development, cell proliferation, cell migration and neuron projection [24]–[27]. Recently, Elongator in mice has been reported to acetylate α-tubulin [27]. However, it is presently unclear whether Elongator in higher eukaryotes functions directly in multiple processes or acts on a small number of targets whose absence leads to pleiotropic defects.
Mutations in the human homologue of yeast ELP1, IKBKAP/hELP1, have been shown to cause Familial Dysautonomia (FD), a genetic disorder primarily affecting the sensory and autonomic nerve systems [28]–[30]. Human IKAP/hELP1 protein is part of a complex of six proteins that also contains the human homologues of yeast Elongator proteins [23]. Whether Elongator in humans or other metazoans promotes tRNA modification has not been reported.
The aim of the present study was to investigate the function of the Elongator homologues, ELPC-1 and ELPC-3 in the nematode, C. elegans. In particular, we were interested to determine first, whether Elongator in metazoans is required for modification of wobble uridines, and second, whether C. elegans could be established as a model to study the role of Elongator in modulating translation within neurons and other tissues. We demonstrate that Elongator is required in C. elegans for the formation of modified nucleosides in tRNA, and that Elongator mutants have defects in neurological and developmental processes associated with reduced translation. We believe our results also have important implications for the etiology of FD disease.
Results
C. elegans elpc-1 and elpc-3 Are Required for Synthesis of mcm5 and ncm5 Side Chains at Wobble Uridines
Searches of the C. elegans protein sequence database with the yeast or human Elp1p and Elp3p sequences revealed that C. elegans contains single Elp1p and Elp3p homologues, named ELPC-1 and ELPC-3, which are encoded by Y110A7A.16 and ZK863.3 respectively [see Materials and Methods for an explanation of gene nomenclature]. To investigate the function of elpc-1 and elpc-3 in C. elegans, we used elpc-1(tm2149) and elpc-3(tm3120), deletion mutants kindly supplied by S. Mitani of the National Bioresource Project, Japan. The elpc-1(tm2149) deletion removes 275 bp of sequence spanning parts of exons 7 and 8 (Figure 2A), whereas the elpc-3(tm3120) removes 356 bp spanning the first half of exon 3 and contains as well an insertion of four nucleotides in the second half of exon 3 (Figure S1A). The elpc-3(tm3120) deletion removes part of a sequence sharing significant homology to the Radical S-adenosylmethionine (SAM) superfamily [31]. Members of this family of proteins contain an FeS cluster and use S-adenosylmethionine (SAM) to catalyse a variety of radical reactions. The Elp3p Radical SAM domain has been found to be required for iron binding in Methanocaldococcus jannaschi [32], and for integrity of the Elongator complex in yeast [33].
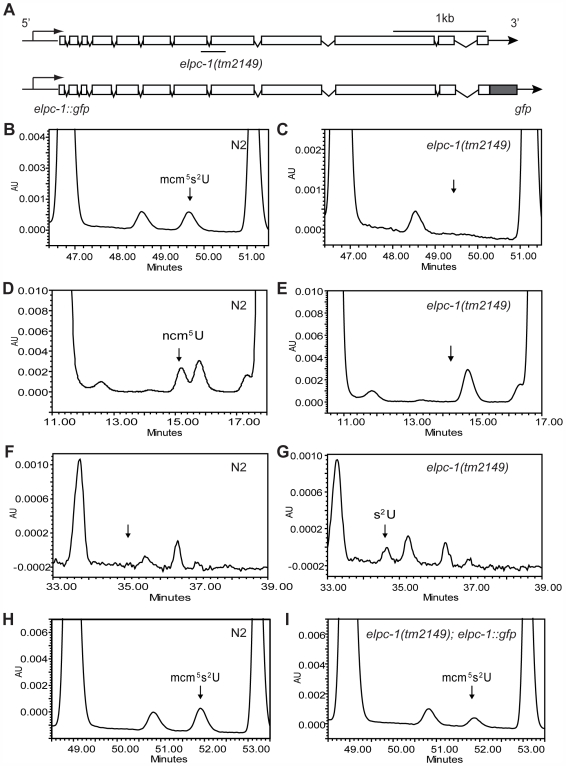
Figure 2. C. elegans elpc-1 is required for mcm5 and ncm5 side chain formation at wobble uridines.
(A) Schematic drawing of the distribution of exons and introns of elpc-1. The exons and introns are depicted as boxes and lines, respectively. At top, the line underneath represents the location of the deletion in elpc-1(tm2149). Below, the structure of the elpc-1::gfp transgene. (B–I) Total tRNA isolated from wild-type and elpc-1(tm2149) worms was analyzed by HPLC. Wild-type (N2) profiles are shown in the left panels. elpc-1(tm2149) and elpc-1(tm2149); elpc-1::gfp profiles are in the right panels. Chromatograms were monitored at 254 nm unless otherwise stated. (B,C) The parts of the chromatograms between retention times 46.4 and 51.5 min are displayed. The arrow in C indicates the expected retention time of mcm5s2U. (D,E) The parts of the chromatograms between retention times 10.5 and 18 min are displayed. The arrow in E indicates the expected retention time of ncm5U. (F,G) The parts of the chromatograms between retention times 33 and 39 min are displayed. The arrow in F indicates the expected retention time of s2U. These chromatograms were monitored at 314 nm. (H,I) The parts of the chromatograms between retention times 48 and 54 min are displayed.
In yeast, Elp1p and Elp3p are required for the formation of mcm5 and ncm5 side chains of modified nucleosides mcm5U, ncm5U, ncm5Um and mcm5s2U present at the wobble position in tRNA [12]. To determine whether their homologues in C. elegans, ELPC-1 and ELPC-3, also function to promote wobble uridine tRNA modification, we examined if the mcm5U, ncm5U or mcm5s2U modified nucleosides were present in tRNA isolated from wild-type and elpc mutants. Total tRNA isolated from wild-type worms contained ncm5U and mcm5s2U nucleosides (Figure 2B and 2D, Figure S1B, S1D). However, no mcm5U was detected (Figure S2D), implying that modification of uridine in C. elegans tRNA differs in at least one respect from that in S. cerevisiae. In contrast to wild-type worms, no mcm5s2U or ncm5U nucleosides were present in tRNA isolated from elpc-1(tm2149) mutants (Figure 2C and 2E). Instead, 2-thio uridine (s2U) was detected in tRNA isolated from the elpc-1(tm2149) mutant but not from wild-type worms (Figure 2F and 2G). This nucleoside arose from a failure in the mutant to add the mcm5 side chain of the mcm5s2U nucleoside. The tRNA modification defect in the elpc-1(tm2149) mutant was rescued by elpc-1 activity provided by a transgene (Figure 2C and 2I). Thus, like yeast Elp1p, C. elegans ELPC-1 is required for the formation of mcm5 and ncm5 side chains in tRNA. Consistent with the tRNA modification defect in the elpc-1(tm2149) mutant, tRNA isolated from elpc-3(tm3120) mutants lacked the mcm5s2U and ncm5U nucleosides and instead contained s2U (Figure S1).
Synthesis of the s2 group of mcm5s2U in yeast requires Tuc1p [15], [34]–[38]. The homologue of Tuc1p in C. elegans is encoded by open reading frame F29C4.6 [39]. In this paper we will refer the F29C4.6 gene as tuc-1. We analyzed tRNA from tuc-1(tm1297) mutant worms by HPLC and confirmed that it lacked the mcm5s2U modification and instead had mcm5U, a nucleoside not normally found in C. elegans tRNA (Figure S2B, S2C, S2D, S2E). Furthermore, a transgene containing wild-type tuc-1 DNA restored formation of mcm5s2U in tRNA (data not shown). Consistently, tRNA isolated from an elpc-1(tm2149); tuc-1(tm1297) double mutant lacked both the 5′- and 2′ side-chains of wobble uridines and no ncm5U or mcm5s2U nucleosides were observed (Figure S3).
An elpc-1::gfp Reporter Gene Is Differentially Expressed
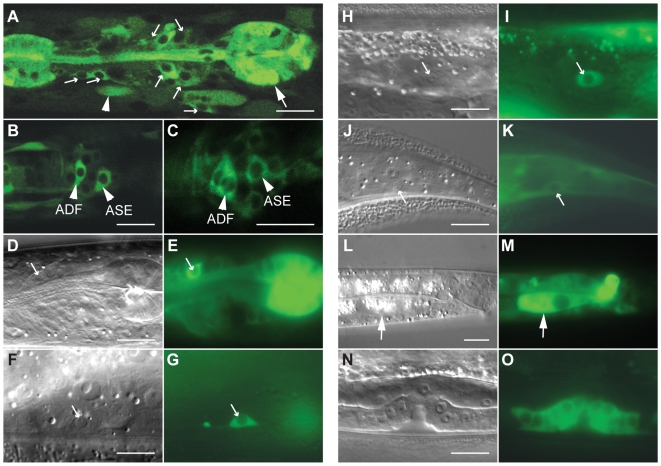
To investigate the expression pattern of C. elegans ELPC-1 in various tissues, we examined worm strains harboring a transgene encoding functional, full length ELPC-1 protein fused to GFP. The transgene contained 435 bp of the promoter region and all 11 introns (Figure 2A). The transgene rescued the tRNA modification defect in the elpc-1 mutants (Figure 2C and 2I). The fusion protein encoded by the transgene was preferentially detected in several tissues including the nervous system (Figure 3). However, its presence was not uniform. Within the nervous system, ELPC-1::GFP was seen predominantly in a pair of neurons that control egg-laying, the HSNs (Figure 3F and 3G), and in chemosensory neurons in the head (Figure 3A–3E). Within the latter class of neurons, the ELPC-1::GFP level was particularly high in the ASE, ADF and ASK pairs of neurons (Figure 3B–3E. For nomenclature of neurons, see Materials and Methods). Expression was seen both within the cell bodies (Figure 3B) and along the entire lengths of the neuronal processes (data not shown). Outside of the nervous system, a strong ELPC-1::GFP signal was seen in the pharynx (the feeding organ) (Figure 3A) and the vulva (Figure 3N and 3O), part of the egg-laying apparatus in the hermaphrodite. In all animals examined, ELPC-1::GFP expression was also seen in the two CAN cells (Figure 3H and 3I), which are associated with the excretory canals and are required for proper function of the excretory system. In all cells in which ELPC-1::GFP was seen, fluorescence was restricted to the cytoplasm (Figure 3A). The ELPC-3::GFP fusion was expressed in the same set of cells (data not shown).
Figure 3. ELPC-1 is differentially expressed in C. elegans.
(A–C) Confocal fluorescence micrographs of an hermaphrodite of the genotype elpc-1(tm2149); svEx557[Pelpc-1::elpc-1::gfp]. The large arrow in A denotes the posterior bulb of the pharynx. The smaller arrows denote sensory neurons in the head. The arrowhead indicates a body muscle. In B and C, specific sensory neurons in the head are indicated. (D–O) Micrographs of elpc-1(tm2149); svEx557[Pelpc-1::elpc-1::gfp] worms viewed with either Nomarski differential contrast (DIC) (D,F,H,J,L,N) or fluorescence (E,G,I,K,M,O) optics. The arrows in D and E indicate an ASK sensory neuron; in F and G, an HSN; in H and I, a CAN cell; in J and K, a PLM neuron; in L and M, a cell in the intestine. The green fluorescence in O is from cells in the developing vulva. Scale bars denote 10 microns.
Wobble Uridine tRNA Modifications Promote Efficient Translation
In S. cerevisiae, defects in wobble uridine tRNA modification are associated with reduced translation efficiency [14]–[16],[40]. The yeast elp3 tuc1 double mutant, in which modifications at both the 5′and 2′positions of the uridine moiety are absent, is lethal [15]. To investigate the influence of wobble uridine modifications on the efficiency of translation in C. elegans, we examined the effect of elpc-1, elpc-3 and tuc-1 mutations on β-galactosidase expression in worms harboring a lacZ transgene driven by heat shock-responsive elements from the hsp16-1 gene. The induction of lacZ mRNA upon heat shock was not reduced in strains lacking wild-type elpc-1, elpc-3 or tuc-1 gene activity, or in elpc-1; tuc-1 double mutant worms (Table 1). However, β-galactosidase activity was 28% lower in protein extracts from heat shocked elpc-1; tuc-1 double mutants than in those from wild-type worms subjected to the same heat shock regime (Table 1). A modest (∼14–18%) but significant reduction in β-galactosidase activity was also seen elpc-1(tm2149), elpc-3(tm3120) or tuc-1(tm1297) single mutant worms (Table 1).
Table 1. Lack of Wobble Uridine tRNA Modification Affects Translation Efficiency.
| Strains | β-gal activity | lacZ mRNA/tbb-2 mRNA |
| N2a | 312.57±17.15 (100.0%) | 1.00±0.09 |
| elpc-1(tm2149) a | 267.84±19.84* (85.7%) | 0.90±0.05 |
| elpc-3(tm3120) a | 257.39±9.37* (82.3%) | 0.92±0.07 |
| tuc-1(tm1297) a | 264.54±14.66* (84.6%) | 0.89±0.12 |
| N2b | 322.43±4.57 (100.0%) | 1.00±0.08 |
| elpc-1(tm2149);tuc-1(tm1297) b | 233.40±6.34** (72.4%) | 1.28±0.22 |
The strains were grown at 20°C before heat-shock at 33°C. The difference of β-gal activity (mean±SD) between N2 and elp-1(tm2149), elp-3(tm3120) or tuc-1(tm1297) was analyzed by student's t test (*p<0.01). For the QRT-PCR data (mean±SD), the lacZ mRNA was normalized against tbb-2 and ubc-2 mRNA (data not shown). In the table are the ratios between lacZ and tbb-2 mRNA. There is no reduction on mRNA level in the mutant strains.
The strains were grown at 15°C before heat-shock at 33°C. The reduction of β-gal activity (mean±SD) in elp-1(tm2149); tuc-1(tm1297) is significant compared to the wild type (**p<0.001). The lacZ mRNA level (mean±SD) was also normalized against tbb-2 and ubc-2 mRNA (data not shown).
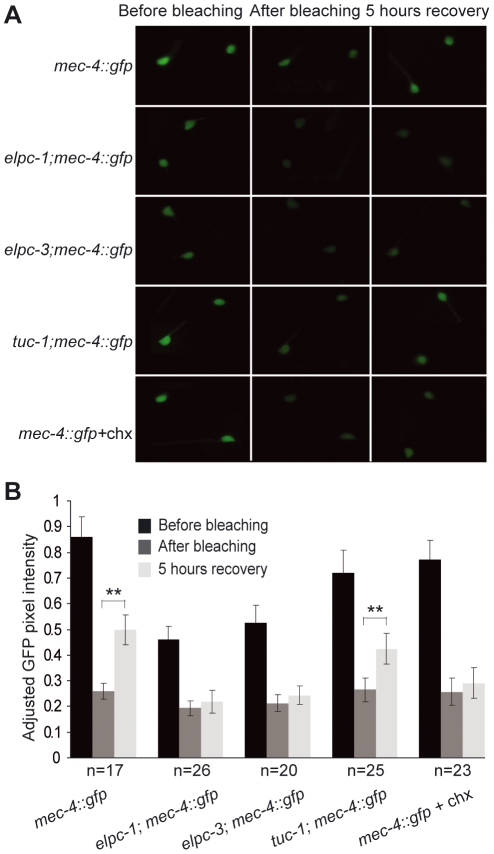
To monitor cell and tissue specific protein synthesis, we used an established technique, fluorescence recovery after photobleaching (FRAP) [41]. The rate of protein synthesis in different cells and tissues was measured using GFP reporters. We used gcy-5::gfp and mec-4::gfp which are expressed in ASER and 6 touch cell neurons respectively, and myo-3::gfp which is expressed in the body wall muscle. In all reporter fusions examined, photobleached wild type animals showed a significant recovery of GFP signal within 5 hours (Figure 4, Figure S4). However, animals with the elpc-1(tm2149) or elpc-3(tm3120) mutations had a slower GFP signal recovery, indicating a reduced rate of protein synthesis (Figure 4, Figure S4). Cycloheximide, an inhibitor of translation, was used to confirm that the recovered GFP signal was due to newly synthesized protein. In animals treated with cycloheximide, no significant recovery of GFP signal was observed (Figure 4, Figure S4). Together, these experiments demonstrate that an absence of uridine modification in tRNA is associated with a reduction in translation efficiency in C. elegans.
Figure 4. The elp-1(tm2149) and elpc-3(tm3120) mutants are defective in fluorescence recovery after photobleaching.
(A) Images of mec-4::gfp reporter in wild type, elpc-1, elpc-3 and tuc-1 backgrounds before photobleaching (left), after photobleaching (middle) and 5 hours recovery (right). The images at the bottom are of the mec-4::gfp reporter strain treated with the cycloheximide. (B) Quantification of GFP pixel intensities before photobleaching, after photobleaching and 5 hours recovery. The number of worms examined of each strain is given under the graph. Error bars represent standard deviations. Two asterisks indicate a significant fluorescence recovery after 5 hours incubation by student's t test (**p<0.001).
elpc-1 and elpc-3 Mutants Show Defects in Neuronal Function
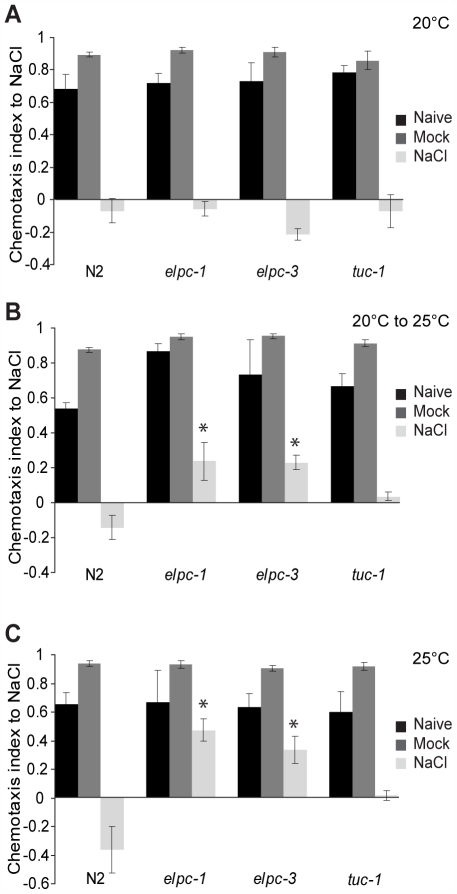
elpc-1 and elpc-3 single mutants were viable and fertile and they were able to move normally on the bacterial lawn. Furthermore, the chemosensory neurons in which ELPC-1::GFP and ELPC-3::GFP are strongly expressed are present at their normal positions and have normal morphology (Figure S5). Among these neurons (Figure 3A–3E), the ASE pair of sensory neurons is required for experience-dependent behaviors elicited by different salt concentrations [42]. Wild-type worms normally chemotax towards NaCl. However, pre-incubation in normal salt concentrations in the absence of nutrients elicits an aversion response to NaCl when worms are subsequently tested in chemotaxis assays [43]. In this salt learning assay, worms that have grown at normal salt concentrations and in the presence of abundant nutrients are first starved for four hours in the presence or absence of salt and then assayed for their chemotactic response to NaCl. Since we observed strong expression of ELPC-1 and ELPC-3 in ASE neurons (Figure 3B and 3C, data not shown), we tested elpc-1 and elpc-3 mutants in a salt learning assay. At 20°C, the mutants behaved as wild type (Figure 5A). At 25°C, wild-type worms exposed to 100 mM NaCl in the absence of nutrients moved away from NaCl, whereas elpc-1 or elpc-3 mutants treated in the same way continued to chemotax towards the NaCl (Figure 5C). In the elpc-1 mutant, this defect was partially rescued by the elpc-1::gfp construct (Figure S6). Thus, C. elegans elpc-1 and elpc-3 are required for an experience-dependent change in behavior. In contrast, in tuc-1 mutant worms no statistically significant changes were observed (Figure 5).
Figure 5. The elp-1(tm2149) and elpc-3(tm3120) mutants show a salt chemotaxis learning defect.
For each strain there are three conditions. ‘NaCl’ indicates worms that were preconditioned on a plate containing 100 mM NaCl for 4 hours prior to the chemotaxis assay. ‘Mock’ indicates that worms were pretreated on NaCl-free plate for 4 hours before assay. ‘Naive’ indicates worms that were assayed without any preconditioning. The chemotaxis index after 30 min of assay is displayed. The assay was repeated four times. Error bars denote standard deviations. Asterisk indicates a significant difference from wild type N2 (*p<0.01 by student's t test). (A) Young adult worms that had been raised at 20°C. (B) Temperature shifted animals. Synchronized eggs were grown at 20°C to the 2nd larval stage (L2). L2 larvae were then shifted to 25°C and cultured until they had become young adults. (C) Young adult worms from a strain that had been maintained at 25°C for several generations.
Ablation of the ASE neurons leads to an inability to chemotax towards certain water-soluble compounds including Na+, Cl−, lysine and biotin [44]. elpc-1 and elpc-3 mutants were able to chemotax both to water soluble and volatile compounds at all temperatures tested (Figure S7). When elpc-1 or elpc-3 mutants were grown at 20°C to the time at which the chemosensory neurons have developed and then shifted to 25°C, salt learning was abnormal (Figure 5B). Together, these observations suggest that the salt learning defect seen in elpc-1 and elpc-3 mutants is not caused by a defect in the development of the ASE chemosensory neurons or in their ability to detect salt.
The Level of a Neuropeptide::GFP Reporter in Neurons Is Reduced in elpc Mutants
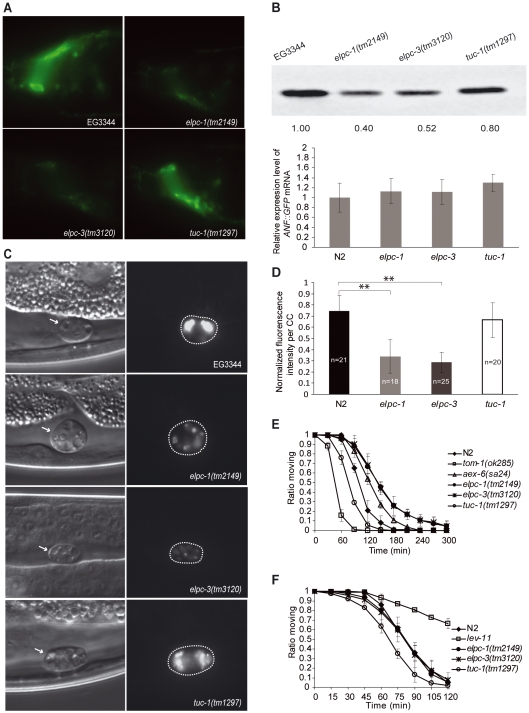
Since neuronal function in metazoans is known to be dependent upon the ability to synthesize and secrete neurotransmitters and neuropeptides, we tested whether these processes were abnormal in C. elegans elpc-1 and elpc-3 mutants. One established assay for examining the synthesis and secretion of neuropeptides involves a heterologous fusion protein, ANF::GFP. The prodomain of a preproANF–GFP fusion protein can be used as a reliable fluorescent reporter of dense-core vesicle transport and exocytosis in rat PC12 cells, as well as in D. melanogaster and C. elegans neurons [45]–[47]. In C. elegans, ANF::GFP is secreted by neurons into the pseudocoelomic space from where it is rapidly cleared by three pairs of coelomocytes [47]. In elpc-1 and elpc-3 mutants, we observed a reduced accumulation of ANF::GFP in coelomocytes (Figure 6C and 6D), which could be caused by either less synthesis or reduced secretion of ANF::GFP from neurons. In both wild-type and elpc mutant worms carrying the ANF::GFP transgene, the fusion protein was visible in neurons, but the GFP signal was weaker in elpc mutants that was also reflected by western blot (Figure 6A and 6B). As there was no significant reduction of ANF::GFP mRNA in elpc mutants, the lower production of ANF::GFP was at the posttranscriptional level (Figure 6A and 6B).
Figure 6. Neurons in the elpc-1(tm2149) and elpc-3(tm3120) mutants show reduced production of neuropeptide.
(A) Fluorescence micrographs showing the nerve ring in worms harboring a transgene encoding ANF::GFP. (B) At top, western blot of protein extracts from worms of the indicated genotypes that contained the ANF::GFP transgene. The same amount of protein was loaded in each lane. The blot was probed with an antibody against GFP. Below, ANF::GFP transcripts were quantified by Real-time PCR. No significant difference was observed in the levels of ANF::GFP mRNA (Student's t test, p>0.05), which were normalized to tbb-2 mRNA. (C) Micrographs showing coelomocytes in worms carrying the ANF::GFP transgene. Those on the left were viewed with DIC optics. Those on the right are of the same worms viewed with fluorescence optics. Dashed lines indicate the locations of the coelomocytes. Note that the intensity of GFP fluorescence in EG3344 and tuc-1(tm1297) coelomocytes is higher than that in elpc-1(tm2149) and elpc-3(tm3120) mutant worms. (D) Graph showing the normalized pixel intensities of confocal images of coelomocytes (CC). The number of coelomocytes measured for each strain is shown on the bar. The strongest pixel intensity per coelomocyte of ANF::GFP in any worm tested was arbitrarily set to 1. Error bars represent standard deviations. Two asterisks indicate the significant difference from control worms by student's t test (**p<0.001). (E) Aldicarb sensitivity assays. The proportion of worms still able to move is plotted against time for the six different genotypes. N2 is the wild-type control; aex-6(sa24) is a strain previously shown to display increased resistance to aldicarb, and tom-1(ok285) is hypersensitive to aldicarb. (F) Levamisol sensitivity assays were performed in the same way as aldicarb assays. N2 is the wild-type control. lev-11 is a strain previously shown to be resistance to levamisol.
To investigate whether elpc-1 and elpc-3 also affected extracellular levels of a neurotransmitter, we examined whether the mutants showed increased resistance to aldicarb, an inhibitor of acetylcholinesterase present in the synaptic cleft. Wild-type worms exposed to aldicarb immediately hypercontract and then die after a few hours because they are unable to reduce synaptic levels of acetylcholine secreted by neurons [48]. Mutants with reduced acetylcholine-mediated signaling are partially or completely resistant to the drug. Aldicarb-resistant mutants fall into two classes, those that have pre-synaptic defects resulting in reduced synthesis or secretion of acetylcholine and those in which the fault lies in the post-synaptic neurons [49]. elpc-1 and elpc-3 mutants showed greater resistance to aldicarb than that displayed by aex-6(sa24) (Figure 6E), which has been described previously as being partially resistant to the drug [50]. elpc-1(tm2149) mutant harboring the elpc-1::gfp transgene on an array behaved as wild type in the aldicarb assay (Figure S8). elpc-1 and elpc-3 mutant worms showed normal response to levamisole (Figure 6F), which activates the post-synaptic acetylcholine receptor [49], suggesting a defect in the pre-synaptic compartment. These results suggest that either less acetylcholine is produced in the neurons or less acetylcholine is released from the neurons in the elpc-1 and elpc-3 mutants.
C. elegans elpc-1 and elpc-3 Mutants Have Normal Levels of Acetylated α-tubulin
Recently it was shown that mouse ELP3 protein can acetylate α-tubulin in vitro [27]. Thus one possibility is that the neural defects seen in mice with reduced Elongator activity is caused by aberrant α-tubulin function. Acetylation of α-tubulins in a wide variety of species occurs on a conserved lysine residue at position 40. In C. elegans, there is a single α-tubulin with a lysine at this position, MEC-12 [51]. To investigate whether Elongator in C. elegans promotes acetylation of α-tubulin, we examined acetylation in elpc-1 or elpc-3 mutants. As previously reported, an antibody that recognizes lysine 40-acetylated α-tubulin in various species, 6-11B-1, could detect the residue in extracts from wild-type worms but not those from the mec-12(e1607) mutant. However, we observed no reduction in the levels of acetylated MEC-12 in elpc-1 or elpc-3 mutants (Figure S9A). Furthermore, unlike elpc-1 or elpc-3 mutants, mec-12(e1607) is not aldicarb resistant (Figure S9B).
Synthetic Effects in elpc-1; tuc-1 Double Mutants Indicate a Role for ELPC-1, ELPC-3, and TUC-1 in Development
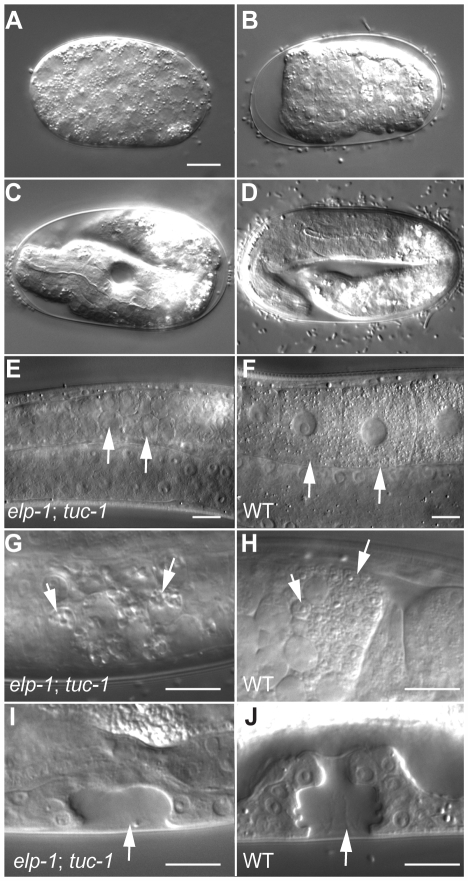
In humans, IKBKAP/hELP1 expression is not confined to the nervous system but is also seen in many other cell types [29],[52],[53]. In C. elegans, we also observed ELPC-1::GFP expression in several non-neuronal tissues (Figure 3). However, in an otherwise wild-type genetic background, although they grow slower than wild-type and had reduced fertility at 25°C (Table 2), the development of elpc-1 or elpc-3 mutants is not grossly disturbed. In yeast, elp1 and elp3 deletion strains are also viable. However, yeast cells lacking both ELP3 and TUC1, which therefore lack both mcm5 and s2 groups of tRNAs having the nucleoside mcm5s2U34, are not viable [15]. In the course of analyzing elpc-1; tuc-1 double mutant worms, we observed that the strain could be propagated at 15°C but not at 25°C. The elpc-1(tm2149); tuc-1(tm1297) double mutant hermaphrodites raised at 15°C for different periods of time were shifted to 25°C and then examined both for their own development and also for their ability to give rise to viable progeny. When 4th larval stage (L4) hermaphrodites were shifted to 25°C, they continued to develop and became fertile adults. However, the eggs they laid arrested development during embryogenesis (Figure 7A–7D). The arrest did not occur at one specific embryonic stage but rather at different stages in different embryos. Some embryos were arrested either prior to enclosure with relatively few cells (<100 cells) (Figure 7A); or at the 3-fold stage (Figure 7D). However, the majority were arrested during or immediately after morphogenesis (Figure 7B and 7C). Similar defects were seen in elpc-3; tuc-1 double mutants (Figure S10). Thus ELPC-1, ELPC-3 and TUC-1 likely function at multiple times during embryogenesis. No synthetic defects were seen in elpc-1; elpc-3 double mutants, suggesting that Elongator function is abolished in both elpc-1 and elpc-3 single mutants.
Table 2. elpc-1(tm2149), elpc-3(tm3120) and tuc-1(tm1297) Worms Display Temperature Sensitive Fecundity and Life Cycle Defects.
| Number of eggs produceda | Length of reproductive cycle (hours)b | |||
| 20°C | 25°C | 20°C | 25°C | |
| N2 | 255±36 (100.0%) | 172±28 (100.0%) | 68.9±1.2 (100.0%) | 54.6±1.1 (100.0%) |
| elpc-1(tm2149) | 303±77 (118.8%) | 42±13** (24.4%) | 80.3±0.9** (116.5%) | 67.8±1.1** (124.2%) |
| elpc-3(tm3120) | 292±48 (114.5%) | 56±16** (32.6%) | 81.2±1.1** (117.9%) | 65.5±2.3** (120.0%) |
| tuc-1(tm1297) | 274±62 (107.5%) | 44±33** (25.6%) | 78.5±1.7** (113.9%) | 64.7±4.7** (118.5%) |
The total numbers of progeny of 20 worms of each genotype were determined. Asterisks represent significant difference from wild type N2 (***p<0.0001 by student's t test).
The time between an egg being laid and the worm it gave rise to producing progeny was measured for 10 worms of each genotype (mean±SD, **p<0.001 by student's t test).
Figure 7. Defects seen in temperature-shifted elpc-1; tuc-1 double mutants.
Micrographs of eggs and larvae viewed with Nomarski DIC optics. (A–D) Embryos arrested prior to (A), during (B,C) or after (D) morphogenesis. (E,F,G,H) Parts of the germline in young adult hermaphrodites. The arrows in E and F indicate oocytes. Note that those in the elpc-1; tuc-1 worm have not matured. The arrows in G and H denote sperm. Those in the elpc-1; tuc-1 worm have grossly abnormal morphology. (I,J) The vulva during the L4 stage. In the elpc-1; tuc-1 double mutant, fewer cells are present and morphogenesis of the vulva to form the tube through which the eggs are laid is abnormal. Scale bars denote 10 microns.
Temperature shift experiments with 1st or 2nd stage larvae (L1 or L2) also indicated a role for ELPC-1, ELPC-3 and TUC-1 in development of the vulva and for generation of germ cells. When L1 or L2 larval hermaphrodites containing both the elpc-1(tm2149) and tuc-1(tm1297) mutations were raised at 15°C and shifted to 25°C, they developed to become small sterile adults. Inspection of the shifted animals at high magnification indicated that vulval development was invariably abnormal (Figure 7I and 7J, Figure S10I, S10J). In wild-type worms, the three progenitors of the vulva, P5.p, P6.p and P7.p are induced to adopt vulval fates: they divide to give rise to 22 cells that together form a tube through which the eggs are laid in adult hermaphrodites. In the temperature-shifted elpc-1; tuc-1 and elpc-3; tuc-1 double mutants, the divisions of P5.p, P6.p and P7.p failed to occur properly and significantly fewer vulval cells were formed (Figure 7I and 7J, Figure S10I, S10J). At the L3 stage, when the vulval developmental fates are induced, expression of the elpc-1::gfp reporter was upregulated in P5.p, P6.p and P7.p as well as in their immediate descendants (Figure S11), suggesting that one or more of the signaling pathways mediating vulval cell fate specification controls elpc-1 expression. Inspection of the gonads of the temperature shifted double mutants revealed that the overall organization of the germline was relatively normal (data not shown). However, the oocytes completely failed to mature (Figure 7E and 7F, Figure S10E, S10F); the sperm were highly vacuolated and grossly abnormal (Figure 7G and 7H, Figure S10G, S10H). These observations imply that elpc-1 and elpc-3 also function in development of non-neuronal tissues.
The developmental defects in the elpc-1; tuc-1 double mutant were rescued by extrachromsomal arrays harboring the elpc-1::gfp transgene. When elpc-1(tm2149); tuc-1(tm1297) double mutant hermaphrodites raised at 15° were allowed to lay eggs at this temperature for two hours and the eggs subsequently shifted to 25°, the progeny invariably arrested either during embryogenesis or during early larval stages (n = 65). However, 60% (n = 40) of elpc-1(tm2149); tuc-1(tm1297); svEx808[elpc-1::gfp Punc-122::gfp] embryos raised grew to become adults with normal vulval development. 15% of these adults gave rise to some live larval progeny indicating partial rescue of both the germline defect and the requirement during early embryogenesis. A second array, svEx806[elpc-1::gfp Punc-122::gfp] also rescued although not quite as efficiently: 40% of embryos grew to become adults.
Discussion
Here we show that the elpc-1 and elpc-3 genes, homologues to yeast ELP1 and ELP3, are required for formation of the ncm5 and mcm5 side chains present in the wobble nucleosides, ncm5U and mcm5s2U in C. elegans tRNA. Worms with mutations in elpc-1 or elpc-3 show a defect in a salt learning assay, associated with reduced expression of neuropeptide and slow accumulation of acetylcholine in the synaptic cleft. elpc-1::gfp and elpc-3::gfp reporters are strongly expressed in certain sensory neurons including ASE, required for salt learning. elpc-1 and elpc-3 mutant phenotypes are strongly exacerbated by mutations in tuc-1, which is required for the formation of 2-thio group in the mcm5s2U modified wobble nucleosides.
The Role of ELPC-1 and ELPC-3 in tRNA Modification
Although a requirement for the Elongator complex for the modification of wobble uridines in yeast tRNA is well documented [12], studies on the role of Elongator in this process in metazoans have not been previously reported. Our results demonstrating that ELPC-1 and ELPC-3 are required for the addition of mcm5 and ncm5 side chains to uridine residues in C. elegans tRNA imply that Elp1p and Elp3p function has been conserved in evolution. Our results also confirm, however, that differences exist in tRNA modification in eukaryotes. In yeast there are 13 tRNA species with a uridine at the wobble position. Of these, eleven contain the nucleoside ncm5U, ncm5Um, mcm5U or mcm5s2U [11]–[14]. In our analysis of C. elegans wild-type tRNAs, we found ncm5U and mcm5s2U but not mcm5U. This observation is consistent with an earlier investigation showing that mcm5U is not present in tRNAs isolated from calf liver [54]. For example, nucleoside 34 in 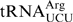 from yeast has mcm5U [55], whereas that from calf liver has mcm5s2U [56]. These findings suggest that mcm5U might be absent from tRNAs in metazoans.
from yeast has mcm5U [55], whereas that from calf liver has mcm5s2U [56]. These findings suggest that mcm5U might be absent from tRNAs in metazoans.
In yeast, Elongator was suggested to participate in three distinct cellular processes: transcriptional elongation, polarized exocytosis and formation of modified wobble uridines in tRNA [12],[21],[22]. Strong genetic evidence was provided that the pleiotropic phenotypes seen in yeast, including those in transcription and exocytosis, were caused by a translational dysfunction due to lack of mcm5 and ncm5 side chains at wobble uridines [16]. This suggests that the physiological relevant role of Elongator complex in this organism is in the formation of modified nucleosides in tRNA, i.e. wobble uridine tRNA modification is crucial for the translation of mRNAs that encode proteins important for transcriptional elongation and polarized exocytosis. Cellular localization studies primarily placed Elongator subunits in the cytosol in yeast, mouse and human cells [22], [23], [27], [57]–[60]. As modifications in the anticodon region normally take place in the cytosol [61], such a localization is consistant with a role in wobble uridine modification. In C. elegans, we did not observe any fluorescence of ELPC-1::GFP in the nucleus suggesting that Elongator in this organism functions in the cytosol.
Translation Is Less Efficient in elpc-1 and elpc-3 Mutants
In elpc-1 and elpc-3 mutants, we observe reduced expression of an ANF::GFP neuropeptide reporter. Given that ANF::GFP mRNA levels are normal in the mutants, the reduction in ANF::GFP accumulation could in principle be explained either by increased degradation of the protein or by decreased translation. Since tRNAs are intimately involved in protein synthesis, we believe it more likely that ELPC-1 and ELPC-3 affect ANF::GFP levels by promoting translation. Further evidence indicating a role for Elongator in translation is that the recovery of GFP signals after photobleaching in strains with gcy-5::gfp, mec-4::gfp and myo-3::gfp reporter genes is slower in Elongator mutants than in wild type. The effect of Elongator on translation is also consistent with the synthetic effects we observe in elpc-1; tuc-1 and elpc-3; tuc-1 double mutants. The reduction in accumulation of β-galactosidase activity in elpc-1 or elpc-3 single mutants (in which the mcm5 side chain of mcm5s2U containing tRNAs is absent) is similar to that seen in tuc-1 single mutants (in which the s2 side chain is absent). In the double mutants (in which both the 2′and 5′modifications are lost) the efficiency of translation is further reduced. An explanation for the reduced efficiency of translation in C. elegans worms lacking elpc-1 or elpc-3 activity is that the modifications of uridine residues at the wobble position aid codon-anticodon interactions [7]–[10]. Experiments in vivo with S. cereverisiae, suggest that the primary function of the mcm5U, ncm5U and mcm5s2U nucleosides is to improve binding to A- and G- ending codons, decoded by tRNAs containing these modified nucleosides [14]–[16]. For tRNAs normally modified at both the 2′and 5′positions, the absence of either modification (or both) did not lead to any obvious misreading of U- or C-ending codons [15],[16]. Thus, presence of modifications at wobble uridines in tRNAs appears to promote the rate of elongation during translation rather than its fidelity.
There are examples of tRNA modification mutants that show temperature sensitive (ts) phenotypes, suggesting a reduced functionality of the hypomodified tRNA at the elevated temperature [16],[62],[63]. In yeast, elp and tuc1 mutations result in hypomodification of 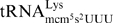 and
and 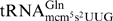 [12],[15]. In the anticodon loop, both tRNAs are rich in uridines that have a low stacking potential, and in
[12],[15]. In the anticodon loop, both tRNAs are rich in uridines that have a low stacking potential, and in 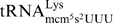 , mcm5 and s2 of U34 are required for a canonical anticodon loop structure [64]. Therefore, we believe that the temperature sensitive phenotype observed in elpc and tuc-1 single mutants and enhanced in elpc-1; tuc-1 and elpc-3; tuc-1 double mutants is caused by destabilization of anticodons in hypomodified tRNAs, resulting in further weakening of codon-anticodon interactions.
, mcm5 and s2 of U34 are required for a canonical anticodon loop structure [64]. Therefore, we believe that the temperature sensitive phenotype observed in elpc and tuc-1 single mutants and enhanced in elpc-1; tuc-1 and elpc-3; tuc-1 double mutants is caused by destabilization of anticodons in hypomodified tRNAs, resulting in further weakening of codon-anticodon interactions.
elpc-1 and elpc-3 Function in the Nervous System
The higher levels of expression of the elpc::gfp reporters within the nervous system of C. elegans is consistent with the finding that the most severe defects of elpc-1 or elpc-3 single mutants are observed in nervous system. It is interesting to note that a strong expression of Elongator subunits was also observed in the nervous system of mice [27]. A possible explanation for the greater requirement for Elongator in neurons is that neurons have markedly higher rates of protein synthesis than most other cell types [41],[65],[66]. It is also striking that in both C. elegans and mice, expression within the nervous system is not uniform. Perhaps different neurons have different rates of translation.
In C. elegans and other metazoans, neuronal function is dependent upon the ability to synthesize and secrete neurotransmitters and neuropeptides. In elpc-1 and elpc-3 mutants, the production of ANF::GFP neuropeptide is reduced at the posttranscriptional level. Thus Elongator mutations might cause the neurological defects by impairing the translation of neuropeptides. In addition, our findings that elpc-1 and elpc-3 mutants appear to have reduced levels of acetylcholine in the synaptic cleft suggest that Elongator is required for the production or secretion of neurotransmitter. Since elpc-1::gfp and elpc-3::gfp are expressed in a set of chemosensory neurons, the salt chemotaxis learning defect displayed by Elongator mutant worms is likely to be a consequence of inefficient communication among various neurons due to low production of neurotransmitters or neuropeptides. It is interesting to note that mutations in the human ELP1 gene, also called IKBKAP, cause the neurodegenerative disease, Familial Dysautonomia (FD) [28],[29]. Furthermore, association studies in humans have revealed that variants at the ELP3 locus confer increased risk for the neurodegenerative disorder Amyotrophic Lateral Sclerosis (ALS) [67]. Neuronal defects are also observed in Drosophila, Zebrafish and mouse with reduced function of ELP3 [27],[67].
Conflicting reports exist concerning the origin of the defects caused in human cells by a reduction in hELP1/IKAP levels [25],[26],[52],[68],[69]. Recently, in mice Elongator was suggested to catalyze α-tubulin acetylation [27]. However, our observations that acetylation is not obviously abnormal in C. elegans elpc-1 or elpc-3 mutants suggest that the neuronal defects observed in Elongator mutants in the worm are not caused by a failure to acetylate α-tubulin.
In contrast to the elpc-1 and elpc-3 mutants, tuc-1 mutants do not display defects in either the salt learning assay or in secretion of ANF::GFP. In yeast, the growth defects of Elongator mutants are more pronounced than those of the tuc mutants [15],[16]. One possible explanation for these differences might be that the absence of the s2 modification has less effect on codon-anticodon interactions than the absence of ncm5- and mcm5-groups. Alternatively, the effects on salt learning might be caused by reduced expression of a protein encoded by an mRNA rich in codons decoded by tRNAs harboring solely the 5′modification.
Synergistic Effects with tuc-1 Indicate Roles for ELPC-1 and ELPC-3 Outside of the Nervous System
Previous studies on ELP1 and ELP3 function in vertebrates have focused on their roles in neurons. While we have shown that ELPC-1 and ELPC-3 are important for nervous system function in the worm, our results clearly demonstrate that they also act in non-neural tissues. Although their expression is far from ubiquitous, the expression of ELPC::GFP reporters is clearly not restricted to neurons. More importantly, the defects in temperature-shifted elpc-1; tuc-1 and elpc-3; tuc-1 double mutants indicate that Elongator is also involved in embryogenesis and vulval development. The phenotypes observed suggest that tRNA modification is a mechanism by which the efficiency of translation is modulated during metazoan development.
Concluding remarks
Our observations suggest that Elongator acts in neurological and developmental processes in C. elegans by modulating translation. An important task in the future is to identify the mRNAs whose translation is dependent on Elongator activity. Identification of these mRNAs might help in the understanding of the molecular mechanisms of Elongator-related human diseases.
Materials and Methods
Nomenclature of Genes and Neurons
The names of genes in the text have been given according to existing nomenclature rules for S. cerevisiae, C. elegans and humans. The yeast ELP1 gene encodes a protein, Elp1p; the equivalent gene in C. elegans, elpc-1 encodes ELPC-1; in humans, IKBKAP/hELP1 encodes IKAP/hELP1. The respective mutant alleles are elp1 (S. cerevisiae) and elpc-1(tm2149) (C. elegans). Neurons in C. elegans have three-letter names e. g., ASE. These names are not acronyms or abbreviations.
Strains
C. elegans worms were cultured and handled as described previously [70]. All strains were maintained at 20°C unless likewise indicated. All are derived from the wild-type strain, Bristol N2 [70]. For routine propagation, worms were maintained on nematode growth medium (NGM) plates [70]. The following mutations were used in this study. Linkage group (LG) I, tom-1(ok285) [71]–[73], aex-6(sa24) [50], lev-11(x11) [74]; elpc-1(tm2149), LG III, mec-12(e1605), mec-12(e1607) [75], LG IV, tuc-1(tm1297) [39]; LG V, elpc-3(tm3120). The transgenes used were ubIn5[hsp16::lacZ] [76], oxIs180[Paex-3::ANF::gfp] [47], svEx557[Pelpc-1::elpc-1::gfp], zdIs5 I[mec-4::gfp lin-15(+)] [77], svEx666[lin-25::HA myo-3::gfp], svEx806[elpc-1::gfp Punc-122::gfp], svEx808[elpc-1::gfp Punc-122::gfp], adEx1262[gcy-5::gfp lin-15(+)] [78]. The elpc-1, elpc-3 and tuc-1 deletion mutants were backcrossed eight times with wild-type N2 before use.
Plasmid Construction
To generate the elpc-1::gfp fusion, the entire elpc-1 coding region together with 435 base pairs of DNA upstream of the start ATG was amplified by using primers 5′-AAAAGCATGCTCCGGTACGGTATGTGGC-3′ and 5′-AAAACTGCAGTGGGAAAACTGAAG CAAATGAA-3′. The PCR product was subcloned into pPD95.77 GFP expression vector between PstI and SphI sites.
DIC and Immunofluorescence Microscopy of Nematodes
A Leica DMRB microscope equipped with both Nomarski differential interference contrast and epifluorescence optics was used to view worms at high magnification. Images were captured with a Deltpix CCD camera and software (Deltapix, Copenhagen). Confocal microscopy was performed on a Leica TCS SP2 confocal microscope. Confocal images were captured using Leica confocal software.
tRNA Isolation and HPLC Analysis
Techniques described by Gaur et al. (2007) were used with minor modifications to isolate and analyze tRNA from C. elegans worms. For each strain, worms from twenty 9 cm culture plates containing mixed-stage populations of worms were used. After extensive washing with M9 buffer, the worm pellets were frozen in the liquid nitrogen and then thawed in the presence of 0.5 volumes of TRIzol (Invitrogen). A tissue-grinder (Kontes) was used to break open the worms. After extraction of the lysate with chloroform, followed by addition of isopropanol, total RNA was sedimented by centrifugation. tRNA was separated from other types of RNA by using methods described previously [79]. Purified tRNA was digested with Nuclease P1 for 16 h at 37°C and then treated with bacterial alkaline phosphatase for 2 h at 37°. The hydrolysate was analyzed by high pressure liquid chromatography with a Develosil C-30 reverse-phase column as described [79]. ncm5U, mcm5U, mcm5s2U and mcm5Um have all been found on wobble uridines in S. cerevisiae tRNA. We did not examine C. elegans tRNA for the presence of ncm5Um because P1 and BAP cannot digest the dinucleotide ncm5UmpX to nucleosides [80].
RNA Isolation and Quantitative RT–PCR
Total RNA was extracted with the aid of a BIO-RAD Aurum total RNA mini kit according to the instruction manual. Real-time PCR was carried out in 25 µl reaction mixes. iScript one-step RT-PCR kit with SYBR green (BIO-RAD) and the iCycler iQ Real-Time PCR Detection System (BIO-RAD) were used. The data were normalized to tbb-2 and ubc-2 mRNA levels. Six independent assays were performed for each strain analyzed.
β-Galactosidase Assay
For each strain analyzed, one 6-cm plate containing a population of well-fed worms was subjected to a 2 h heat shock at 33°. The worms were washed from the plate with M9 salt solution, sedimented, washed once in M9 and then once in breaking buffer (100 mM Tris-HCl, 1 mM DTT, 20% glycerol). After resuspension in 250 µl of breaking buffer containing Roche protease inhibitor cocktail, the worms were broken open by sonication. Five 2 sec pulses at maximum effect were used. The extracts were transferred to microcentrifuge tubes and worm debris was sedimented by centrifugation at 13,000 rpm for 15 min. β-galactosidase activity in the cleared extracts was measured using standard protocols [81].
Fluorescence Recovery after Photobleaching (FRAP)
The assay was performed as described in detail by Kourtis and Tavernarakis [41]. Worms carrying the gcy-5::gfp, mec-4::gfp or myo-3::gfp reporters were mounted on the agarose pad in the presence of levamisol and photobleached with light from an HBO 103W/2 mercury lamp (OSRAM). A 63× objective was used for photobleaching gcy-5::gfp and mec-4::gfp strains, a 20× objective for myo-3::gfp strains.
Chemotaxis Assay
Salt chemotaxis assays were performed as described by Ward [82] and Bargmann and Horvitz [44]. All the assays were carried out at room temperature (ca. 21.5°C) on 9 cm agar plates containing 5 mM KH2PO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4 and 2% agar. N2, elpc-1(tm2149), elpc-3(tm3120) and tuc-1(1297) strains were maintained at 25°C for at least three generations prior to being assayed. The salt gradient with a peak 0.5 cm from one edge of the plate was formed overnight by placing a block of agar measuring approximately 5 mm in each dimension and containing 100 mM NaCl, 5 mM KH2PO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4 and 2% agar. In each single test, 80–100 young adult worms were washed three times in 5 mM KH2PO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4 and then placed in the center of the assay plates. Before the worms were placed on the assay plate, 1 µl of 0.5 M sodium azide was spotted both at the salt gradient peak and at the opposite side of the plate to capture the worms moving to those areas. The numbers of worms at different positions on the plate were counted every 10 min after the start of the assay. The formula 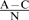 was used to calculate the chemotaxis index. In this equation, A was the number of worms at the attractant area, C the number of worms at the control spot, and N the total number of worms placed on the plates. Each experiment was repeated at least 4 times. For chemotaxis assays with isoamyl alcohol, the odorant was dropped on the assay plate immediately prior to the addition of worms to the plate.
was used to calculate the chemotaxis index. In this equation, A was the number of worms at the attractant area, C the number of worms at the control spot, and N the total number of worms placed on the plates. Each experiment was repeated at least 4 times. For chemotaxis assays with isoamyl alcohol, the odorant was dropped on the assay plate immediately prior to the addition of worms to the plate.
Salt Learning Assay
The assay was performed as described [43], with minor modifications. For each assay, adult worms were washed off the culture plates with chemotaxis washing buffer (5 mM KH2PO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4) and then washed three times in the same buffer. For the naive condition, worms were washed and then assayed immediately without further incubation. The other worms were conditioned respectively on nematode growth medium (NGM) plates containing 100 mM NaCl, or on NaCl-free NGM plates for 4 hours. After conditioning, worms were collected again and placed on the assay plates. After 30 min, the numbers of worms in the NaCl spot (A) and the control region (C) were counted. The index was calculated using the formula, 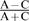 .
.
Western Blot
ANF::GFP levels were measured by western blotting using an anti-GFP antibody (Clontech, JL-8). 50 L4 larvae of each genotype were collected, boiled in SDS sample buffer for 5 min and loaded onto a 10% SDS PAGE. Quantification of imaging pixel intensity was performed by NIH image J. To measure acetylated α-tubulin levels by western blot, protein was extracted from young adult worms. To avoid protein degradation, worms were suspended in ice-cold extraction buffer containing proteinase inhibitors and rapidly frozen in liquid nitrogen. The frozen pellets were ground to a powder in a mortar. 20 µg protein was loaded on the gel in each lane. The dilution of anti-lys40-acetylated-α-tubulin antibody (abcam, 6-11B-1) was 1∶1000, and of anti-α-tubulin antibody (Sigma, B-5-1-2) was 1∶2000.
Measurment of Aldicarb and Levamisol Sensitivity
The assays were performed as described by Mahoney et al. [49]. 25–30 worms were used for each genotype. The assay was performed blind in triplicate at room temperature (ca. 21.5°C). The worms were cultivated at 25°C prior to being assayed.
Quantification of ANF::GFP Fluorescence in Coelomocytes
The assay was performed as described by Speese et al. [47]. Fluorescence confocal micrographs were made of coelomocytes. The intensity of GFP fluorescence in captured images in grey scale was measured with the aid of the NIH ImageJ software.
Supporting Information
C. elegans elpc-3 is required for mcm5 and ncm5 side chain formation at wobble uridines. (A) The diagram shows the genomic structures of elpc-3 and ZK863.4, which is suggested to be in the same operon. The exons and introns are depicted as boxes and lines respectively. At top, the line underneath represents the location of the deletion in elp-3(tm3120). Below, a representation of predicted motifs in the ELPC-3 protein: the Radical S-adenosyl methionine (Radical-SAM) [31], and histone acetyltransferase (HAT) domains [83]. The region deleted in elpc-3(tm3120) is indicated by a line beneath. (B–G) Chromatograms of total tRNA isolated from wild-type and elpc-3(tm3120) worms analyzed by HPLC. Wild-type (N2) profiles are shown in the left panels; elpc-3(tm3120) profiles are shown in the right panels. Chromatograms were monitored at 254 nm, unless otherwise stated. (B,C) The parts of chromatograms between retention times 46 and 51.5 min are displayed. The arrow in C indicates the expected retention time of mcm5s2U. (D,E) The parts of the chromatograms between retention times 11 and 18 min are displayed. The arrow in E indicates the expected retention time of ncm5U. (F,G) The parts of the chromatograms between retention times 31 and 37 min are displayed. The arrow in F indicates the expected retention time of s2U. Chromatograms were monitored at 314 nm.
(0.55 MB TIF)
tuc-1 in C. elegans is required for 2-thio wobble uridine tRNA modification. (A) The schematic structure of tuc-1. Exons and introns are represented by boxes and lines, respectively. The line underneath indicates the region deleted in tuc-1(tm1297). (B–E) Chromatograms showing total tRNA isolated from wild-type (N2) and tuc-1(tm1297) worms analyzed by HPLC. N2 profiles are shown in the left panels; tuc-1(tm1297) profiles are shown in the right panels. Chromatograms were monitored at 254 nm. (B,C) The parts of the chromatograms between retention times 46.2 and 51.6 min are displayed. The arrow in C indicates the expected retention time of mcm5s2U. (D,E) The parts of the chromatograms between retention times 35 and 42 min are displayed. The arrow in D indicates the expected retention time of mcm5U.
(0.43 MB TIF)
Formation of ncm5, mcm5 and s2 side chains is abolished in elpc-1 ; tuc-1 double mutants. (A–F) Total tRNA isolated from wild type, elpc-1(tm2149) or elpc-1(tm2149); tuc-1(tm1297) worms was analyzed by HPLC. Wild-type (N2) and elpc-1(tm2149) profiles are shown in left panels. elpc-1(tm2149); tuc-1(tm1297) profiles are in right panels. Chromatograms were monitored at 254 nm, unless otherwise stated. (A,B) The parts of chromatograms between retention times 48.5 and 53.5 min are displayed. The arrow in the right panel indicates the expected retention time of mcm5s2U. (C,D) The parts of the chromatograms between retention times 11 and 18 min are displayed. The arrow in the right panel indicates the expected retention time of ncm5U. (E,F) The parts of the chromatograms between retention times 33 and 39 min are displayed. The arrow in the right panel indicates the expected retention time of s2U. Chromatograms were monitored at 314 nm.
(0.45 MB TIF)
The elp-1(tm2149) and elpc-3(tm3120) mutants are defective in fluorescence recovery after photobleaching. Quantification of fluorescence signals in worms carrying gcy-5::gfp (A) or myo-3::gfp (B) reporters. The pixel intensities in wild type, elpc-1, elpc-3 and tuc-1 backgrounds before photobleaching, after photobleaching, and after 5 hours recovery are shown. In ‘gcy-5::gfp+CHX’, fluorescence recovery was measured in the presence of cycloheximide (CHX). The number of worms examined of each strain is denoted under the graph. Error bars represent standard deviations.
(0.67 MB TIF)
Neuronal morphology in elpc-1 , elpc-3 and tuc-1 mutant worms is normal. (A, C, E, G, I, K) Micrographs of hermaphrodite worms fed with DiI viewed with Nomarski DIC optics. The arrows denote three amphid neurons, ASI, ADL and ASK. (B, D, F, H, J, L) The same worms viewed with fluorescence optics. Note that DiI efficiently labels the neurons in the mutant worms, indicating that the outgrowth of the neuronal processes was normal.
(5.94 MB TIF)
The salt chemotaxis learning defect of elpc-1(tm2149) is rescued by an elpc-1::gfp construct. Worms were synchronized and raised at 25°C to the young adult stage. The chemotaxis index after 30 min of assay is displayed. The assay was repeated four times. Error bars denote standard deviations. Two asterisks indicate a significant difference between elpc-1 and elpc-1; elpc-1::gfp (**p<0.001 by student's t test).
(0.20 MB TIF)
elpc-1 , elpc-3 , and tuc-1 chemotax to both NaCl and isoamylalcohol. (A, B) Chemotaxis to NaCl (A) and isoamyl alcohol (B) is shown. The chemotaxis indices were plotted against time for four different genotypes. For each genotype, 80–100 young adult worms that had been raised at 25°C were placed on a plate equidistant from the attractant and a control spot. The numbers of worms at the NaCl (or isoamylalcohol) and the control spots were counted every 10 minutes for 1 hour. Each assay was repeated for 4 times.
(0.26 MB TIF)
The increased aldicarb resistance of the elpc-1(tm2149) mutant is complemented by elpc-1::gfp . The proportion of worms still able to move is plotted against time. 25–30 worms were used for each genotype. The assay was performed blind in triplicate at room temperature (ca. 21.5°C). The worms were cultivated at 25°C prior to being assayed.
(0.22 MB TIF)
Acetylated α-tubulin levels are not decreased in Elongator mutants. (A) Western blot of whole animal lysates for wild type (N2), elpc-1, elpc-3, mec-12(e1605) and mec-12(e1607). Both acetylated α-tubulin and α-tubulin migrated just above 50 KDa. Top, blotted with anti-lys40-acetylated-α-tubulin antibody at a dilution of 1∶1000. Bottom, blotted with anti-α-tubulin antibody at a dilution of 1∶2000. Lys40 acetylated α-tubulin signals were normalized to that of α-tubulin, and the amount of lys40 acetylated α-tubulin was expressed relative to the corresponding value in the wild type strain, which was set to 1. NA, not applicable. (B) mec-12(e1607) worms are not resistant to aldicarb. The proportion of worms still able to move is plotted against time. 25–30 worms were used for each genotype. The assay was performed blind in triplicate at room temperature (ca. 21.5°C). The worms were cultivated at 25°C prior to being assayed.
(0.66 MB TIF)
Defects Seen in Temperature-shifted elpc-3 ; tuc-1 Double Mutants. Micrographs of eggs and larvae viewed with Nomarski DIC optics. (A–D) Embryos arrested prior to (A), during (B,C) or after (D) morphogenesis. (E,F,G,H) Parts of the germline in young adult hermaphrodites. The arrows in E and F indicate oocytes. Note that those in the elpc-3; tuc-1 worm have not matured. The arrows in G and H denote sperm. Those in the elpc-3; tuc-1 worm have grossly abnormal morphology. (I,J) The descendants of P5.p, P6.p and P7.p during the L4 stage. The arrows in I denote the descendants of P5.p, P6.p and P7.p. In the animal shown, these three cells adopted the 3° cell fate and divided just once. In wild-type worms, P6.p adopts the 1° cell fate whereas P5.p and P7.p adopt the 2° fate. The 1° and 2° fates involve three rounds of cell division; the descendants of P5.p, P6.p and P7.p together form a tube through which the eggs are laid. The arrow in J denotes the tube as it is forming. Scale bars denote 10 microns.
(2.42 MB TIF)
ELPC-1::GFP is expressed during vulval cell fate specification. Micrographs of an L3 hermaphrodite worm of the genotype elpc-1(tm2149); svEx557[Pelpc-1::elpc-1::gfp] viewed with either Nomarski differential contrast (DIC) (A) or fluorescence (B) optics. The arrows denote the six descendants of P5.p, P6.p and P7.p.
(1.58 MB TIF)
Acknowledgments
We thank S. Mitani of the National Bioresource Project, Japan, for the elpc-1, elpc-3 and tuc-1 deletion mutants. We are grateful to E. Jorgensen for the ANF::GFP strain and to M. Chalfie for mec-12(e1605) and mec-12(e1607) strains. We thank Gunilla Jäger for performing the HPLC analysis of tRNA. We are grateful to the Caenorhabditis Genetics Center and the C. elegans gene knockout consortium for strains and to A. Fire for plasmids. Members of the Byström and Tuck labs are gratefully acknowledged for discussions. We thank G. Björk, J. Anderson, and G. Kao for comments on the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants from the Swedish Cancer Foundation (07 0600 to ST and 07 0637 to ASB) (http://www.cancerfonden.se), Swedish Science Research council (521-2006-2872 to ST and 621-2006-4269 to ASB) (http://www.vr.se), and Bernard and Signe Bäckström Foundation (223-438-07 to ASB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 2.Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, et al. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Raught B, Gingras A-C. Signaling to translation initiation. In: Mathews MB, Sonenberg N, Hersey JWB, editors. In Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- 5.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 6.Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim VI. Analysis of action of wobble nucleoside modifications on codon-anticodon pairing within the ribosome. J Mol Biol. 1994;240:8–19. doi: 10.1006/jmbi.1994.1413. [DOI] [PubMed] [Google Scholar]
- 9.Takai K, Yokoyama S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003;31:6383–6391. doi: 10.1093/nar/gkg839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, et al. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985;82:4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Johansson MJ, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Huang B, Esberg A, Johansson MJ, Byström AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson MJ, Esberg A, Huang B, Björk GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008 doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esberg A, Huang B, Johansson MJ, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, et al. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem. 2001;276:32743–32749. doi: 10.1074/jbc.M105303200. [DOI] [PubMed] [Google Scholar]
- 18.Krogan NJ, Greenblatt JF. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jablonowski D, Frohloff F, Fichtner L, Stark MJ, Schaffrath R. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol Microbiol. 2001;42:1095–1105. doi: 10.1046/j.1365-2958.2001.02705.x. [DOI] [PubMed] [Google Scholar]
- 21.Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 22.Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 24.Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, et al. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci U S A. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Johansen LD, Naumanen T, Knudsen A, Westerlund N, Gromova I, et al. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J Cell Sci. 2008;121:854–864. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 27.Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, et al. Elongator Controls the Migration and Differentiation of Cortical Neurons through Acetylation of alpha-Tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 28.Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gold-von Simson G, Axelrod FB. Familial dysautonomia: update and recent advances. Curr Probl Pediatr Adolesc Health Care. 2006;36:218–237. doi: 10.1016/j.cppeds.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 33.Greenwood C, Selth LA, Dirac-Svejstrup AB, Svejstrup JQ. An iron-sulfur cluster domain in ELP3 important for the structural integrity of elongator. J Biol Chem. 2008 doi: 10.1074/jbc.M805312200. [DOI] [PubMed] [Google Scholar]
- 34.Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai Y, Nakai M, Hayashi H. Thio-modification of Yeast Cytosolic tRNA Requires a Ubiquitin-related System That Resembles Bacterial Sulfur Transfer Systems. J Biol Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 36.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008;105:18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009 doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 38.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, et al. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci U S A. 2008;105:5459–5464. doi: 10.1073/pnas.0709404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kourtis N, Tavernarakis N. Cell-specific monitoring of protein synthesis in vivo. PLoS ONE. 2009;4:e4547. doi: 10.1371/journal.pone.0004547. doi:10.1371/journal.pone.0004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, et al. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J Exp Biol. 2001;204:1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- 44.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 45.Burke NV, Han W, Li D, Takimoto K, Watkins SC, et al. Neuronal peptide release is limited by secretory granule mobility. Neuron. 1997;19:1095–1102. doi: 10.1016/s0896-6273(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 46.Husain QM, Ewer J. Use of targetable gfp-tagged neuropeptide for visualizing neuropeptide release following execution of a behavior. J Neurobiol. 2004;59:181–191. doi: 10.1002/neu.10309. [DOI] [PubMed] [Google Scholar]
- 47.Speese S, Petrie M, Schuske K, Ailion M, Ann K, et al. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci. 2007;27:6150–6162. doi: 10.1523/JNEUROSCI.1466-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen M, Alfonso A, Johnson CD, Rand JB. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics. 1995;140:527–535. doi: 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahoney TR, Luo S, Nonet ML. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat Protoc. 2006;1:1772–1777. doi: 10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- 50.Mahoney TR, Liu Q, Itoh T, Luo S, Hadwiger G, et al. Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell. 2006;17:2617–2625. doi: 10.1091/mbc.E05-12-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukushige T, Siddiqui ZK, Chou M, Culotti JG, Gogonea CB, et al. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J Cell Sci. 1999;112 (Pt 3):395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- 52.Holmberg C, Katz S, Lerdrup M, Herdegen T, Jaattela M, et al. A novel specific role for I kappa B kinase complex-associated protein in cytosolic stress signaling. J Biol Chem. 2002;277:31918–31928. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- 53.Cuajungco MP, Leyne M, Mull J, Gill SP, Lu W, et al. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am J Hum Genet. 2003;72:749–758. doi: 10.1086/368263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high performance liquid chromatography. In: Gehrke CW, Kuo KCT, editors. In Chromatography and Modification of Nucleosides Part A Analytical Methods for Major and Modified Nucleosides J Chromatography Library. Amsterdam: Elsevier; 1990. pp. A3–A71. [Google Scholar]
- 55.Keith G, Dirheimer G. Reinvestigation of the primary structure of brewer's yeast tRNA 3 Arg. Biochem Biophys Res Commun. 1980;92:116–119. doi: 10.1016/0006-291x(80)91527-2. [DOI] [PubMed] [Google Scholar]
- 56.Keith G. The primary structures of two arginine tRNAs (anticodons C-C-U and mcm5a2U-C-psi) and of glutamine tRNA (anticodon C-U-G) from bovine liver. Nucleic Acids Res. 1984;12:2543–2547. doi: 10.1093/nar/12.5.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JH, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci U S A. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 59.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 60.Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJ, et al. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol. 2003;49:1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x. [DOI] [PubMed] [Google Scholar]
- 61.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 62.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 63.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44:8078–8089. doi: 10.1021/bi050343f. [DOI] [PubMed] [Google Scholar]
- 65.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 66.Rennie MJ, Smith K, Watt PW. Measurement of human tissue protein synthesis: an optimal approach. Am J Physiol. 1994;266:E298–307. doi: 10.1152/ajpendo.1994.266.3.E298. [DOI] [PubMed] [Google Scholar]
- 67.Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen L, Henzel WJ, Baeuerle PA. IKAP is a scaffold protein of the IkappaB kinase complex. Nature. 1998;395:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 69.Krappmann D, Hatada EN, Tegethoff S, Li J, Klippel A, et al. The I kappa B kinase (IKK) complex is tripartite and contains IKK gamma but not IKAP as a regular component. J Biol Chem. 2000;275:29779–29787. doi: 10.1074/jbc.M003902200. [DOI] [PubMed] [Google Scholar]
- 70.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gracheva EO, Burdina AO, Holgado AM, Berthelot-Grosjean M, Ackley BD, et al. Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol. 2006;4:e261. doi: 10.1371/journal.pbio.0040261. doi:10.1371/journal.pbio.0040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McEwen JM, Madison JM, Dybbs M, Kaplan JM. Antagonistic regulation of synaptic vesicle priming by Tomosyn and UNC-13. Neuron. 2006;51:303–315. doi: 10.1016/j.neuron.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 73.Dybbs M, Ngai J, Kaplan JM. Using microarrays to facilitate positional cloning: identification of tomosyn as an inhibitor of neurosecretion. PLoS Genet. 2005;1:e2. doi: 10.1371/journal.pgen.0010002. doi: 10.1371/journal.pgen.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lewis JA, Wu CH, Berg H, Levine JH. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 76.Stringham EG, Candido EP. Targeted single-cell induction of gene products in Caenorhabditis elegans: a new tool for developmental studies. J Exp Zool. 1993;266:227–233. doi: 10.1002/jez.1402660309. [DOI] [PubMed] [Google Scholar]
- 77.Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130:3781–3794. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- 78.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, et al. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glasser AL, el Adlouni C, Keith G, Sochacka E, Malkiewicz A, et al. Presence and coding properties of 2′-O-methyl-5-carbamoylmethyluridine (ncm5Um) in the wobble position of the anticodon of tRNA(Leu) (U*AA) from brewer's yeast. FEBS Lett. 1992;314:381–385. doi: 10.1016/0014-5793(92)81510-s. [DOI] [PubMed] [Google Scholar]
- 81.Burke D, Dawson D, Stearns T, editors. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 82.Ward S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci U S A. 1973;70:817–821. doi: 10.1073/pnas.70.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. elegans elpc-3 is required for mcm5 and ncm5 side chain formation at wobble uridines. (A) The diagram shows the genomic structures of elpc-3 and ZK863.4, which is suggested to be in the same operon. The exons and introns are depicted as boxes and lines respectively. At top, the line underneath represents the location of the deletion in elp-3(tm3120). Below, a representation of predicted motifs in the ELPC-3 protein: the Radical S-adenosyl methionine (Radical-SAM) [31], and histone acetyltransferase (HAT) domains [83]. The region deleted in elpc-3(tm3120) is indicated by a line beneath. (B–G) Chromatograms of total tRNA isolated from wild-type and elpc-3(tm3120) worms analyzed by HPLC. Wild-type (N2) profiles are shown in the left panels; elpc-3(tm3120) profiles are shown in the right panels. Chromatograms were monitored at 254 nm, unless otherwise stated. (B,C) The parts of chromatograms between retention times 46 and 51.5 min are displayed. The arrow in C indicates the expected retention time of mcm5s2U. (D,E) The parts of the chromatograms between retention times 11 and 18 min are displayed. The arrow in E indicates the expected retention time of ncm5U. (F,G) The parts of the chromatograms between retention times 31 and 37 min are displayed. The arrow in F indicates the expected retention time of s2U. Chromatograms were monitored at 314 nm.
(0.55 MB TIF)
tuc-1 in C. elegans is required for 2-thio wobble uridine tRNA modification. (A) The schematic structure of tuc-1. Exons and introns are represented by boxes and lines, respectively. The line underneath indicates the region deleted in tuc-1(tm1297). (B–E) Chromatograms showing total tRNA isolated from wild-type (N2) and tuc-1(tm1297) worms analyzed by HPLC. N2 profiles are shown in the left panels; tuc-1(tm1297) profiles are shown in the right panels. Chromatograms were monitored at 254 nm. (B,C) The parts of the chromatograms between retention times 46.2 and 51.6 min are displayed. The arrow in C indicates the expected retention time of mcm5s2U. (D,E) The parts of the chromatograms between retention times 35 and 42 min are displayed. The arrow in D indicates the expected retention time of mcm5U.
(0.43 MB TIF)
Formation of ncm5, mcm5 and s2 side chains is abolished in elpc-1 ; tuc-1 double mutants. (A–F) Total tRNA isolated from wild type, elpc-1(tm2149) or elpc-1(tm2149); tuc-1(tm1297) worms was analyzed by HPLC. Wild-type (N2) and elpc-1(tm2149) profiles are shown in left panels. elpc-1(tm2149); tuc-1(tm1297) profiles are in right panels. Chromatograms were monitored at 254 nm, unless otherwise stated. (A,B) The parts of chromatograms between retention times 48.5 and 53.5 min are displayed. The arrow in the right panel indicates the expected retention time of mcm5s2U. (C,D) The parts of the chromatograms between retention times 11 and 18 min are displayed. The arrow in the right panel indicates the expected retention time of ncm5U. (E,F) The parts of the chromatograms between retention times 33 and 39 min are displayed. The arrow in the right panel indicates the expected retention time of s2U. Chromatograms were monitored at 314 nm.
(0.45 MB TIF)
The elp-1(tm2149) and elpc-3(tm3120) mutants are defective in fluorescence recovery after photobleaching. Quantification of fluorescence signals in worms carrying gcy-5::gfp (A) or myo-3::gfp (B) reporters. The pixel intensities in wild type, elpc-1, elpc-3 and tuc-1 backgrounds before photobleaching, after photobleaching, and after 5 hours recovery are shown. In ‘gcy-5::gfp+CHX’, fluorescence recovery was measured in the presence of cycloheximide (CHX). The number of worms examined of each strain is denoted under the graph. Error bars represent standard deviations.
(0.67 MB TIF)
Neuronal morphology in elpc-1 , elpc-3 and tuc-1 mutant worms is normal. (A, C, E, G, I, K) Micrographs of hermaphrodite worms fed with DiI viewed with Nomarski DIC optics. The arrows denote three amphid neurons, ASI, ADL and ASK. (B, D, F, H, J, L) The same worms viewed with fluorescence optics. Note that DiI efficiently labels the neurons in the mutant worms, indicating that the outgrowth of the neuronal processes was normal.
(5.94 MB TIF)
The salt chemotaxis learning defect of elpc-1(tm2149) is rescued by an elpc-1::gfp construct. Worms were synchronized and raised at 25°C to the young adult stage. The chemotaxis index after 30 min of assay is displayed. The assay was repeated four times. Error bars denote standard deviations. Two asterisks indicate a significant difference between elpc-1 and elpc-1; elpc-1::gfp (**p<0.001 by student's t test).
(0.20 MB TIF)
elpc-1 , elpc-3 , and tuc-1 chemotax to both NaCl and isoamylalcohol. (A, B) Chemotaxis to NaCl (A) and isoamyl alcohol (B) is shown. The chemotaxis indices were plotted against time for four different genotypes. For each genotype, 80–100 young adult worms that had been raised at 25°C were placed on a plate equidistant from the attractant and a control spot. The numbers of worms at the NaCl (or isoamylalcohol) and the control spots were counted every 10 minutes for 1 hour. Each assay was repeated for 4 times.
(0.26 MB TIF)
The increased aldicarb resistance of the elpc-1(tm2149) mutant is complemented by elpc-1::gfp . The proportion of worms still able to move is plotted against time. 25–30 worms were used for each genotype. The assay was performed blind in triplicate at room temperature (ca. 21.5°C). The worms were cultivated at 25°C prior to being assayed.
(0.22 MB TIF)
Acetylated α-tubulin levels are not decreased in Elongator mutants. (A) Western blot of whole animal lysates for wild type (N2), elpc-1, elpc-3, mec-12(e1605) and mec-12(e1607). Both acetylated α-tubulin and α-tubulin migrated just above 50 KDa. Top, blotted with anti-lys40-acetylated-α-tubulin antibody at a dilution of 1∶1000. Bottom, blotted with anti-α-tubulin antibody at a dilution of 1∶2000. Lys40 acetylated α-tubulin signals were normalized to that of α-tubulin, and the amount of lys40 acetylated α-tubulin was expressed relative to the corresponding value in the wild type strain, which was set to 1. NA, not applicable. (B) mec-12(e1607) worms are not resistant to aldicarb. The proportion of worms still able to move is plotted against time. 25–30 worms were used for each genotype. The assay was performed blind in triplicate at room temperature (ca. 21.5°C). The worms were cultivated at 25°C prior to being assayed.
(0.66 MB TIF)
Defects Seen in Temperature-shifted elpc-3 ; tuc-1 Double Mutants. Micrographs of eggs and larvae viewed with Nomarski DIC optics. (A–D) Embryos arrested prior to (A), during (B,C) or after (D) morphogenesis. (E,F,G,H) Parts of the germline in young adult hermaphrodites. The arrows in E and F indicate oocytes. Note that those in the elpc-3; tuc-1 worm have not matured. The arrows in G and H denote sperm. Those in the elpc-3; tuc-1 worm have grossly abnormal morphology. (I,J) The descendants of P5.p, P6.p and P7.p during the L4 stage. The arrows in I denote the descendants of P5.p, P6.p and P7.p. In the animal shown, these three cells adopted the 3° cell fate and divided just once. In wild-type worms, P6.p adopts the 1° cell fate whereas P5.p and P7.p adopt the 2° fate. The 1° and 2° fates involve three rounds of cell division; the descendants of P5.p, P6.p and P7.p together form a tube through which the eggs are laid. The arrow in J denotes the tube as it is forming. Scale bars denote 10 microns.
(2.42 MB TIF)
ELPC-1::GFP is expressed during vulval cell fate specification. Micrographs of an L3 hermaphrodite worm of the genotype elpc-1(tm2149); svEx557[Pelpc-1::elpc-1::gfp] viewed with either Nomarski differential contrast (DIC) (A) or fluorescence (B) optics. The arrows denote the six descendants of P5.p, P6.p and P7.p.
(1.58 MB TIF)