Abstract
Metastasis is the primary cause of death in patients with breast cancer. Overexpression of c-myc in humans correlates with metastases, but transgenic mice only show low rates of micrometastases. We have generated transgenic mice that overexpress both c-myc and vascular endothelial growth factor (VEGF) (Myc/VEGF) in the mammary gland, which develop high rates of pulmonary macrometastases. Gene expression profiling revealed a set of deregulated genes in Myc/VEGF tumors compared to Myc tumors associated with the increased metastatic phenotype. Cross-comparisons between this set of genes with a human breast cancer lung metastasis gene signature identified five common targets: tenascin-C (TNC), matrix metalloprotease-2, collagen-6-A1, mannosidase-α-1A and HLA-DPA1. Signaling blockade or knockdown of TNC in MDA-MB-435 cells resulted in a significant impairment of cell migration and anchorage-independent cell proliferation. Mice injected with clonal MDA-MB-435 cells with reduced expression of TNC demonstrated a significant decrease (P < 0.05) in (1) primary tumor growth; (2) tumor relapse after surgical removal of the primary tumor and (3) incidence of lung metastasis. Our results demonstrate that VEGF induces complex alterations in tissue architecture and gene expression. The TNC signaling pathway plays an important role in mammary tumor growth and metastases, suggesting that TNC may be a relevant target for therapy against metastatic breast cancer.
Keywords: metastasis, vascular endothelial growth factor, transgenic mice, mammary cancer, tenascin-C
Introduction
Metastasis is the primary cause of mortality from breast cancer (BC), resulting in over 40 000 deaths per year in the United States (Jemal et al., 2007). A general lack of knowledge about the molecular mechanisms involved in the metastatic process has greatly hampered the development of therapies against this form of advanced disease. Therefore, a major challenge remains to identify genes responsible for the acquisition of the metastatic phenotype, with the ultimate goal of inhibiting the metastatic disease.
Multiple steps are involved in the metastatic process including angiogenesis, invasiveness, migration through the extracellular matrix and stroma, intravasation into blood or lymphatic vessels, and extravasation and growth of new tumor masses (Folkman, 2002; Gupta and Massague, 2006). Invasion and tumor vascularization imply degradation of connective tissue, a process that requires the activity of extracellular matrix (ECM) proteases, such as matrix metalloproteases (MMPs) and serine proteases (Noel et al., 2004).
Many studies have shown that vascular endothelial growth factor (VEGF) is a key proangiogenic factor that is overexpressed in virtually all solid tumors, acting through its receptors VEGFR1 (Flt-1) and VEGFR2 (Flk-1/KDR). VEGF is overexpressed in BC, and a strong correlation between VEGF expression and microvessel density has been demonstrated (De Paola et al., 2002). VEGF plasma levels also correlate with presence of metastasis in patients with BC (Adams et al., 2000). In addition, we and others have demonstrated that VEGF may have a direct autocrine effect on tumor cells (Huh et al., 2005; Schoeffner et al., 2005), suggesting that VEGF may stimulate tumor growth and metastases through nonvascular mechanisms.
Amplification and overexpression of c-myc is frequently found in human BC and is associated with metastasis to the lymph nodes and lungs (Deming et al., 2000). However, the MMTV-Myc (Myc) mouse mammary tumor model is poorly metastatic (Amundadottir et al., 1995; Hundley et al., 1997). Therefore, the fact that these transgenic animals are unable to fully mimic the metastatic progression of human c-myc-associated breast tumor metastasis suggests that c-myc may act in concert with other factors to produce the metastatic phenotype.
Considering the potentially important role of VEGF in BC metastasis, and that Myc mammary tumors are poorly vascularized (Amundadottir et al., 1995), we hypothesized that targeting VEGF expression to the mammary epithelium of Myc transgenic mice would promote a metastatic phenotype. To test this hypothesis, we have generated compound transgenic mice where both VEGF and c-myc are co-overexpressed in the mammary gland, and demonstrate that these mice develop a high rate of lung macrometastases. Gene expression profiling of these primary tumors indicates that the overexpression of VEGF results in the robust induction of genes involved in angiogenesis, ECM proteins and processing enzymes, several of which have been implicated in human metastatic disease. Functional analysis demonstrates that one such gene, tenascin-C (TNC), is involved in the acquisition of the metastatic phenotype.
Results
Myc/VEGF transgenic mice develop highly angiogenic and metastatic mammary tumors
The individual phenotypes of MMTV-Myc (Amundadottir et al., 1995; Hundley et al., 1997) and MMTV-VEGF (Schoeffner et al., 2005) transgenic mice have been previously reported. Although Myc female mice develop mammary tumors, the mammary glands of MMTV-VEGF female mice appear morphologically similar to wild types and do not develop tumors (Schoeffner et al., 2005).
Virgin Myc/VEGF transgenic females developed mammary tumors with a lower incidence than Myc mice: 27% (13 of 48) vs 67% (36 of 54), possibly related to reduced levels of Myc expression in Myc/VEGF tumors compared to Myc tumors (see below). Tumor onset was also delayed in Myc/VEGF mice (76.1 ± 29 weeks) compared to Myc mice (37.3 ± 9), though tumor multiplicity was similar between both genotypes (1.7 tumors per mouse, on average). All Myc tumors were adenocarcinomas with a tubulopapillary pattern (Figure 1a), as previously described (Amundadottir et al., 1995). However, Myc/VEGF tumors were pleomorphic adenocarcinomas (Figure 1b), and some tumors exhibited an adenosquamous appearance (Figure 1c). In addition, Myc/VEGF tumors demonstrated a general increase in fibrosis and ECM content compared to Myc tumors. Mammary tumors arising in either Myc or Myc/VEGF mice were characterized by the presence of appropriate transgene-specific transcripts (Supplementary Figure 1).
Figure 1.
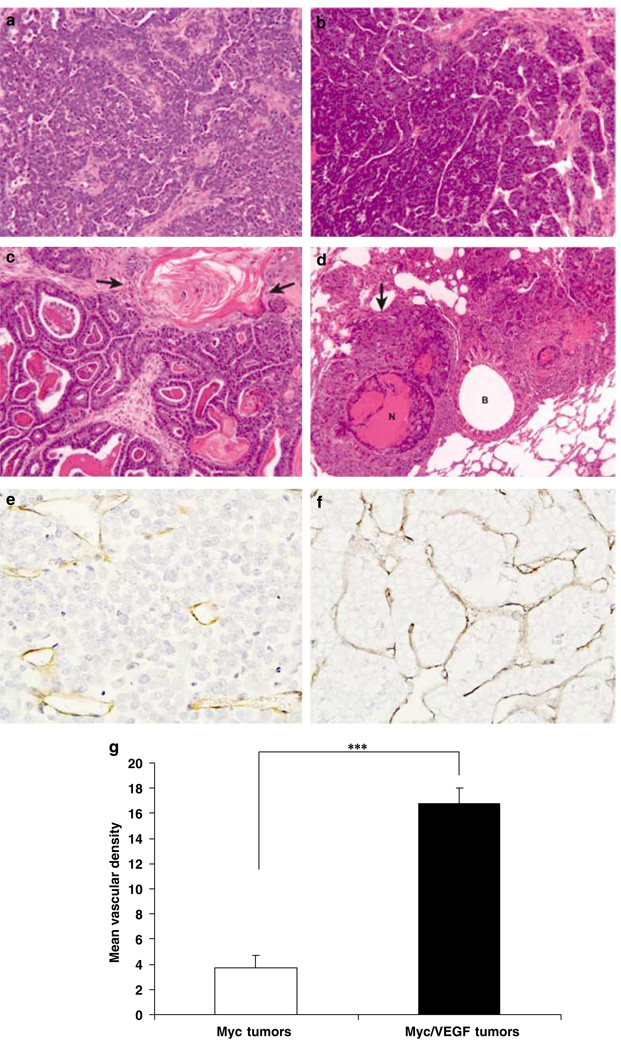
Morphology of primary Myc and Myc/VEGF mammary tumors. (a) Adenocarcinoma of a Myc transgenic mouse. (b) Adenocarcinoma of a Myc/VEGF mouse. (c) Adenosquamous (metaplastic) carcinoma of a Myc/VEGF mouse. Arrows indicate stratified squamous epithelium with keratinization. (d) Large pulmonary metastasis (arrow) with central necrosis (N). Metastasis surrounds a bronchus (B). (e) Immunostaining for CD-31 in a Myc tumor. (f) Myc/VEGF tumor showing higher vascularization (anti-CD-31 staining) than Myc tumors. (g) Comparison of the mean vascular density between Myc and Myc/VEGF tumors. ***P < 0.0004.
Real time RT-PCR of angiogenic markers (Supplementary Table 1) revealed that VEGF was increased 2.5-fold (P < 0.01) in Myc/VEGF tumors compared to Myc tumors. VEGFR1 (Flt-1) and VEGFR2 (Flk-1) were also significantly increased (P = 0.001). CD-31 and vascular endothelial Cadherin (VE-cadherin), specific markers for endothelial cells, were significantly higher (P = 0.015 and P = 0.036, respectively) in Myc/VEGF tumors than in Myc tumors (Supplementary Table 1). Quantification of vascular density by immunostaining for CD-31 (Figures 1e and f) demonstrated that Myc/VEGF tumors had increased vascularity compared to Myc tumors. The mean vascular density (MVD) was 3.71 ± 1 (percent of total tumor) for Myc tumors vs 16.8 ± 1 for Myc/VEGF tumors (P < 0.0004) (Figure 1g).
Lungs from a subset of mice that developed tumors were evaluated for metastases by detailed histological analyses (Supplementary Table 2). Microscopic metastases were observed in 7 of 19 (36.9%) of Myc mice, with only 2 of 19 (10.5%) of mice developing macroscopic metastases. Overall, 8 of 19 (42.1%) had metastases of any size. In contrast, 16 of 18 (88.8%) of Myc/VEGF mice developed microscopic metastases and 8 of 18 (44%) of Myc/VEGF mice developed macroscopic metastases. Overall, 16 of 18 (88.9%) had metastases of any size. Thus, Myc/VEGF mice were more than twice as likely as Myc mice to develop lung metastases of any size, and four times as likely to develop macroscopic metastases. In cross-sectional areas, the size of Myc/VEGF lung metastases was more than 20 times larger (on average) than those of Myc metastases (Supplementary Table 2).
To further investigate metastatic potential, 1 mm3 pieces of either Myc or Myc/VEGF tumors were implanted subcutaneously into the flanks of female mice. All tumor implants grew following transplant and the resultant Myc and Myc/VEGF tumors grew at approximately the same rate. Eighty percent (8 of 10) of the Myc/VEGF transplant recipients (in both nude and FVB/N background) developed macroscopic pulmonary metastases over a 3 to 4-week period. In contrast, no macroscopic lung metastases were observed for transplanted Myc tumors (0 of 10).
Genetic portraits of Myc/VEGF metastatic primary tumors reveal numerous changes in genes related to ECM remodeling, invasion and adhesion
High-throughput gene expression profiles of Myc tumors were compared to those of Myc/VEGF tumors, and 722 genes with a twofold differential expression (P < 0.01) were identified (506 corresponding to upregulated genes and 216 to downregulated genes). Expression changes in all five Myc/VEGF tumors were similar with a correlation coefficient of 0.71 and similar patterns of expression. A selected list of genes with particular relevance to the acquisition of the metastatic phenotype is presented in Table 1.
Table 1.
Selected list of genes with a potential role in the acquisition of the metastatic phenotype found in Myc/VEGF tumors
| Gene name/gene category | Average ratio | Gene function/gene expression in cancer |
|---|---|---|
| Associated with metastasis/invasion/adhesion | ||
| CD-44 | 3.1±0.1 | Upregulated in many metastatic tumors, including BC |
| ECM-1 | 9.7±2.2 | Angiogenesis and bone formation. Upregulated in metastatic BC |
| Metastasis-associated gene 1 (MTA-1) | 2.0±0.4 | Metastasis signature |
| GLYCAM-1 | 15.8±2.7 | Adhesion molecule. Upregulated in metastatic BC |
| Fibronectin-1 | 6.8±1.4 | Metastatic signature in many types of tumors, including BC |
| Laminin-5-γ-2 | 10.6±1.1 | Metastatic signature in many types of tumors, including BC |
| Rho-related GTPases | Cytoskeletal reorganization through actin interaction | |
| Rho-C | 2.0±0.2 | Metastatic signature in melanoma |
| Rac-1 | −2.0±0.6 | |
| Rho-GDI-γ | −1.8±0.1 | |
| Rap | −2.0±0.1 | |
| Pak-1 | 1.7±0.3 | |
| Integrin α3 | 2.6±0.2 | Metastatic signature for some types of tumors |
| P-cadherin | 2.0±0.5 | Upregulated in BC in relation to ER loss and malignancy |
| CEACAM-1 | −3.3±0.1 | Downregulated in several types of tumors including BC |
| CEACAM-2 | −2.5±0.2 | Unknown expression in tumors |
| Tenascin-C | 5.5±2.3 | Cell-ECM adhesion. Upregulated in many types of tumors |
| ECM | ||
| Procollagen I, α-1 | 5.0±2.1 | Fibers of connective tissue. Metastatic signature |
| Procollagen III, α-1 | 4.8±1.7 | Fibers of connective tissue |
| Procollagen V, α-2 | 6.5±1.8 | Fibers of connective tissue |
| Procollagen VI, α-1 | 3.2±1.0 | Fibers of connective tissue |
| Procollagen XIV, α-1 | 3.2±1.0 | Fibers of connective tissue |
| Angiogenesis | ||
| Hypoxia-induced gene-1 | 2.4±0.0 | Activates expression of pro-angiogenic factors |
| Neuropilin | 2.1±0.3 | VEGF receptor. Promotion of tumor angiogenesis |
| Proteinases | ||
| MMP-2 | 5.2±1.4 | Promotes ECM degradation and activation of growth factors. Upregulated in many types of carcinomas, including BC |
| MMP-23 | 2.2±0.5 | Unknown substrate and function |
| Cathepsin-L | 3.0±1.0 | Promotes ECM degradation. Upregulated in BC |
| Cathepsin-C | 2.0±0.1 | Promotes ECM degradation. Unknown status in cancer |
| ADAMs | Family of proteins which interact with integrins, implicated in proteolytic cleavage of cell surface molecules |
|
| ADAM-8 | 3.7±0.7 | |
| ADAM-9 | 2.0±0.2 | |
| ADAM-12 | 3.4±1.2 | |
| ADAM-19 | 2.2±0.6 | |
| Cystatin-C | 2.7±0.1 | Protease inhibitor |
| Cystatin-B | 3.8±0.4 | Protease inhibitor |
| Cytoskeleton | ||
| Keratins | Family of proteins related to cytoskeleton | |
| Krt1-19 | 9.3±0.7 | |
| Krt2-6a | 23.5±2.9 | |
| Krt1-13 | 7.1±1.4 | |
| Krt2-4 | 2.7±0.1 | |
| Dystonin | 3.8±0.7 | Interacts with actin and microtubules |
| Cell signaling | ||
| AXL-receptor tyrosine kinase | 4.6±1.0 | Involved in cell signaling related to cell adhesion |
| Myristoylated alanine-rich C-kinase substrate |
4.7±0.3 | Participates in cytoskeletal remodeling |
| Growth factors | ||
| TGF-β induced 68 kDa | 4.8±2.4 | Cell growth of stromal components. Promotion of tumor angiogenesis. Interaction with ECM components |
| BMP-1 | 3.3±0.8 | Bone formation. Promotion of angiogenesis |
| Cell cycle/transcription factors | ||
| c-Myc | −10.0±0.1 | Transcription factor involved in cell cycle regulation and apoptosis. Overexpressed in a high percentage of BC |
| Myb | −2.9±0.4 | Transcription factor involved in cell cycle regulation, overexpressed in several types of tumors including some CB |
| Cyclin B | −2.5±0.8 | Cell cycle regulator. It binds to cdc2 |
| Cdk-1 | −2±0.1 | Cell cycle regulator. It binds to cyclin B |
| Cyclin D1 | 2.1±0.2 | Increased expression in BC, especially in metastasis. Collaboration with Rho GTPases in cell migration signaling |
| Cyclin G | 3.4±0.2 | Cell cycle regulator at the S, G2/M checkpoint |
Abbreviations: BC, breast cancer; BMP, bone morphogenetic protein; ECM, extracellular matrix; MMP, matrix metalloprotease; TGF, transforming growth factor-β; VEGF, vascular endothelial growth factor.
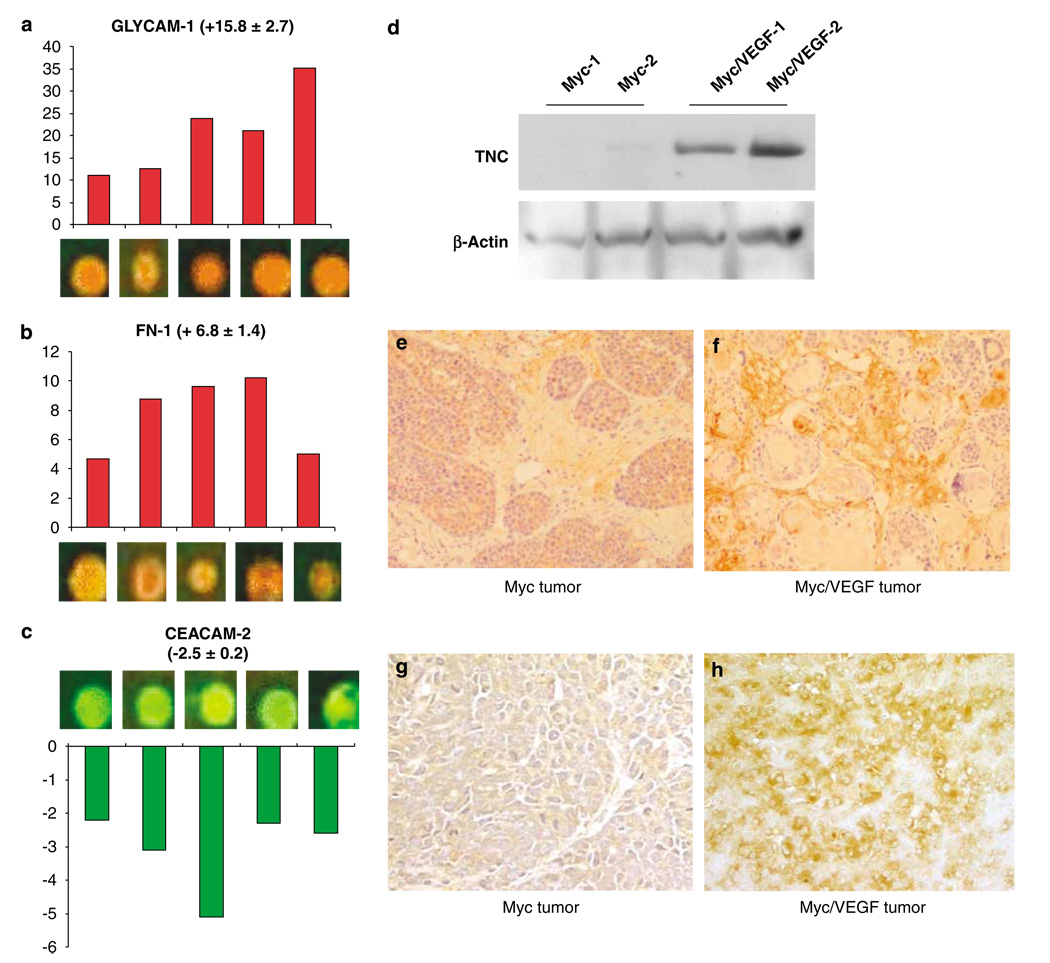
Examples of genes highly upregulated in primary tumors with high metastatic propensity were fibronectin-1 and glycosylation-dependent cell adhesion molecule-1 (GLYCAM-1), whose expression was increased by 6.8 ± 1.4- and 15.8 ± 2.7-fold, respectively (Figures 2a and b). An example of a downregulated gene in tumors from mice overexpressing both Myc and VEGF was cell adhesion molecule-2 (CEACAM-2) (Figure 2c). TNC mRNA was found to be upregulated 5.5 ± 2.3-fold in Myc/VEGF tumors as compared to Myc tumor and confirmed by western blot and immunohistochemistry (Figures 2d–f). Immunostaining was mainly found in the ECM and, in a lesser extent, in tumor cells. The stromal compartment of the blood vessel walls was also labeled, but not the endothelial cells. Myc/VEGF tumors exhibited a stronger pattern of TNC staining (Figures 2e and f). Laminin-5-γ-2, a gene whose expression has been linked to BC metastasis (Pyke et al., 1994), was upregulated by 10.6 ± 1.1-fold in tumors overexpressing both Myc and VEGF. Immunostaining for laminin-5 was observed in tumor cells and the ECM, and Myc/VEGF tumors showed more intense staining (Figures 2g and h).
Figure 2.
Changes in gene expression between Myc and Myc/VEGF tumors. (a) Microarray spots for GLYCAM-1 after competitive hybridization of Myc/VEGF tumors with Myc tumors. Bars represent fold-change increase in Myc/VEGF tumors compared to Myc tumors. (b) Microarray spots and fold-change increase for fibronectin-1 (FN-1) in Myc/VEGF tumors, as compared to Myc cancers. (c) CEACAM-2 is downregulated in bitransgenic tumors in comparison with Myc malignant tissues. (d) Validation of the upregulation of TCN by western blot. (e) Myc tumor stained for TNC. As seen by immunohistochemistry, TNC was mainly found in the extracellular matrix (ECM) and, in a lesser extent, in tumor cells. (f) Myc/VEGF tumor stained for TNC. Immunostaining for TNC is stronger in Myc/VEGF than in Myc tumors. (g) Myc tumor stained for laminin-5. Staining is observed in tumor cells and in the ECM. (h) Myc/VEGF tumor stained for laminin-5. A stronger staining was found for these types of tumors.
The VEGF-dependent metastatic phenotype was characterized by numerous changes in the expression of ECM proteins including collagens, ECM proteases and cell adhesion proteins (Table 1). Of particular interest are type 1 collagen α-1 (Colla1), fibronectin-1 and tenascin-C, as these genes have been identified in other microarray metastatic signatures (Ramaswamy et al., 2003; Minn et al., 2005). The critical role of proteases in modifying the ECM and promoting cell tumor invasion and metastasis has been well established (Stamenkovic, 2000). Upregulation of numerous proteases in Myc/VEGF tumors was observed (Table 1), including MMPs-2 and -23; cathepsins (-L and -C); cystatins (-B and -C), and disintegrin and metalloproteinases (ADAMs)-8, -9, -12 and -19. We also identified deregulated expression of genes belonging to the Rho GTPase signaling pathway in the tumors with high metastatic propensity, including Rho-C, Rac-1, Rap and Pak-1 (Table 1) that follow a concordant pattern of expression with reports of metastatic tumors published elsewhere (Schmitz et al., 2000). Therefore, VEGF over-expression strongly modifies the tumor ECM, which likely elicits cell migration and invasion of the primary tumor.
A significant reduction in c-Myc (−10.0 ± 0.1) expression was observed in the Myc/VEGF transgenic tumors compared to Myc tumors (Table 1) and confirmed by western blot (Supplementary Figure 2). Reduction in c-Myc levels might explain why Myc/VEGF mice have a lower incidence and delayed onset of mammary tumors compared to Myc transgenic mice.
Microarray data generated for this study are available at http://www.ncbi.nlm.nih.gov/geo/.
Comparisons of mouse and human breast cancer lung metastasis signatures
In order to identify a set of candidate genes with a potentially high likelihood for influencing the metastatic phenotype, we employed a cross-species comparison of gene expression profiles identified in this study with array data from human mammary tumors implicated in lung metastasis using a publicly available microarray data from metastatic BC patients retrieved from GEO (GSE2603) http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE2603. A set of 54 genes, corresponding to a BC lung metastatic signature was cross-compared with the set of mouse genes identified in this study. Five genes with a similar pattern of expression in both species were identified: upregulated genes TNC, MMP-2, collagen-6-A1, mannosidase-α-1A, and the downregulated gene HLA-DPA1. The functional role of TNC in BC lung metastasis was further investigated as it has been implicated in tumor cell invasion (Tsunoda et al., 2003) and remodeling of angiogenic tissues (Zagzag and Capo, 2002).
Tenascin-C blockade decreases migration of metastatic mammary cells
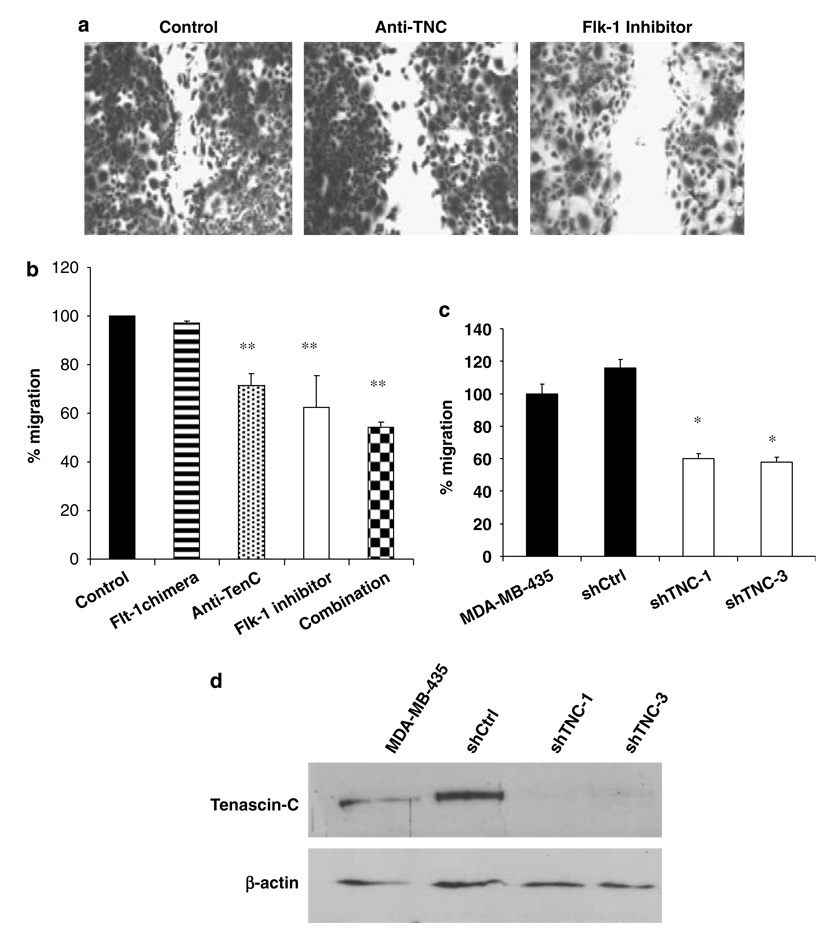
The cell line MV630, established from Myc/VEGF mammary tumors, expresses high levels of VEGF, Flt-1, Flk-1 and TNC (data not shown). Anti-TNC blocking antibodies significantly (P < 0.01) inhibited cell migration of MV630 cells (Figures 3a and b). A control IgG antibody did not inhibit migration (result not shown). Although blockade of Flt-1 did not inhibit MV630 cell migration, blockade of Flk-1 signaling or the combination of anti-TNC blocking antibody and the Flk-1 inhibitor resulted in a significant reduction in cell migration (P < 0.01) (Figures 3a and b). Flk-1 blockade caused inhibition of MV630 cell growth as well, as demonstrated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (data not shown). MDA-MB-435 cells, a human metastatic cancer cell line that constitutively expresses TNC (Figure 3d), was used for knockdown experiments using a retroviral vector expressing shRNA to TNC (Figure 3d). TNC expression was reduced by 90–93% in the clone shTNC-1 and 83.3% in the shTNC-3 with respect to shCtrl. Migration experiments were performed using either parental cells, the control clone shCtrl, or shTNC-1 and shTNC-3 clones. Migration assays further confirmed that downregulation of TNC causes a significant reduction (P < 0.05) in the ability of MDA-MB-435 cells to migrate (Figure 3c). These results demonstrate that tumor cell migration can be decreased by targeting TNC- and Flk-1-dependent signaling pathways.
Figure 3.
Migration assays. (a) Migration of M630 cells is impaired by TCN and Flk-1 signaling blockade. (b) Quantification of migration inhibition in M630 cells. Treatment with an Flt-1 antagonist did not change cell migration. Treatment with an Flk-1 inhibitor results in a significant reduction of migration (P < 0.01). Treatment with an anti-TNC blocking antibody also causes significant inhibition of cell migration (P < 0.01). The combination treatment using anti-TNC plus Flk-1 inhibitor produces the strongest inhibitory effect (P < 0.01). (c) Migration assays in MDA-MB-435 cells and cell clones shCtrl, shTNC-1 and shTNC-3. Downregulation of TNC in clones shTNC-1 and shTNC-3 reduces significantly (P < 0.05) cell migration when compared to MDA-MB-435 cells and the control clone shCtrl. (d) Western blot analysis to determine TNC protein expression in MDA-MB-435, shCtrl, shTNC-1 and shTNC-3 cells. *P < 0.05; **P < 0.01.
Interestingly, VEGF did not directly regulate the expression of TNC in tumor cells. Neither the addition of exogenous VEGF nor the reduction of VEGF expression through the addition of siRNA for VEGF altered the level of TNC in MDA-MB-435 cells (results not shown). This result suggests that overexpression of VEGF does not directly increase TNC levels in tumor cells, but likely acts through indirect mechanisms involving the stromal compartment.
Downregulation of TNC results in a significant decrease in mammary tumor growth, tumor relapse and lung metastases
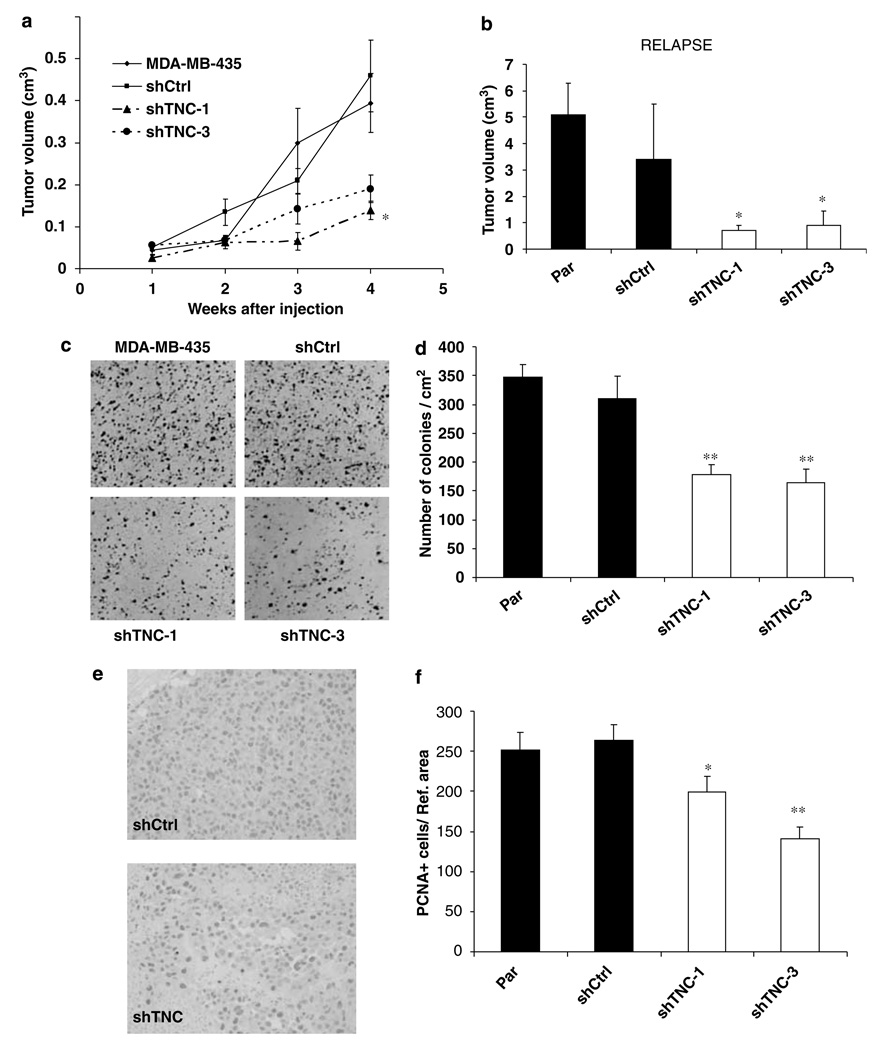
In vivo experiments were performed to determine the impact of TNC downregulation on tumor growth and lung metastasis development. Female nude mice were injected with either 106 MDA-MB-435 parental cells, or shCtrl, shTNC-1 and shTNC-3 cell clones, into the mammary fat pads (Figure 4a). Average tumor volume in MDA-MB-435 and shCtrl groups was similar, whereas a significant decrease in tumor volume (approximately 50% reduction, P < 0.05) was observed for both the shTNC-1 and shTNC-3 clones. Primary tumors were surgically removed when they reached 1.5 cm in diameter (at 4–5 weeks postinjection) and animals were then monitored for relapse until week 12. Figure 4b shows that tumors that relapsed in both the control groups were significantly larger than relapsed tumors from the clones with reduced expression of TNC. Therefore, tumors with high expression of TNC are more prone to relapse following surgical resection than tumors with low expression of this protein.
Figure 4.
Decrease in primary tumor growth and cell proliferation in MDA-MB-435 cells with a downregulated expression of TNC. (a) Primary tumor volume is significantly lower (P < 0.05) in shTNC clones than in controls. (b) Relapsed tumor volume is significantly lower (P < 0.05) in cell clones lacking TCN. (c) Clonogenic assay in anchorage-independent conditions shows a lower number of colonies in shTNC clones, compared to controls. (d) Quantification of the number of colonies demonstrates that reduction in TNC levels results in lower clonogenic potential (P < 0.01) than that found for controls. (e) Immunohistochemistry for PCNA shows a decrease in PCNA-positive cells in shTNC tumors, compared to controls. (f) Quantification of PCNA-positive cells in primary tumors demonstrates that MDA-MB-435 and shCtrl tumors have similar rate of proliferating cells, which is higher than that found for shTCN-1 and shTNC-3 clones. *P < 0.05; **P < 0.01.
To determine whether reduced TNC altered cell proliferation, which could be responsible for the decrease in tumor growth of both the primary and relapsed tumors, we conducted anchorage-dependent and -independent proliferation assays, and quantified PCNA-positive cells in the tumor samples. No differences in rates of anchorage-dependent proliferation were found between control cells and shTNC-1 or shTNC-3 clones as analysed by MTT assays of cells attached to the flask (results not shown). However, anchorage-independent cell growth was significantly reduced (P < 0.01) in cell clones with reduced expression of TNC compared to control cells (Figures 4c and d).
We also analysed the number of proliferating cells (PCNA-positive), and the total number of tumor cells, in sections of primary tumors (Figures 4e and f). The number of PCNA-positive cells was significantly reduced in shTNC-1 (P < 0.05) and shTNC-3 (P < 0.01) tumors compared to controls. The number of PCNA-positive cells was reduced by 46.7% in shTNC-3 tumors, as compared to shCtrl tumors. In addition, the total number of tumor cells per reference area was found to be reduced in shTNC-1 tumors compared to control tumors (P < 0.05) but did not reach the significance for the shTNC-3 tumors: 388 ± 33 for control MDA-MB-435, 348.2 ± 16 for shCtrl, 285 ± 14 for shTNC-1 and 313 ± 13 for shTNC-3.
To determine whether downregulation of TNC was impairing tumor angiogenesis, we quantified CD-31 levels by immunohistochemistry and image analysis, and Q-RT-PCR, in primary tumors. No decrease in CD-31 mRNA or protein levels was seen in shTNC-1 or shTNC-3 clones compared to MDA-MB-435 or shCtrl cells (results not shown). Therefore, alterations in angiogenesis do not appear to be involved in TNC-dependent reduction of tumor size.
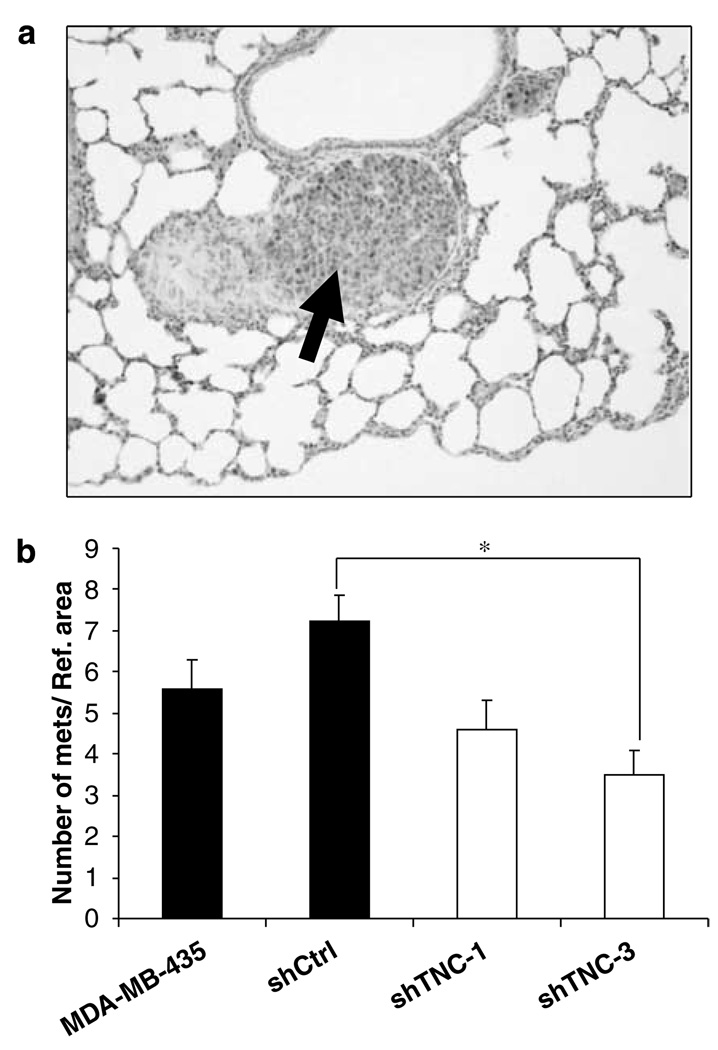
The number of lung metastatic foci was quantified in mice 2 weeks after injection of 2 × 104 cells into the tail vein. At this time, lungs only showed microscopic metastatic foci (Figure 5a). This time point was selected to avoid the difficulty in counting foci that become confluent at later times in the TNC high-expressing MDA-MB-435 cells. Mice injected with shCtrl cells had the highest number of metastatic foci (Figure 5b), correlating to their high TNC expression (Figure 3d). The number of metastatic foci was significantly reduced when comparing the shCtrl group with the shTNC-3 group (P < 0.05), and almost reached a significant difference when compared with the shTNC-1 group (P = 0.05). Comparison between combined data from both the control groups with combined data from shTNC groups showed a significant reduction (P = 0.025) in the number of metastatic foci.
Figure 5.
Downregulation of TNC in MDA-MB-435 cells reduces the number of lung metastatic foci. (a) Metastatic nodule (arrow) in the lung of a mouse injected with shCtrl cells. (b) Quantification of the number of lung metastatic foci. The average number of metastatic nudules was lower in animals injected with shTNC-3 cell clones than in mice injected with shCtrl cells (P < 0.05). No statistically significant differences were found when comparing data from shTNC-1 and shTNC-3 cells with parental cells or when comparing data from shTNC-1 with the shCtrl group (*P = 0.05 for the latter comparison).
Discussion
A major component of the complex metastatic process involves the generation of new blood vessels within the tumor through angiogenesis, where VEGF plays a key role (Ferrara, 1999). We have demonstrated that the targeted overexpression of VEGF in the context of Myc overexpression drastically alters the biologic behavior of mammary tumors leading to a high rate of lung metastases. As predicted, Myc/VEGF tumors are highly angiogenic, with increased vascularity and expression of angiogenic markers (VEGF, Flk-1, Flt-1, CD-31 and VE-cadherin).
Earlier studies from our group and others have demonstrated that VEGF may also act as an autocrine growth factor for breast tumor cells (Huh et al., 2005; Schoeffner et al., 2005). In this regard, the observation that Myc/VEGF compound transgenic tumors had an increased latency and decreased incidence, compared to tumors developed solely by targeted overexpression of Myc, was unexpected. Although the mechanism for this paradoxical effect remains unclear, the reduced tumor incidence and increased latency appear to correlate with a reduction in Myc levels in the Myc/VEGF tumors compared to Myc tumors. As both the transgenes operate off the MMTV promoter, it is possible that the transgenes compete for the same transcription factors, thus reducing the level of Myc transcription. Alternatively, it may be possible that VEGF overexpression inhibits transgene transcription or alters steady-state levels of Myc, or indeed that VEGF overexpression renders the mammary epithelial cells to be more refractory to the tumorigenic effects of Myc overexpression.
Presumably many molecular alterations in the primary tumors identified through our gene expression profiling studies occur through both direct and indirect effects of VEGF and are potentially involved in the increased metastatic phenotype observed in Myc/VEGF tumors. The molecular characterization of this Myc/VEGF model of metastases further credentials it as a relevant system in which to understand metastatic progression. Many differences in gene expression of ECM-related genes, such as collagens, proteases and adhesion proteins (fibronectin, laminins, integrins and tenascin-C) were observed between Myc/VEGF and Myc. Deregulation of ECM-related genes has also been reported as a key feature of metastatic signatures in several other microarray studies (Ramaswamy et al., 2003; Eckhardt et al., 2005; Minn et al., 2005; Gupta et al., 2007). Interestingly, it has been shown that VEGF requires interactions with ECM components to exert a proliferative effect on endothelial cells (Miralem et al., 2001).
The Myc/VEGF tumors exhibited a fivefold increase in type 1 collagen α-1 (Colla1) which was also found to be one of the 17 signature genes in a microarray meta-analysis predictive of metastasis (Ramaswamy et al., 2003). Colla1 was also identified as a gene associated with high metastatic potential in MMTV-PyMT mammary mouse tumors (Qiu et al., 2004). Laminins are a family of ECM proteins that are involved in adhesion and migration of a variety of cells (Miyazaki, 2006). Of the many laminin isoforms, laminin-5 (α3β3γ2), upregulated in the Myc/VEGF mammary tumors, is important for the acquisition of tumor cell invasive properties (Giannelli and Antonaci, 2000; Yamamoto et al., 2001).
In order to utilize the array expression data generated from the mouse metastasis model as a filter to identify genes whose deregulated expression is also altered in metastatic human BC, we compared the dataseis from this study with a lung metastasis gene signature for human BC (Minn et al., 2005). Five genes were similarly deregulated in the Myc/VEGF mouse tumor metastatic signature and in the lung metastasis signature from primary human BC: TNC, MMP-2, collagen-6-A1, mannosidase-α-1A and HLA-DPA1. Previous reports demonstrated that TNC modulates tumor and endothelial cell migration (Zagzag et al., 2002; Chiquet-Ehrismann and Chiquet, 2003), and is expressed in metastatic BC (Chiquet-Ehrismann and Chiquet, 2003). Moreover, TNC upregulates MMP-9 cooperatively with transforming growth factor-β (TGF-β), in mammary cancer cells (Kalembeyi et al., 2003). Therefore, we performed functional assays to determine the role of TNC in tumor growth and metastasis. We demonstrate that blockade of TNC strongly impairs cell migration, anchorage-independent cell proliferation and tumor growth. In addition, down-regulation of TNC causes a significant reduction in the ability of cancer cells to disseminate and grow in the lungs.
Knock down of TNC does not modify cell proliferation rates of attached cells in culture, but does decrease the clonogenic potential of cells cultured under anchorage-independent conditions in soft agar, and inhibits tumor cell proliferation in vivo. This suggests that the effects of TNC on proliferation require cellular interaction with the ECM. Our data indicate that reduced angiogenesis is not responsible for the decreased tumor size in shTNC tumors, as CD-31 levels were similar between shTNC tumors and control tumors. In addition, tumor recurrence following surgical removal is significantly more rapid in mice whose original tumors expressed high levels of TNC compared to tumors with low TNC expression. This may be due to a higher proliferative rate of remaining tumor cells following resection, or that cells expressing TNC are more invasive and less amenable to resection.
Importantly, we observed that the number of lung metastases correlates to the level of TNC expression in MDA-MB-435 and derivative cells. A significant (37.6%) decrease in the number of metastatic nodules was found in the lungs of mice injected with shTNC expressing cells compared to mice injected with the control cells. This is in contrast to a previous study in which loss of TNC expression in MMTV-PyMT mice did not decrease either the primary tumor growth or the rate of lung metastasis. It is likely that PyMT activates other metastatic pathways not dependent upon TNC (Talts et al., 1999). Indeed, our previous microarray analysis of MMTV-PyMT primary tumors failed to find TNC as one of the metastatic signature genes in this model (Qiu et al., 2004), suggesting that mechanisms of metastatic progression may depend upon earlier oncogenic events and the concerted effects of multiple genes and epigenetic phenomena. Thus, TNC may significantly contribute to metastatic progression in certain contexts. Therefore, an approach targeting several genes involved in metastasis may be necessary to reduce metastatic BC.
Materials and methods
Animals
MMTV-Myc mice, obtained from Charles Rivers Laboratory and MMTV-VEGF165 mice (Schoeffner et al., 2005) were in the FVB/N background. Mouse genotypes were determined by PCR from tail DNA using standard conditions (Jhappan et al., 1990). Animals were observed weekly to evaluate tumor onset and growth and were treated in accordance with the guidelines of the Animal Care and Use of Laboratory Animals (NIH Publication No. 86-23, 1985) under an approved animal protocol.
Real time RT-PCR, northern blot and microarray analyses
Five tumors from either Myc or Myc/VEGF transgenic animals were used to quantify mRNA levels of VEGF, Flk-1/KDR, Flt-1, PECAM/CD-31, VE-cadherin and cyclophilin (internal control) by real time RT-PCR as previously described (Shih et al., 2002). Northern blots were conducted according to previous publications (Amundadottir et al., 1995). Comparative gene expression profiles between five Myc and five Myc/VEGF primary tumors were assayed by microarray analysis using the Incyte Genomics mouse GEM1 set of cDNA clones as previously described (Calvo et al., 2002) (see Supplementary Methods).
Histology, immunohistochemistry and western blot
Tumors and lungs were processed for histology, or snap frozen for molecular assays. The number and size of macrometastases (> 100 µm2) and micrometastases (< 100 µm2) were quantified in whole sections of lungs. The following primary antibodies were used for immuno-histochemistry: Anti-Fit-1 and anti-Flk-1 (Santa Cruz, Santa Cruz, CA, USA), anti-tenascin-C (Millipore, Temecula, CA, USA), anti-CD-31 (Pharmingen, San Diego, CA, USA), anti-laminin-5 (Santa Cruz) and anti-PCNA (Dako, Capenteria, CA, USA). Western blots were performed using standard procedures (Calvo et al., 2002). Anti-tenascin-C antibody (Chemicon), anti-c-Myc (Santa Cruz) and anti-β-actin (Sigma, St Louis, MO, USA) were used at a 1:1500–1:2000 dilutions. Quantification of the MVD in CD-31-stained tumors was conducted by image analysis (see Supplementary Methods).
Cell culture, proliferation assays and in vivo assays
Cell lines from transgenic mice were isolated and characterized following published protocols (Pei et al., 2004). MDA-MB-435 cells were purchased from ATCC (Manassas, VA, USA). Cells were cultured in RPMI 1640 with 10% fetal calf serum (FCS) medium. Proliferation assays were performed using the MTT assay (Roche, Palo Alto, CA, USA) following the manufacturer's protocol. For orthotopic assays, 106 cells were used, and for lung metastasis experiments, 2 × 104 cells were injected in the tail vein (see Supplementary Methods).
Generation of shRNA constructs for TNC
A 19-nucleotide (CAGTTACAGAATTAAGTAT) targeting TNC was cloned into the pMSCV-Δ3′LTR vector to generate pRS-TNC. A control-sequence vector (pRS-Ctrl), employing the sequence GTTCAGTGGTTCGTAGGGC (not present in the human or mouse transcriptome) was constructed. Vectorcontaining MDA-MD-435 cells were selected with 2 mg/ml puromycin.
Migration and anchorage-independent clonogenic assay
Migration assays were conducted as previously described (Suyama et al., 2003). Briefly, cells were grown until confluence and then, a p20 pipette tip was used to scratch the confluent cells. Twenty-four hours later, cells were fixed, stained with crystal violet and analysed using Image Analysis software. Six wells per condition were used. Soft agar assays using standard protocols were performed to evaluate the clonogenic potential of MDA-MB-435 cells, and clones shCtrl, shTNC-1 and shTNC-3. Data are shown as number of colonies per cm2 (see Supplementary Methods).
Statistical analysis
Kruskal-Wallis test was used to determine significant differences between the treatment groups. The Mann–Whitney U-test was used to compare pairs of groups when appropriate.
Supplementary Material
Acknowledgements
We would like to thank Ms Mercedes Calvo for manuscript preparation, Victor Segura and Elizabeth Guruceaga (CEIT and Tecnun, Pamplona, Spain) for their help win bioinformatic analysis, GlaxoSmithKline for the GW654652 compound, Dr René Bernards (The Netherlands Cancer Institute) for the pMSCV-Δ31LTR vector, Dr Linda Metheny-Barlow (at the LCCC) for helpful discussions, Edward C Rosfjord and Sandra L Deming and Gloria Chepko for contributions to the initial Myc/VEGF crossbreeding study. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, with funds from NIH(to GM) under Contract No. NO1-CO-12400, a grant from the Susan G Komen Foundation (to RBD), a grant from the DOD Breast Cancer Program DAMD17-01-1-0255 (to MDJ), NIHR01 CA72460 (to RBD), NIH2R01 CA104963 (to RBD) ISCIII-RETIC (RD06/0020, to AC) and a Fulbright-MEC fellowship (AC); O G-M was supported by a Spanish MEC fellowship, and RBD by an NIHgrant 2 R01 AG14963-06.
References
- Adams J, Carder PJ, Downey S, Forbes MA, MacLennan K, Aligar V, et al. Vascular endothelial growth factor (VEGF) in breast cancer: comparison of plasma, serum, and tissue VEGF and microvessel density and effects of tamoxifen. Cancer Res. 2000;60:2898–2905. [PubMed] [Google Scholar]
- Amundadottir LT, Johnson MD, Merlino G, Smith GH, Dickson RB. Synergistic interaction of transforming growth factor alpha and c-myc in mouse mammary and salivary gland tumorigenesis. Cell Growth Differ. 1995;6:737–748. [PubMed] [Google Scholar]
- Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, et al. Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res. 2002;62:5325–5335. [PubMed] [Google Scholar]
- Chiquet-Ehrismann R, Chiquet M. Tenascins: regulation and putative functions during pathological stress. J Pathol. 2003;200:488–499. doi: 10.1002/path.1415. [DOI] [PubMed] [Google Scholar]
- De Paola F, Granato AM, Scarpi E, Monti F, Medri L, Bianchi S, et al. Vascular endothelial growth factor and prognosis in patients with node-negative breast cancer. Int J Cancer. 2002;98:228–233. doi: 10.1002/ijc.10118. [DOI] [PubMed] [Google Scholar]
- Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–1695. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt BL, Parker BS, van Laar RK, Restall CM, Natoli AL, Tavaria MD, et al. Genomic analysis of a spontaneous model of breast cancer metastasis to bone reveals a role for the extracellular matrix. Mol Cancer Res. 2005;3:1–13. [PubMed] [Google Scholar]
- Ferrara N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney Int. 1999;56:794–814. doi: 10.1046/j.1523-1755.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29(6 Suppl 16):15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- Giannelli G, Antonaci S. Biological and clinical relevance of Laminin-5 in cancer. Clin Exp Metastasis. 2000;18:439–443. doi: 10.1023/a:1011879900554. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- Huh JI, Calvo A, Stafford J, Cheung M, Kumar R, Philp D, et al. Inhibition of VEGF receptors significantly impairs mammary cancer growth in C3(1)/Tag transgenic mice through antiangiogenic and non-antiangiogenic mechanisms. Oncogene. 2005;24:790–800. doi: 10.1038/sj.onc.1208221. [DOI] [PubMed] [Google Scholar]
- Hundley JE, Koester SK, Troyer DA, Hilsenbeck SG, Barrington RE, Windle JJ. Differential regulation of cell cycle characteristics and apoptosis in MMTV-myc and MMTV-ras mouse mammary tumors. Cancer Res. 1997;57:600–603. [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH, Merlino GT. TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990;61:1137–1146. doi: 10.1016/0092-8674(90)90076-q. [DOI] [PubMed] [Google Scholar]
- Kalembeyi I, Inada H, Nishiura R, Imanaka-Yoshida K, Sakakura T, Yoshida T, et al. Tenascin-C upregulates matrix metalloproteinase-9 in breast cancer cells: direct and synergistic effects with transforming growth factor betal. Int J Cancer. 2003;105:53–60. doi: 10.1002/ijc.11037. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralem T, Steinberg R, Price D, Avraham H. VEGF(165) requires extracellular matrix components to induce mitogenic effects and migratory response in breast cancer cells. Oncogene. 2001;20:5511–5524. doi: 10.1038/sj.onc.1204753. [DOI] [PubMed] [Google Scholar]
- Miyazaki K. Unique biological activity and role in tumor growth and invasion. Cancer Sci. 2006;97:91–98. doi: 10.1111/j.1349-7006.2006.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel A, Maillard C, Rocks N, Jost M, Chabottaux V, Sounni NE, et al. Membrane associated proteases and their inhibitors in tumour angiogenesis. J Clin Pathol. 2004;57:577–584. doi: 10.1136/jcp.2003.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei XF, Noble MS, Davoli MA, Rosfjord E, Tilli MT, Furth PA, et al. Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In vitro Cell Dev Biol Anim. 2004;40:14–21. doi: 10.1290/1543-706X(2004)40<14:ECOPMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Pyke C, Romer J, Kallunki P, Lund LR, Ralfkiaer E, Dano K, et al. The gamma 2 chain of kalinin/laminin 5 is preferentially expressed in invading malignant cells in human cancers. Am J Pathol. 1994;145:782–791. [PMC free article] [PubMed] [Google Scholar]
- Qiu TH, Chandramouli GV, Hunter KW, Alkharouf NW, Green JE, Liu ET. Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res. 2004;64:5973–5981. doi: 10.1158/0008-5472.CAN-04-0242. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Schmitz AA, Govek EE, Bottner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- Schoeffner DJ, Matheny SL, Akahane T, Factor V, Berry A, Merlino G, et al. VEGF contributes to mammary tumor growth in transgenic mice through paracrine and autocrine mechanisms. Lab Invest. 2005;85:608–623. doi: 10.1038/labinvest.3700258. [DOI] [PubMed] [Google Scholar]
- Shih SC, Robinson GS, Perruzzi CA, Calvo A, Desai K, Green JE, et al. Molecular profiling of angiogenesis markers. Am J Pathol. 2002;161:35–41. doi: 10.1016/S0002-9440(10)64154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- Suyama E, Kawasaki H, Nakajima M, Taira K. Identification of genes involved in cell invasion by using a library of randomized hybrid ribozymes. Proc Natl Acad Sci USA. 2003;100:5616–5621. doi: 10.1073/pnas.1035850100. Epub 28 April 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talts JF, Wirl G, Dictor M, Muller WJ, Fassler R. Tenascin-C modulates tumor stroma and monocyte/macrophage recruitment but not tumor growth or metastasis in a mouse strain with spontaneous mammary cancer. J Cell Sci. 1999;112(Pt 12):1855–1864. doi: 10.1242/jcs.112.12.1855. [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Inada H, Kalembeyi I, Imanaka-Yoshida K, Sakakibara M, Okada R, et al. Involvement of large tenascin-C splice variants in breast cancer progression. Am J Pathol. 2003;162:1857–1867. doi: 10.1016/S0002-9440(10)64320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Itoh F, Iku S, Hosokawa M, Imai K. Expression of the gamma(2) chain of laminin-5 at the invasive front is associated with recurrence and poor prognosis in human esophageal squamous cell carcinoma. Clin Cancer Res. 2001;7:896–900. [PubMed] [Google Scholar]
- Zagzag D, Capo V. Angiogenesis in the central nervous system: a role for vascular endothelial growth factor/vascular permeability factor and tenascin-C. Common molecular effectors in cerebral neoplastic and non-neoplastic ‘angiogenic diseases’. Histol Histopathol. 2002;17:301–321. doi: 10.14670/HH-17.301. [DOI] [PubMed] [Google Scholar]
- Zagzag D, Shiff B, Jallo GI, Greco MA, Blanco C, Cohen H, et al. Tenascin-C promotes microvascular cell migration and phosphorylation of focal adhesion kinase. Cancer Res. 2002;62:2660–2668. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.