Abstract
The highly variable clinical phenotype observed in patients homozygous for the C282Y mutation of the hereditary hemochromatosis gene (HFE) is likely due to the influence of non-HFE modifier genes. The primary functional abnormality causing iron overload in hemochromatosis is hyper-absorption of dietary iron. We found that iron absorption in inbred mice varies in a strain-specific manner, as does the pattern of iron distribution to the liver and spleen. A/J mice absorbed approximately twice the amount of 59Fe delivered by gavage compared to the C57BL/6 strain. Genetic comparisons between A/J and C57BL/6 were facilitated by the availability of consomic chromosome substitution strains (CSS). Each CSS has an individual chromosome pair from A/J on an otherwise C57BL/6J background. We found that iron absorption and iron content in liver and in spleen were continuous variables suggesting that each trait is under multigenic control. No trait co-segregated among the CSS. Chromosome 5 from A/J, however, imparted the highest iron absorption phenotype and multiple CSS had absorption levels equivalent to A/J. Chromosomes 9 and X were associated with high spleen iron content. These data suggest that multiple genes contribute to the regulation of iron absorption and that individual organ iron phenotypes are independently regulated.
Keywords: Iron, Phenotype, Mapping
Introduction
Most patients with hereditary hemochromatosis are homozygous for a single mutation in the hemochromatosis gene (HFE) that results in the conversion of a cysteine to a tyrosine residue at position 282 in the HFE protein (C282Y). Homozygosity for the C282Y mutation alone, however, does not result in a constant clinical phenotype. Penetrance of the clinical phenotype varies widely between homozygotes and this variation has been attributed to the existence of non-HFE modifier genes [1, 2]. We found a difference in the incidence of disease-related morbidity in C282Y homozygous siblings when pedigrees with clinically affected probands were compared to pedigrees in which healthy probands were detected through screening programs. Homozygous siblings of clinically affected probands were approximately three times more likely to have signs or symptoms of disease-related morbidity than homozygous siblings of healthy probands [1]. This finding supports the concept that other genes modify penetrance of the C282Y mutation. Identification of such modifier genes could facilitate risk assessment and provide potential gene targets for the treatment of iron overload diseases.
Analysis of different inbred mouse strains demonstrates the influence of genetic background on iron metabolism. For example, parameters such as percent transferrin saturation, hepatic iron content and the rate at which hepatic iron accumulates with increased dietary iron vary widely between strains and do not co-segregate [3]. Crossbreeding experiments suggest that hepatic iron content is regulated by multiple loci [4-6] and that iron accumulation by the liver may be controlled differently than iron accumulation in other tissues [3, 6]. Importantly, the genetic background influences the degree of liver iron loading associated with mutations in the Hfe gene [7, 8].
Because of the genetic and biological complexities associated with organ iron content, we focused on the primary phenotype of gastrointestinal iron absorption. We found that A/J mice absorbed approximately twice as much iron as C57BL/6 mice. We utilized a set of mouse Chromosome Substitution Strains (CSS) [9] to identify A/J chromosomes that modify iron absorption in the C57BL/6 background strain. In comparison to the host low-absorption C57BL/6 background strain, we found that chromosome 5 from donor strain A/J (high-absorption strain) imparted the highest absorption value although several other chromosome substitutions also resulted in high absorption phenotypes. Iron absorption did not segregate with liver or spleen iron content. Our data suggest that regulation of each of these parameters is multigenic in nature.
Materials and Methods
Animals
Wild-type C57BL/6, A/J, DBA/2, and (C57BL/6 × A/J) F1 mice were obtained from Jackson Laboratories (Bar Harbor, ME). Wild type 129/SvEv mice were obtained from Dr. Nancy Andrews (Children's Hospital, Boston, MA). Chromosome substitution strains (CSS) were originally obtained as a gift from Dr. Joseph Nadeau (Department of Genetics, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106). Each CSS is a homozygous inbred strain that is C57BL/6J with one chromosome pair from A/J. There are 21 CSSs in this complete set covering the 19 autosomes and two sex chromosomes [9]. Each CSS is designated by the A/J chromosome present on the C57BL/6 background (e.g. B6.A1, B6.A2, etc.). Mice were subsequently maintained as inbred strains at the University of Washington and test animals transferred to the University of Utah for analysis. All mice were shipped from the University of Washington at 6 wks of age and maintained in the isolation facility at the University of Utah for exactly two weeks before analysis. Mice were maintained on a standard rodent diet containing approximately 350 mg iron per kg (Harlan Teklad TD 8640, Madison, WI) and maintained in a temperature controlled (25°C) facility with a strict 12 h light/dark cycle and given free access to food and water. The Animal Care and Use Committees of the University of Washington and University of Utah approved this project. CSS are now commercially available through The Jackson Laboratories (www.jax.org).
Iron absorption
Male mice, 8-9 wks of age, were fasted 3 h prior to gavage but allowed water ad libitum. Both A/J and C57BL/6 mice consumed approximately 4 g of food per day, an amount consistent with published values [10]. Mice ingested approximately 1.4 mg iron per day as the Harlan Teklad 8640 diet contains approximately 350 mg iron per kg. Approximately 2.5 μCi (9.25 ×104 Bq) 59Fe-HCl was diluted into 0.2 mL of a solution of 0.5 M ascorbic acid, 0.15 M NaCl, and 5 ug of iron as FeSO4. Non-radioactive iron was added as a carrier to ensure that substrate did not become limiting during the assay.
Each dose was counted prior to gavage to ensure that equivalent amounts were administered. The iron mixture was administered to each animal with an olive-tipped gavage needle. Mice were placed in a metabolic cage, supplied with water, fasted for 3 h more, and then allowed food overnight. Food consumption did not vary between mice. Approximately 24 h after gavage, mice were euthanized by anesthesia overdose and dissected. The gastrointestinal tract, head, and lungs were removed and radioactivity in blood (adjusted for volume), major organs, and carcass was determined in a gamma counter (Perkin Elmer Instruments, Shelton, CT). Iron absorption was calculated as: (59Fe in blood, organs, and carcass/59Fe administered by gavage × 100). The gastrointestinal tract was not included in the measure of absorption to avoid counting non-absorbed radioactivity due to incomplete flushing of luminal contents.
Iron measurements
Organ iron content was determined by acid digestion followed by iron quantification with atomic absorption spectroscopy [11]. Transferrin saturation was determined using a colorimetric assay (Wako Chemicals, USA).
Statistical Analysis
Data are expressed as mean ± SEM. Student's unpaired t-test followed by Bonferroni corrections for multiple samples was used for comparisons between groups. Spearman's rank correlation test was used for some analyses.
Results
Strain-specific differences in iron parameters
Various measures of iron metabolism including hepatic iron content and transferrin (Tf) saturation differ between inbred mouse strains [3]. We found that iron absorption is also strain-dependent. Four strains were compared for their ability to absorb the measured dose of 59Fe. Strains 129/SvEv, A/J and DBA/2 absorbed approximately 11%, 17% and 15% of the administered 59Fe, respectively, while C57BL/6 absorbed only 6% (Figure 1a). Hepatic iron content and percent Tf saturation were also compared between these strains. The highest hepatic iron content was found in 129/SvEv (211 ug/g wet wt), A/J had an intermediate value of 91 ug/g, and C57BL/6 and DBA/2 were lowest with 70 and 66 ug/g, respectively (Figure 1b).
Figure 1. Iron measures in different strains of mice.
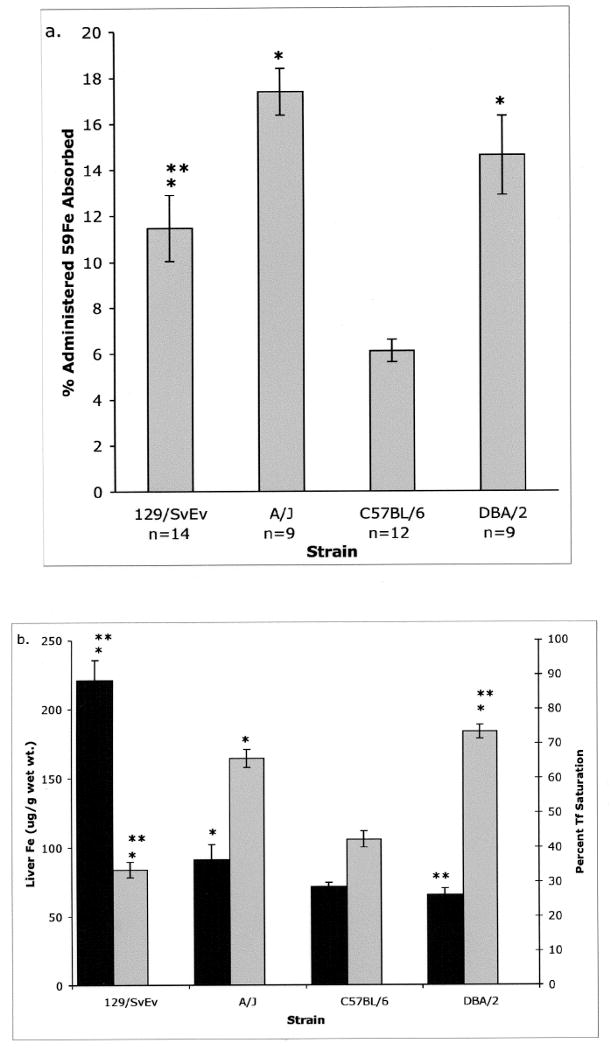
a. Mice were fasted and given a measured dose of 59Fe by gavage. Twenty-four h later, mice were sacrificed, dissected and the percent of the administered radiolabel remaining with the carcass minus the head and GI tract was measured. b. The hepatic iron content (black bars) and the percent Tf saturation (grey bars) were determined for the four strains. N values are the same for animals in Figures a and b. Error bars represent SEM.
* P< 0.05 vs. C57BL/6. ** P< 0.05 vs. A/J.
A Spearman's rank correlation test was performed comparing iron absorption, liver iron and percent Tf saturation to determine if any of the iron parameters were correlated. There was a significant negative correlation between hepatic iron content and Tf saturation (P < 0.003, Correlation Coefficient = -0.391). Correlations were not seen between iron absorption and the other two traits suggesting that iron absorption is regulated independently from hepatic iron content and Tf saturation.
Differences in iron distribution between A/J and C57BL/6
We focused on A/J and C57BL/6 strains because they demonstrated the greatest difference in iron absorption. There was also a significant difference in hepatic iron content between these two strains (Figure 1b). When the distribution of absorbed 59Fe was examined, a greater percentage of administered radiolabel was retained in the duodenum of C57BL/6 mice (Table 1). Further analysis revealed that A/J mice distributed 17.45% of total absorbed 59Fe to the liver as compared to 12.23% in C57BL/6 mice. A/J mice also distributed more iron to the spleen. In contrast, C57BL/6 mice had a greater percentage of counts remaining with the carcass (bones, muscle, skin and fur).
Table 1. Percent of absorbed 59Fe distributed to blood, organs and carcass.
| A/J* (n=19) | C57BL/6J* (n=57) | ||
|---|---|---|---|
| Organ | Mean (±SEM) | Mean (±SEM) | p A/J vs. C57BL/6J |
| Blood | 7.32 ± 0.95 | 5.0 ± 1.1 | 0.35 |
| Liver | 17.45 ± 0.87 | 12.23 ± 0.3 | < 0.0009 |
| Spleen | 3.13 ± 0.33 | 1.65 ± 0.08 | < 0.0009 |
| Heart | 1.04 ± 0.11 | 1.13 ± 0.07 | 0.48 |
| Kidney | 2.8 ± 0.14 | 2.05 ± 0.06 | < 0.0009 |
| Duodenum | 19.7 ± 1.39 | 26.89 ± 1.06 | < 0.001 |
| Carcass | 46.7 ±1.13 | 52.5 ±0.79 | <0.0009 |
All mice were 8-9 week old males.
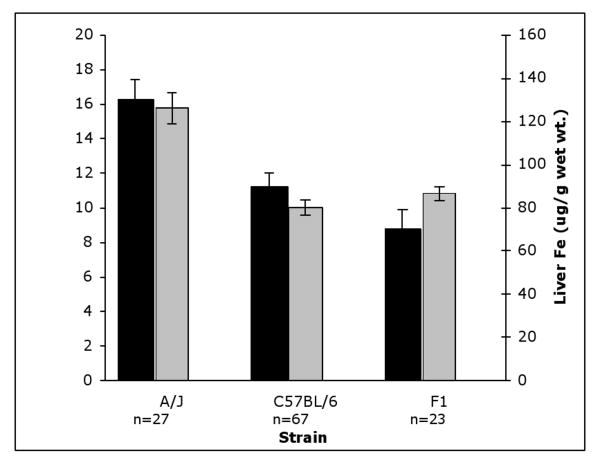
Analysis of (C57BL/6J × A/J) F1 mice
Iron absorption and liver iron content for (C57BL/6 × A/J) F1 mice were compared to A/J and C57BL/6. Both absorption of 59Fe and hepatic iron content were similar to the wild type parent C57BL/6 strain, indicating that the low iron phenotype is a dominant trait (Figure 2).
Figure 2. Comparison of iron parameters in F1 mice.
Black bars: Absorption of 59Fe. Absorption was measured in A/J, C57BL/6 and F1 mice. Gray bars: Hepatic iron content in A/J, C57BL/6 and F1 mice. All mice were 8-9 week old males. Error bars represent SEM.
Chromosome Substitution Strains
We utilized chromosome substitution strains (CSS) to map genes responsible for strain-specific variations in the iron phenotype to specific chromosomes (see Materials and Methods). The CSS permit a determination of the influence of alleles on a single A/J chromosome in an otherwise C57BL/6J genetic background. Six to 17 mice from each substitution strain were compared for iron absorption and iron content of liver and spleen. A total of 330 CSS mice were tested along with 89 C57BL/6 and 50 A/J mice as controls. An average of 10 mice were tested per day. Absorption values for CSS mice were obtained from at least two different experiments.
Iron absorption varied two-fold among the CSS with extreme values significantly outside parental values. Strain B6.A5 exhibited the highest iron absorption which was 50% greater than seen for A/J, and 2-fold greater than for C57BL/6. B6.A18 mice had the lowest absorption level at 30% of the absorption exhibited by C57BL/6. Strains A/J, B6.A5, B6.A6, B6.A15, B6.A17, B6.A19 and B6.AY had absorption values statistically greater than C57BL/6J (Figure 3a). These data suggest that multiple chromosomes harbor loci contributing to the modulation of iron absorption. These contributions could represent negative effects of C57BL/6 alleles, positive effects of A/J alleles or both. A further indication that iron absorption is genetically complex is the continuous distribution of iron absorption values among this collection of strains.
Figure 3. Iron measures in CSS mice.
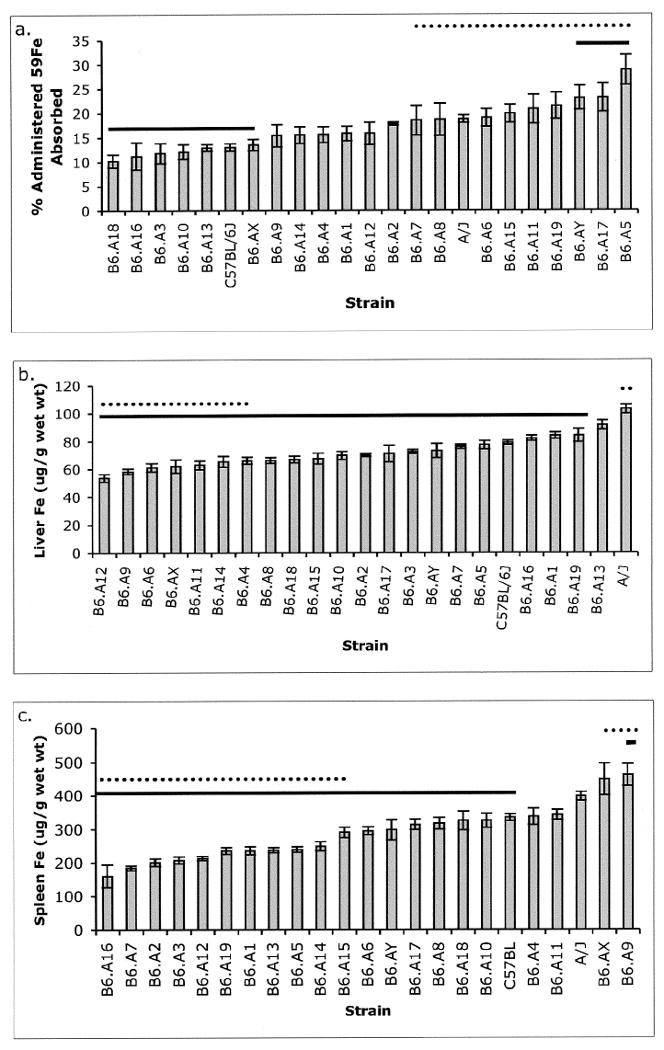
a. 59Fe absorption was measured in CSS and ordered by mean percent of administered radiolabel absorbed. b. Liver iron concentration was measured and ordered by increasing value of the mean. c. Spleen iron was measured and ordered by increasing values of the mean. Student's T-test was applied to compare C57BL/6J to all other strains. Error bars represent SEM.
* Mean values statistically different from C57BL/6 (dotted line) and A/J (solid line) when adjusted for the number of groups (p < 0.0024).
A continuous distribution in tissue iron content was also seen for hepatic and spleen iron levels. Hepatic iron content varied by approximately 2-fold, with the highest level exhibited by parental strain A/J (Figure 3b). Spleen iron values also varied by approximately 2-fold but only A/J and B6.A9 mice had spleen iron values statistically greater than C57BL/6 (Figure 3c). Multiple CSS exhibited absorption or organ iron values significantly lower than the parent strains suggesting that the low iron phenotype in these mice is likely due to the affect of multiple alleles on different chromosomes.
Values for 59Fe absorption, hepatic iron content, spleen iron content and Tf saturation were analyzed for all animals studied (A/J, C57BL/6 and the 21 CSS; n=469). A Spearman's rank analysis indicated a strong positive correlation between iron content in liver and spleen, 59Fe absorption and % Tf saturation, and a weak correlation between liver iron content and 59Fe absorption (Table 2). The distribution between macrophage and parenchymal cell iron was not determined. When individual strains were compared correlations could be detected between various iron parameters in the parental A/J and C57BL/6 strains and in 13 of the CSS. No parameter, however, was consistently correlated with iron absorption (not shown). The distribution of absorbed 59Fe was also measured for each strain. Iron absorption in the 21 CSS did not correlate with the distribution of absorbed radio-iron for any organ. Values for iron parameters in A/J and C57BL/6 mice bred at the University of Washington and shipped to Utah did not differ significantly from mice bred at the University of Utah and have been included in the data set.
Table 2. Correlation analysis of all strains examined.
| Phenotype 1 | Phenotype 2 | Correlation Coeffcient | p value |
|---|---|---|---|
| Liver Fe | Spleen Fe | 0.230** | 1.46 E-05 |
| Liver Fe | 59Fe Absorption | 0.146* | 0.0103 |
| Liver Fe | % Tf Saturation | - 0.049 | 0.5735 |
| Spleen Fe | 59Fe Absorption | - 0.042 | 0.4586 |
| Spleen Fe | % Tf Saturation | 0.060 | 0.4197 |
| 59Fe Absorption | % Tf Saturation | 0.261** | 0.0046 |
Correlation is significant at the 0.01 level (2-tailed).
Correlation is significant at the 0.05 level (2-tailed).
Candidate Modifier Genes
Multiple CSS exhibited values for iron absorption and/or organ iron content that varied significantly from C57BL/6. These CSS were divided according to iron phenotypes (high or low) relative to C57BL/6 (Table 3), and known iron-related genes mapping to each chromosome identified. Several genes including transferrin receptor 2, erythropoietin (chr 5) and the tetratricopeptide repeat domain 7 (mutated in the hereditary erythroblastic anemia, chr 17), were associated with high iron absorption. Neogenin, a protein that was found to interact with hemojuvelin [12], is located on chromosome 9 which was found to have the highest spleen iron content.
Table 3. A/J derived chromosomes controlling the phenotypes of iron absorption and tissue iron levels that differed significantly from the C57BL/6 parent strain. Each response is designated as ‘low’ or ‘high’ to reflect the response as compared with C57BL/6. Suggested candidate genes known to reside on each chromosome are also listed.
| Phenotype: | Absorption | Liver Iron | Spleen Iron | Candidate Genes** |
|---|---|---|---|---|
| Chromosome | ||||
| 1 | low | Fpn, Bmpr2, Steap3 | ||
| 2 | low | Cybrd1, Bmp2 | ||
| 3 | low | Hjv, Bmpr1b | ||
| 4 | low | Irp1 | ||
| 5 | high | low | Trfr2, Epo, Ppargc1a | |
| 6 | high | low | Fancd2 | |
| 7 | low | Hamp1 | ||
| 9 | low | high | Neo1 | |
| 11 | (high)* | low | Leap2, Hga, Atox1 | |
| 12 | low | low | Fancm | |
| 13 | low | Hfe | ||
| 14 | low | low | Bmp4, Bmpr1a | |
| 15 | high | low | Dmt1 | |
| 16 | low | Trfr1 | ||
| 17 | high | Ttc7, Sod2 | ||
| 18 | low | Smad4 | ||
| 19 | high | low | ||
| X | low | (high)* | Heph | |
| Y | high |
(high), did not reach statistical significance from C57BL/6 but was bracketed by other significant chromosomal substitution strains.
Gene names are as: Fpn, Ferroportin; Bmpr, Bone morphogenetic protein receptor; Cybrd1, Cytochrome b reductase 1; Steap3, Six-transmembrane epithelial antigen of the prostate 3; Bmp, Bone morphogenetic protein; Hjv, Hemojuvelin; Irp1, Iron responsive element binding protein; Trfr2, transferrin receptor 2; Epo, Erythropoietin; Ppargc1a, Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha; Fanconi anemia, complementation group D2; Hamp1, Hepcidin; Neo1, Neogenin; Leap2, Liver-expressed antimicrobial peptide 2, Hga, ; Atox1, antioxidant protein 1 homolog 1; Hfe, hemochromatosis; Dmt1, Divalent metal transporter 1; Trfr1, Transferrin receptor 1; Ttc7, tetratricopeptide repeat domain 7; Sod2, Superoxide dismutase 2; Smad4, MAD homolog; Heph, Hephaestin
Discussion
Iron homeostasis in mammals is tightly regulated at the level of absorption by the duodenal enterocyte. Excessive tissue iron accumulation, characteristic of hemochromatosis, is the result of hyperabsorption of dietary iron and failure to adequately down regulate absorption in the face of elevated iron stores, as seen in humans and mouse models with Hfe defects [13]. No naturally occurring mutations have been identified in the mouse Hfe gene located on chromosome 13. The CSS with the highest hepatic iron content was B6.A13, but the value was not statistically different from C57BL/6. Quantitative trait locus (QTL) analyses seeking loci involved in the regulation of hepatic iron content have not identified Hfe as a candidate locus [4, 14]. Further evidence for non-Hfe modifier genes is also evident in human pedigrees where homozygotes for the HFE C282Y mutation may exhibit vastly different clinical phenotypes compared to their homozygous siblings [1]. In this study, we show that iron absorption is a polygenic trait and we have identified specific chromosomes controlling the extent of iron absorption and tissue iron levels.
Strain-specific differences in iron parameters have been recognized in a number of studies. Specific measures for organ iron content and Tf saturations can vary significantly between studies and even for the same mouse strain [15]. The relative differences between iron measures, however, remain consistent between strains. We first observed that the low iron phenotypes seen for C57BL/6 were dominant in (C57BL6 × A/J) F1 mice indicating that homozygosity for A/J alleles is required for the high iron phenotypes. We selected the “low” iron strain C57BL/6 and the “high” iron strain A/J for these studies due to the availability of the CSS panel. The CSS panel enabled us to evaluate the effects of homozygosity for alleles contained on a single A/J chromosome in an otherwise C57BL/6 genetic background. An alternate approach to localize modifier genes would be to utilize QTL analysis. QTL analysis has the advantage of tracking a phenotype with no bias with respect to gene function through a mouse “family” but extensive genotyping of each animal is required. The CSS approach, however, permits assignment of a locus of interest to a specific chromosome without the need for further genotyping and CSS provide a renewable resource of genetically characterized mice in which numerous measures of iron metabolism can be obtained.
A major finding in our analysis of CSS strains is that the B6.A5 strain showed the greatest absorption of 59Fe. Thus, one or more genes on chromosome 5 are permissive for high iron absorption. Because iron absorption of the A/J strain was markedly lower than that seen for B6.A5, additional unlinked genes (not on chromosome 5) contribute opposing effects which act to reduce overall iron absorption in the A/J strain. In contrast, C57BL/6 alleles of these unlinked genes either do not inhibit, or inhibit to a lesser degree, the action directed by the chromosome 5 allele(s). Thus, iron absorption is a polygenic trait characterized by epistasis acting between a major gene on chromosome 5 and other modifier genes on additional chromosomes. We are performing additional genetic studies to identify the chromosome 5 gene(s).
Strains B6.A6, B6.A15, B6.A19, B6.A17 and B6.AY also had absorption values significantly greater than C57BL/6J, but were comparable to A/J. These data are interesting for two reasons. First, they support the observation that iron absorption is polygenic in nature since the high iron absorption phenotype was seen for multiple chromosome substitutions. Second, it is sufficient for C57BL/6 mice to carry any one A/J allele (chromosome) to sustain the high iron absorption phenotype. Thus, crosses between each of these CSS strains and C57BL/6 can be utilized to eventually identify high absorption alleles on each of these chromosomes. This also suggests that in the A/J mouse, the combination of these high iron alleles must not be additive in nature and/or be balanced by additional alleles which maintain the A/J iron absorption phenotype.
In addition to iron absorption, tissue iron stores are also polygenic traits as evidenced by the patterns of continuous variation among the CSS for liver and spleen iron levels. Because the A/J strain had the highest liver iron concentration, homozygosity for multiple A/J alleles is required to achieve the extreme of high liver iron content. This is also likely true for genetic determinants of spleen iron for which A/J shows high levels, although distinct gene(s) on chromosome 9 and possibly chromosome X yield singular high spleen iron contents on the C57BL/6 background. Two independent QTL approaches also identified a locus on chromosome 9 associated with high splenic iron content and also found that spleen and liver iron content were controlled independently [6, 16]. Independent genetic control of tissue iron is also shown by our data as B6.A9 had high spleen iron but low liver iron content (Figure 3). Thus, further genetic studies of chromosome 9 are warranted in order to reveal genetic determinants of tissue iron levels.
Our data demonstrate that high iron absorption does not predict increased iron in the liver or spleen and that these three traits are associated with genes located on different chromosomes. For example, the high iron absorption strain B6.A5 showed liver and spleen iron contents that were similar to C57BL/6. Taken together with the data in Figure 1 and previous studies among mouse strains [3, 14], each of the primary measures of iron metabolism (iron absorption, liver iron, spleen iron, percent transferrin saturation) are modulated by unique sets of genes. Identification of these gene sets is critical in order to eventually unravel iron homeostasis relationships in clinical settings.
The total amount of iron delivered by gavage (5 ug) was only 0.36% of the daily consumption by a mouse (see Methods), making it unlikely our absorptive assay created a non-physiological challenge. The lack of correlation between absorption and organ iron content in the CSS was somewhat unexpected and suggests that absorption is not regulated in response to organ iron content under normal dietary and health circumstances. The absence of a correlation between iron absorption and organ iron content was also observed in a study of genetic variation in basal iron status. DBA/2 mice showed a nearly two-fold increase of mucosal iron transfer compared to the SWR strain, yet SWR mice had approximately two-fold greater hepatic iron content and ferritin values compared to DBA/2 [15].
The availability of CSS provided us with the opportunity to assign iron modifier loci to single chromosomes through a series of relatively simple phenotyping procedures. Moving from a chromosomal assignment to a more restricted region can also be accomplished with simple phenotyping procedures. Recombinant congenic strains have been derived from A/J and C57BL/6 through a systematic inbreeding scheme [17]. These strains are now commercially available (Emerillon Therapeutics, Montreal, Canada) and have been used to map genes of interest to chromosomal regions as small as 3 cM. This greatly simplifies gene localization by restricting the region in which fine mapping is required. We are now pursuing this and other approaches to isolate the high absorption locus on chromosome 5.
Acknowledgments
CSS strains were a kind gift from Dr. Joseph Nadeau (Department of Genetics, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106). This work was supported in part by NIH grants RO1 DK062106 and P30 DK072437 (RSA and JPK) and NIH grant R01 DK063159 (RCL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bulaj ZJ, Ajioka RS, Phillips JD, et al. Disease-related conditions in relatives of patients with hemochromatosis. N Engl J Med. 2000;343(21):1529–35. doi: 10.1056/NEJM200011233432104. [DOI] [PubMed] [Google Scholar]
- 2.Le Gac G, Ferec C. The molecular genetics of haemochromatosis. Eur J Hum Genet. 2005;13(11):1172–85. doi: 10.1038/sj.ejhg.5201490. [DOI] [PubMed] [Google Scholar]
- 3.LeBoeuf RC, Tolson D, Heinecke JW. Dissociation between tissue iron concentrations and transferrin stauration among inbred mouse strains. J Lab Clin Med. 1995;126:128–36. [PubMed] [Google Scholar]
- 4.Bensaid M, Fruchon S, Mazeres C, et al. Multigenic control of hepatic iron loading in a murine model of hemochromatosis. Gastroenterology. 2004;126(5):1400–8. doi: 10.1053/j.gastro.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Sproule TJ, Jazwinska EC, Britton RS, et al. Naturally variant autosomal and sex-linked loci determine the severity of iron overload in beta 2-microglobulin-deficient mice. Proc Natl Acad Sci U S A. 2001;98(9):5170–4. doi: 10.1073/pnas.091088998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant GR, Robinson SW, Edwards RE, et al. Multiple polymorphic loci determine basal hepatic and splenic iron status in mice. Hepatology. 2006;44(1):174–85. doi: 10.1002/hep.21233. [DOI] [PubMed] [Google Scholar]
- 7.Dupic F, Fruchon S, Bensaid M, et al. Inactivation of the hemochromatosis gene differentially regulates duodenal expression of iron-related mRNAs between mouse strains. Gastroenterology. 2002;122(3):745–51. doi: 10.1053/gast.2002.31877. [DOI] [PubMed] [Google Scholar]
- 8.Fleming RE, Holden CC, Tomatsu S, et al. Mouse strain differences determine severity of iron accumulation in Hfe knockout model of hereditary hemochromatosis. Proc Natl Acad Sci U S A. 2001;98(5):2707–11. doi: 10.1073/pnas.051630898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadeau JH, Singer JB, Matin A, et al. Analysing complex genetic traits with chromosome substitution strains. Nat Genet. 2000;24(3):221–5. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- 10.Bachmanov AA, Reed DR, Beauchamp GK, et al. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet. 2002;32(6):435–43. doi: 10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards CQ, Carroll M, Bray P, et al. Hereditary hemochromatosis. Diagnosis in siblings and children. N Engl J Med. 1977;297(1):7–13. doi: 10.1056/NEJM197707072970102. [DOI] [PubMed] [Google Scholar]
- 12.Zhang AS, West AP, Jr, Wyman AE, et al. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J Biol Chem. 2005;280(40):33885–94. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]
- 13.Ajioka R, Levy J, Andrews N, et al. Regulation of iron absorption in Hfe mutant mice. Blood. 2002;100(4) doi: 10.1182/blood-2001-11-0037. [DOI] [PubMed] [Google Scholar]
- 14.Robinson SW, Clothier B, Akhtar RA, et al. Non-ahr gene susceptibility Loci for porphyria and liver injury induced by the interaction of ‘dioxin’ with iron overload in mice. Mol Pharmacol. 2002;61(3):674–81. doi: 10.1124/mol.61.3.674. [DOI] [PubMed] [Google Scholar]
- 15.Clothier B, Robinson S, Akhtar RA, et al. Genetic variation of basal iron status, ferritin and iron regulatory protein in mice: potential for modulation of oxidative stress. Biochem Pharmacol. 2000;59(2):115–22. doi: 10.1016/s0006-2952(99)00306-8. [DOI] [PubMed] [Google Scholar]
- 16.Custodio AO. Medical Sciences/Genetics. Harvard University; Cambridge, MA: 2005. Analysis of Genetic Modifiers of Murine Iron Homeostasis; p. 136. [Google Scholar]
- 17.Fortin A, Diez E, Rochefort D, et al. Recombinant congenic strains derived from A/J and C57BL/6J: a tool for genetic dissection of complex traits. Genomics. 2001;74(1):21–35. doi: 10.1006/geno.2001.6528. [DOI] [PubMed] [Google Scholar]