Abstract
The biochemical sequelae to chloroethyl mustard exposure correspond very well to toxic processes initiated by free radicals. Additionally, mustard solutions contain spontaneously formed cyclic onium ions which produce carbon free radicals when reduced electrochemically. Therefore, we hypothesized that the onium ions of sulfur or nitrogen mustards might produce carbon free radicals upon being reduced enzymatically, and that these radicals might constitute a metabolic activation. We set out to document radical production using an in vitro metabolic system and electron paramagnetic resonance (EPR). Our system consisted of NADPH, one of several pyridine nucleotide-driven flavoprotein reductases, cytochrome c as a terminal electron acceptor, various sulfur or nitrogen mustards and the spin trap α-[4-pyridyl-1-oxide]-N-tert-butylnitrone in buffer. Reactions were started by adding the reductase to the other materials, vortexing and immediately transferring the mixture to a 10 mm EPR flat cell. Repeated scans on a Bruker ESP 300E EPR spectrometer produced a triplet of doublets with hyperfine splitting constants of aN = 15.483 G and aH = 2.512 G. The outcome supported our hypothesis that carbon-centered free radicals are produced when mustard-related onium ions are enzymatically reduced. The EPR results varied little with the chloroethyl compound used or with porcine or human cytochrome P450 reductase, the reductase domain of rat brain neuronal nitric oxide synthase or rat liver thioredoxin reductase. Our results offer new insight into the basis for mustard-induced vesication and the outcome of exposure to different mustards. The free radical model provides an explanation for similarities in the lesions arising from mustard exposure and energy-based lesions such as those from heat, ultraviolet and nuclear radiation as well as damage across tissue types such as skin, eyes or airway epithelium.
Keywords: Chemical warfare, Flavoenzyme, Chloroethyl mustards, Onium ion, Enzymatic reduction, Electron transport, Free radical, EPR
Introduction
Sulfur mustard (2, 2′-chloroethyl sulfide [CAS 505-60-2], mustard gas, NATO Standard Agreement designation; HD) and related chloroethyl compounds (Fig. 1) are alkylating agents and potent vesicants. It was last employed in combat in the conflict between Iraq and Iran in the 1980s. Erythema, blistering, and necrosis follow dermal contact. Conjunctivitis and/or corneal opacity result from exposure of the eyes. Inhalation can lead to pneumonia, chronic bronchitis and asthma, while systemic symptoms have radiomimetic characteristics. Injury is dose dependent and initially painless (Papirmeister et al., 1991; Somani, 1992). It is interesting that the dermal reaction to sulfur mustard exposure in humans is different from that in most other species; no animal blisters like man (Marshall et al., 1919). Recent in vitro work (Sabourin et al., 2000; Smith et al., 2001) has revealed the initiation of proinflammatory cytokine production at the cellular level starting several hours after exposure. However, the initial biochemical lesion, the event that ultimately sets off cytokine production and characteristic symptoms, remains unidentified.
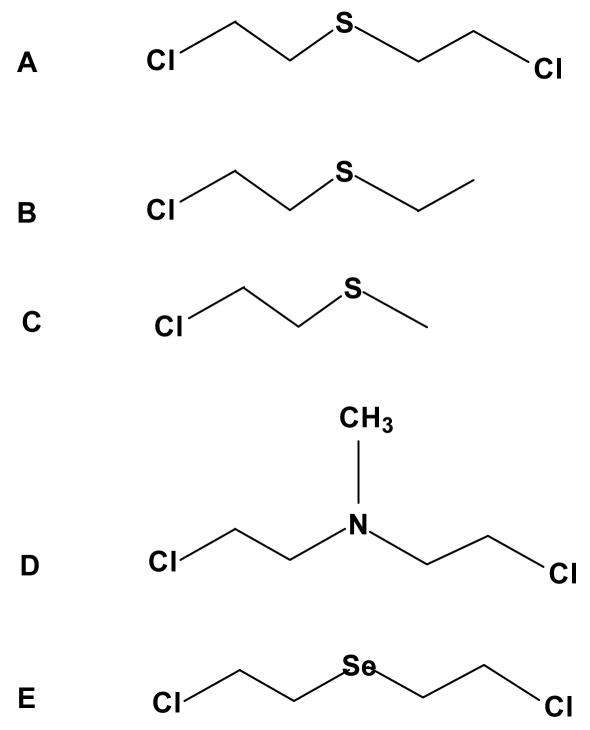
Figure 1. Compounds spontaneously forming “onium” ions.
A. Sulfur mustard, bis-(chloroethyl) sulfide (HD), B. Chloroethyl ethylsulfide (CEES), C. Chloroethyl methylsulfide (CEMS), D Mechlorethamine (HN2), a nitrogen mustard, E. bis-(chloroethyl) selenide.
Several lines of inquiry have indicated that this initial lesion might involve free radicals. For example, reports in the literature show a strong parallel between the cellular and biochemical effects of sulfur mustard exposure and the effects of free radical damage (Papirmeister et al., 1991; Somani, 1992; Halliwell and Gutteridge, 1989). Additionally, when an electron rich center such as a sulfur, nitrogen or selenium is in a vicinal relationship to a halide such as one finds with these chloroethyl compounds (Fig. 1), it favors intramolecular cyclization to form an energetically strained, reactive three-membered cyclic onium ion (Fig. 2) (Yang et al., 1988). Onium ions, cyclic and acyclic, form spontaneously in solutions of mustards (Kang and Spears, 1987; Ross, 1962; Bartlett et al., 1949). Electrochemical studies have shown that the one electron reduction of onium ions results in free radical production (Saeva and Morgan, 1984). Reports of the direct electrochemical reduction of sulfonium ions in aqueous systems (Chambers, 1978) indicated the mechanistic feasibility of this process in vivo and raised the possibility of enzymatic reduction as the source of mustard free radicals.
Figure 2. The rapid Sn1 mediated internal cyclization of sulfur mustard with loss of chlorine to form the cyclic sulfonium ion.
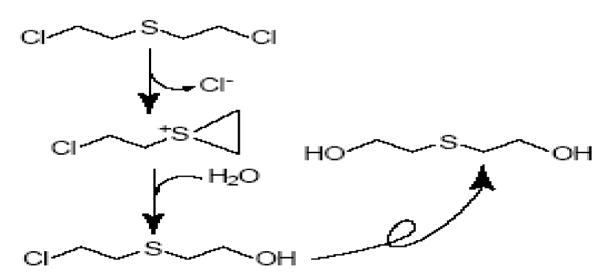
This scheme shows nucleophilic attack by water to form first the chlorohydrin and then, after the formation of a second onium ion and another reaction with water, the doubly hydrolysed thiodiglycol.
Additional evidence of free radical participation in mustard toxicity arose as part of a study designed to evaluate the potential for interaction among military deployment-related chemicals at the level of metabolism. We investigated the effect of sulfur mustard on the microsomal cytochrome P450 drug metabolizing system using NADP, an NADPH generating system and mouse liver microsomes induced with phenobarbital. The results indicated that mustard inhibited the O-demethylation of p-nitroanisole by cytochrome P450 (Brimfield and Hodgson, 2005). To clarify where mustard might be exerting its effect, we simplified the mixture from the previous work by using purified NADPH-cytochrome P450 reductase (CYPOR) in place of microsomes, and cytochrome c as a terminal electron acceptor in place of the microsomal cytochrome P450. Employing this system we were able to follow electron transport spectrophotometrically by measuring cytochrome c reduction (Brimfield and Hodgson, 2005). The rate of cytochrome c reduction was slowed in the presence of mustard just as the rate of p-nitroanisole O-demethylation had been. That effect provided an indication that the flavoprotein reductase itself (the only remaining enzyme in the test system) was the probable site of mustard interaction. This phenomenon has also been reported with hydrazines which are known to impair the oxidative metabolism of co-administered substrates in the microsomal drug metabolizing system and also to be enzymatically activated to carbon free radicals (Muakkassah et al., 1981; Reed, 1976; Lee and Lucier, 1976).
Taken together those results suggested that mustard was inhibiting all cytochrome P450 activity by uncoupling electron transport at the flavoenzyme reductase and diverting electrons directly to xenobiotic reduction. This mode of toxicant transformation can lead to free radical formation (Testa, 1995) and lends weight to our earlier speculation about enzymatic onium ion reduction. Ubiquitous pyridine nucleotide-driven flavoenzyme reductases such as NADPH-cytochrome P450 reductase [EC1.6.2.4] (Tew, 1993), the reductase domain of neuronal nitric oxide synthase [EC 1.14.13.39] (Steuhr et al., 1991), the cytosolic thioredoxin-disulfide reductase [EC 1.8.1.9] (Gray et al., 2007) and others (Halliwell and Gutteridge, 1999) appear to act as good electron sources for this one-electron reduction process.
It has been known for some time that onium compounds such as diphenyliodonium ion and the dipyridylium ions of the herbicides paraquat and diquat are capable of uncoupling extramitochondrial electron transport and being reduced in place of cytochrome P450 or other terminal electron acceptors (Testa, 1995; Halliwell and Gutteridge, 1999). The diphenyliodonium cation offers an especially interesting example. When it undergoes single electron enzymatic reduction by either cytochrome P450 reductase or the reductase domain of nitric oxide synthase it rapidly fragments to yield iodobenzene and a phenyl free radical (Tew, 1993; Steuhr et al., 1991) capable of forming radical adducts (O’Donnell et al., 1994). This provided additional evidence that free radical production might be an early step in the mechanism of mustard toxicity. As a result, we hypothesized that a process mechanistically analogous to that seen with diphenyliodonium and paraquat led to the cytochrome P450 reductase (CYPOR) inhibition that we had observed and explained the parallel biochemical effects of free radical and mustard exposure. Spontaneously formed onium ions could be enzymatically reduced to yield carbon-centered free radicals which reacted with and damaged cellular macromolecules leading to toxicity.
The detection of free radical formation from a mustard in the presence of NADPH and a flavoenzyme reductase in vitro would prove the feasibility of this mechanism. Since an electron paramagnetic resonance (EPR) signal is the preferred method for free radical detection, we proceeded with the development of a spin trapping technique able to detect carbon-centered free radicals emanating from the enzymatic one electron reduction of onium ions originating with the mustards.
Materials and Methods
Reagents
Recombinant porcine CYPOR and the reductase domain of neuronal nitric oxide synthase (nNOSR) from rat brain were a kind gift from Professor Bettie Sue Masters of the Department of Biochemistry, University of Texas Health Sciences Center at San Antonio. Recombinant human cytochrome P450 reductase (HCYPOR) was purchased from BD Gentest, Bedford, MA. Rat liver thioredoxin reductase (TRXR), β-NADPH + H+, the spin trap α-(4-pyridyl-1-oxide)-N-t-butylnitrone (4-POBN), calmodulin from bovine brain, cytochrome c from bovine heart, cantharidin, Chelex-100 and CaCl2 were purchased from Sigma Chemical Co., St. Louis, MO. The nitrogen mustard mechlorethamine (methylbis (chloroethyl amine), HN2), the monofunctional sulfur mustards chloroethyl ethyl sulfide (CEES) and chloroethyl methyl sulfide (CEMS), the spin trap 2-methyl-2-nitrosopropane dimer (MNP) and trimethylsulfonium iodide were purchased from Aldrich Chemical Company, Milwaukee, WI. The alkyl sulfonium ions triethylsulfonium iodide and triethylsulfonium bromide were synthesized in the Department of Chemistry, Florida Institute of Technology, Melbourne, FL using methods described by Lowe (1981). Sulfur mustard (98% purity) and technical grade mustard were obtained from the U.S. Army Edgewood Chemical Biological Center, Aberdeen Proving Ground, MD. Buffer components were from standard sources and were reagent grade or better. Water used in conjunction with EPR studies was deionized to 18.2 MΩ/cm using a Milli-Q Plus Deionizing System (Millipore Inc., Billerica, MA).
The enzymatic spin-trapping systems
Simple enzymatic assays served as the basis for the development of the spin trapping systems and varied somewhat from enzyme to enzyme. The concentration of each enzyme was kept constant after we established a workable assay. No effort was made to refine the systems further.
HCYPOR and CYPOR
The HCYPOR and CYPOR systems contained 0.016 nM recombinant enzyme as described by Strobel and Dignam (1978). Sulfur mustard (up to 4.0 mM), CEES, CEMS or HN2 at desired mM levels were introduced with 40 μM cytochrome c in 0.1 M KPO4 buffer pH 7.5 made 0.25 M with respect to NaCl. The concentration of the spin trap 4-POBN was 1.03 M and the total volume was 500 μL.
nNOSR
The enzymatic system used to detect free radicals formed by rat brain nNOSR was modeled on that described by Professor Betty Sue Masters (personal communication). The mixture contained 0.13μM enzyme, 0.096 μM calmodulin and 12.2 mM calcium chloride. It was made up in 0.05 M tris pH 7.4, containing 0.1 M NaCl, in place of the KPO4 buffer described above, to avoid precipitation of calcium phosphate. Mustard concentrations were as mentioned above. The concentration of 4-POBN was 1.03 M and the total volume was 500 μL.
TRXR
EPR studies using TRXR (0.034μM) for enzymatic activation were run on a Bruker EMX Plus EPR spectrometer equipped with a 4119HS cavity, and employed an enzyme system based on the one described by Elias et al. (1999). Briefly, the incubation mixture consisted of 2.4 mM NADPH, 1.03 M 4-POBN, up to 4.0 mM sulfur mustard or concentrations of the other mustards necessary to yield a free radical signal, in Chelex-100 treated 0.1 M KPO4 buffer, pH 7.5. Total volume was 500 μL.
Electron Paramagnetic Resonance Spectroscopy
With the exception of TRXR (see above), the EPR studies were done on a Bruker ESP 300E EPR spectrometer (Bruker Biospin, Billerica, MA) equipped with a 4103 TM cavity operating at 9.80 GHz. The in vitro systems used to evaluate the reduction of sulfur and nitrogen mustards are described above. The reaction was started by adding the desired volume of a solution of the reductase of interest to a tube containing the other materials at room temperature, vortexing and immediately transferring the mixture to a 10 mm flat cell (Wilmad LabGlass, Buena, NJ). In the interest of consistency, cytochrome c was included in the systems incorporating CYPOR, HCYPOR and nNOSR as a carry-over from the assay development stage in which the reaction was followed by measuring the reduction of cytochrome c spectrophotometrically. Cytochrome c was not used in the TRXR incubation mixture. Unless otherwise indicated, the instrument settings were: microwave power 10mW, modulation amplitude 1.0 Gauss (G), time constant 327.68 ms, sweep width 100 G, center field 3480 G and a sweep time of 1342.2 s. Each spectrum was the accumulation of three scans. Simulations were generated using Win-Sim software created at NIEHS.
Identification of the mustard free radical structure
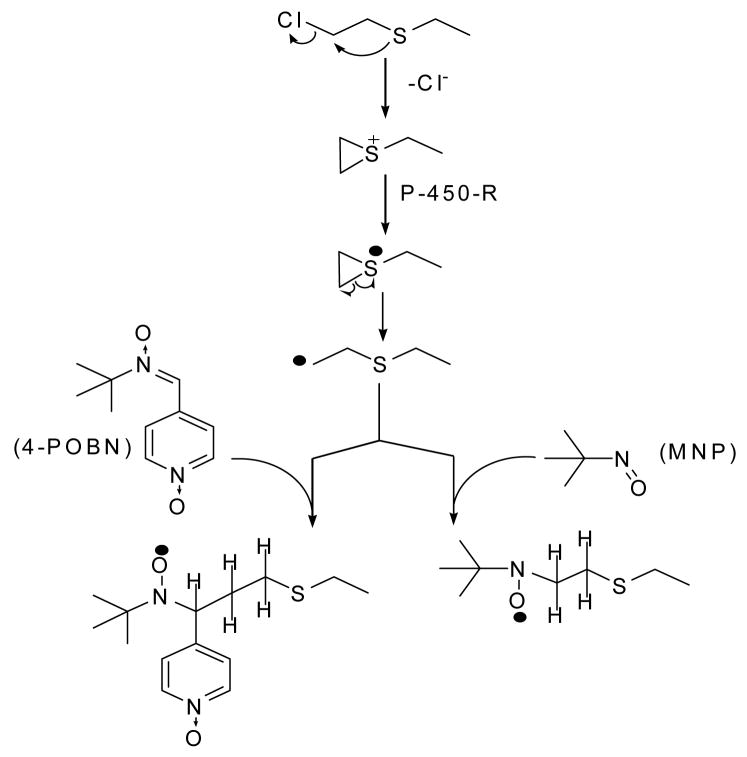
With the use of the correct spin trap one can deduce the structure of a radical from the fine splitting pattern of its spin trap adduct. However, because 4-POBN is a nitrone, the adduct formed occurred at the carbon adjacent to the imino nitrogen of the spin trap, too many bonds distant from the protons on the chloroethyl group to allow interaction with magnetic nuclei in the free radical to generate a useful hyperfine splitting pattern (see Fig. 7 for structures).
Figure 7. The proposed pathway for the production of free radicals from chloroethyl mustards.
Figure 7 gives the structure of the free radicals formed from the interaction of the flavoenzyme reductases with chloroethyl mustards, and the structure of the spin adducts formed with the spin traps 4-POBN or MNP. The compound shown is chloroethylethyl sulfide. However, the process would be the same with any of the chloroethyl mustards including sulfur mustard.
The nitroso spin trap MNP forms adducts at the nitrogen which creates a close enough association between the magnetic nuclei in the radical and the nitroso nitrogen of the MNP to provide the detail needed to allow deduction of the radical’s structure. We used CEES in place of sulfur mustard for the structural determination because the work was done at NIEHS rather than at the Institute of Chemical Defense. In reality, the non-sulfur mustard part of the radical was too remote from the spin trap to interfere. So it made no difference in the outcome whether we used CEES or sulfur mustard.
MNP (3 mg) was dissolved in 1 mL of Chelex 100-treated phosphate buffer (0.1 M, pH 7.4) overnight in a Thermomixer® (Eppendorf, Germany), set at 30 °C and 1400 rpm. The MNP was protected from light to prevent degradation. The complete reaction system was prepared by adding 84 mM CEES (neat, 2 μL from stock) to 200 μL of the MNP solution which contained 3 mM NADPH. This mixture was vortexed for 1 minute. The mixture was made 64 nM with respect to HCYPOR to initiate the reaction and was then transferred to a quartz flat cell and mounted in the spectrometer.
Control experiments were carried out without HCYPOR, without CEES or without NADPH and the reductase. The spectra were recorded with a Bruker EMX spectrometer (Billerica, MA) equipped with an ER 4122 SHQ cavity operating at 9.78 GHz and 100 KHz modulation field at room temperature with the following parameters: power = 20 mW, scan rate = 0.47 G/s, modulation amplitude = 0.4 G, receiver gain = 1.00×105. Each spectrum was the accumulation of four scans (355 s/scan, time constant = 327 ms). Simulations were generated using Win-Sim software created at NIEHS.
Molecular modeling
Electron density calculations for the determination of relative theoretical onium ion reducibility were performed with Spartan ’04 software (Wavefunction, Inc., Irvine, CA) using B3LYP density functional theory with the 6-31G* basis set and with geometry optimization in the aqueous phase (Lee et al., 1988; Becke, 1993).
Results
The toxic effects produced by sulfur mustard and those produced by toxic processes known to act via free radical generation are similar (Papirmeister et al., 1991; Somani, 1992; Halliwell and Gutteridge, 1989). Additionally, onium ions form spontaneously in solutions of mustard (Kang and Spears, 1987; Bartlett et al., 1949; Ross, 1962) and reduction of onium ions leads to carbon-centered free radical production (Chambers, 1978, Stasko et al., 1993, Saeva and Morgan, 1984). On that basis we hypothesized that mustard onium ions might be enzymatically reduced by flavoenzyme reductases to produce carbon-centered free radicals. In order to validate that hypothesis we sought a means to detect free radical production without ambiguity by using EPR spectrometry with spin trapping.
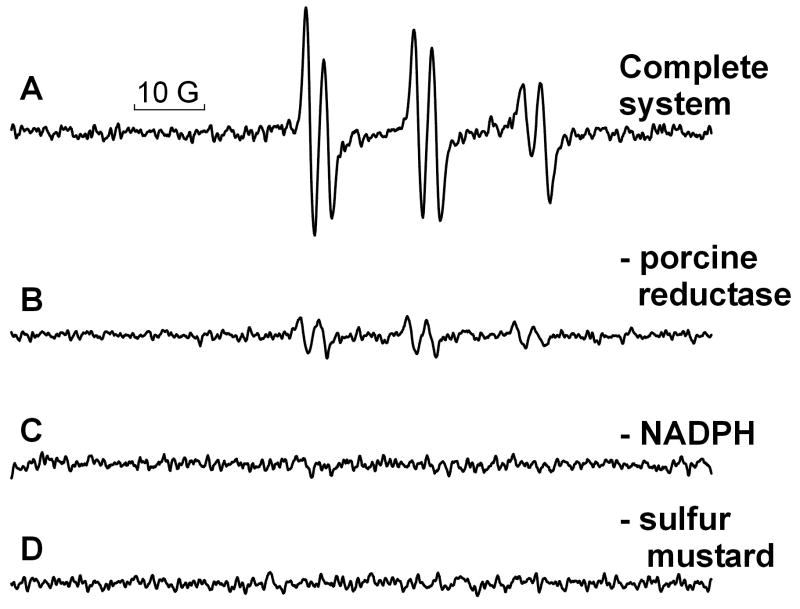
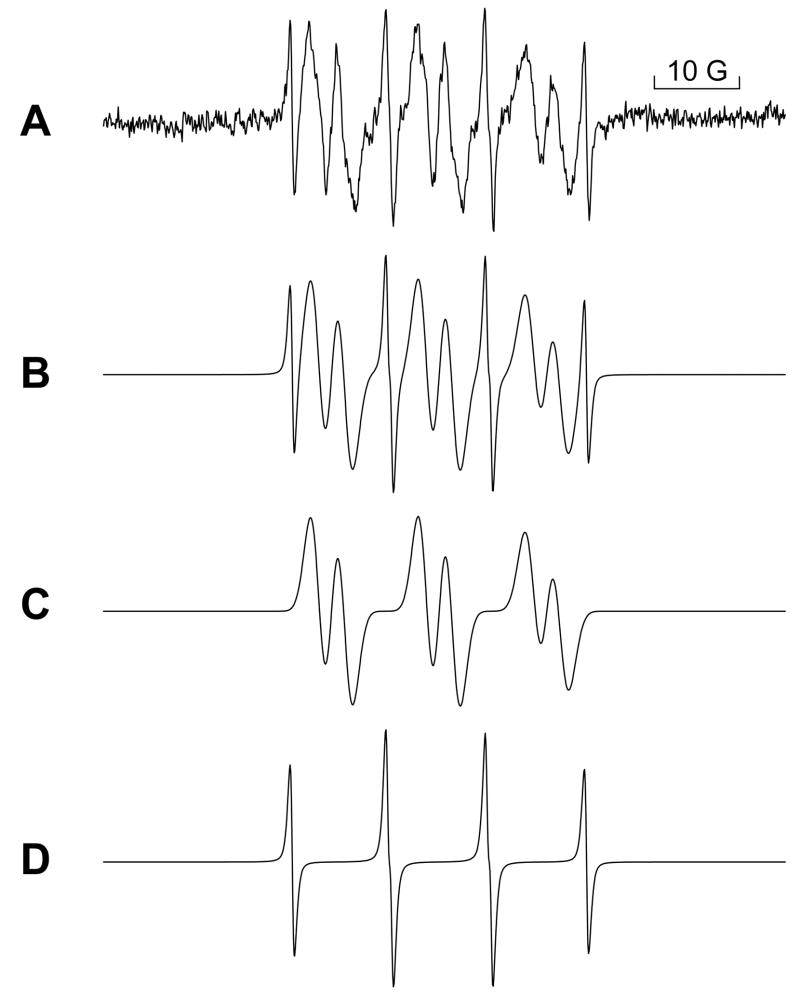
Inclusion of the nitrone 4-POBN in our incubation mixture at the molar level enabled us to observe a six-line 4-POBN-radical adduct spectrum when recombinant CYPOR was incubated with NADPH, sulfur mustard, the spin trap and cytochrome c (Fig. 3A). To test the proposition that each constituent was required for activity, we repeated the EPR analysis in the absence of each component. As shown in Fig. 3(B,C and D) omitting reductase, NADPH or mustard from the otherwise complete in vitro mixture caused the production of free radicals to cease or nearly cease, providing proof that each component is required for free radical generation.
Figure 3. EPR spectra generated using the in vitro system.
A. Complete room temperature incubation containing porcine NADPH – cytochrome P450 reductase (0.016 μM), 2.4 mM NADPH, 4.0 mM sulfur mustard, 1.03 M 4-POBN and 0.21 mM cytochrome c in 0.1M KPO4 buffer containing 0.25 M NaCl, pH 7.5. B. Same as in A, but with the porcine reductase replaced with an equal volume of buffer. C. Same as A, but without the NADPH. D. Same as A, but without sulfur mustard. These spectra were recorded on a Bruker ESP 300E spectrometer equipped with a 4103 TM cavity operating at 9.80 GHz, microwave power: 10 mW, sweep width: 100 Gauss, midscale: approximately 3488 G, number of scans: 3, receiver gain: 1.25e5, time constant: 327 ms, conversion time: 655 ms, modulation amplitude: 1.0 G. All spectra are presented at the same gain. The 10 Gauss bar is included to define the scale.
The inclusion of cytochrome c in the incubation mixture originated with the cytochrome c reductase assay of Strobel and Dignam (1978) in which we we first discovered the mustard-flavoenzyme interaction and which subsequently served as a model for the free radical generating system. Also, from his investigation of CYPOR inhibition by diphenyliodonium cation, Tew (1993) found that turnover was a prerequisite for inhibition of the reductase, and therefore, for free radical generation. We did not find that to be the case with the onium species from the mustards. Running the enzymatic reaction using NADPH, CYPOR and sulfur mustard, but neglecting to include cytochrome c in the mixture had no effect on the quality of the EPR signal produced (results not shown). In an unrelated experiment, replacing mustard with the natural vesicant cantharidin in the incubation mixture (results not shown) produced no free radical signal. This may be an indication of an alternative mechanism for cantharidin-induced vesication.
Dependence of Activity on Onium Ion Structure
Both strained, three-membered cyclic sulfonium (Yang et al., 1988) and immonium ions (Bartlett et al., 1949) as well as acyclic alkyl, unstrained cyclic and polymeric onium ions (St. Quintin et al., 2003; Black et al., 1992) form in mustard solutions. We did not differentiate among these in our original hypothesis regarding onium ion reduction. We felt that free radicals would be formed by enzymatic reduction of onium ions regardless of their structure.
However, free radicals were only detected when sulfur mustard (Fig. 1A), CEES (Fig. 1B), CEMS (Fig. 1C), HN2 (Fig. 1D) or technical sulfur mustard (a complex mixture of mustard and related material not shown) were included in the reaction mixture. Our hypothesis predicted free radical formation by enzymatic reduction of all onium ions. To test this idea, we tested stable, non-cyclic alkyl onium ions such as trimethylsulfonium iodide, triethylsulfonium iodide and triethylsulfonium bromide. None of the compounds lacking the chloroethyl group and incapable of forming the strained, three-membered cyclic onium ions gave rise to a free radical signal by EPR using our in vitro system (data not shown).
In an attempt to explain this difference in enzymatic reducibility we applied density functional theory modeling (Lee et al., 1988; Becke, 1993). This technique allowed us to evaluate molecular features to provide a theoretical framework by which to interpret the variability in our results arising from ion structure. The calculations for sulfonium ions in an aqueous environment indicated that the energy of the lowest unoccupied molecular orbital (LUMO) in the strained ring system was lower (−0.93eV) than the energy of the LUMO of acyclic alkyl sulfonium ions (−0.81eV). In theory, at least, this means the cyclic ions can accept an electron more readily than the acyclic onium ions can. Therefore, an enzymatic system having a less robust reduction potential than direct electrochemical reduction or a chemical reducing agent is capable of donating an electron to the strained cyclic three-membered ring and less capable of reducing the more stable alkyl onium ions.
Dependence of Activity on Reductase
Recombinant CYPOR, HCYPOR, rat brain nNOSR and TRXR from rat liver each supported free radical production in the presence of NADPH, mustard, and 4-POBN. Each of these enzymes functions as part of an extramitochondrial electron transport chain distributing reducing equivalents from pyridine nucleotide coenzymes to metabolic endpoints. Each passes electrons to a terminal electron acceptor one at a time via flavin prosthetic groups. The CYPOR, HCYPOR and nNOSR accomplish this by employing both flavin-adenine dinucleotide (FAD) and flavin mononucleotide (FMN) (Wang et al., 1997; Vasquez-Vivar et al., 1999). In each case FAD accepts two electrons from NADPH and transfers them to FMN which passes the electrons one at a time to a terminal electron acceptor providing the sequential individual electron transfers necessary for the reduction of cyclic onium ions arising from chloroethyl mustards and the production of free radicals.
TRXR is a cytosolic enzyme that catalyzes the NADPH-dependent reduction of the active site disulfide in oxidized thioredoxin, a powerful protein disulfide reductase catalyzing either electron transport to ribonucleotide reductase and other reductive enzymes or redox regulation of enzymes and transcription factors. It is part of a mechanistically different family of enzymes, pyridine nucleotide-disulfide oxidoreductases, having only one flavin prosthetic group. In this case, electrons are passed from NADPH via FAD to an active site disulfide, which again reduces the electron acceptor through a one-electron transfer (Mustacich and Powis, 2000). The end result is the same. There are additional reductases in this enzyme family that may also be capable of reducing the cyclic onium ions.
Table 1 shows the hyperfine splitting constants calculated from the computer-based simulations of the spin trap adduct spectra resulting from the reduction of each of the chloroethyl vesicants by each of the four enzymes tested. The data given for the reduction of sulfur mustard by CYPOR are from the spectrum shown in Fig. 3A. Along with the generation of a detectable EPR signal by each of the reductases, the similarity among the hyperfine splitting constants from the spectra of the 4-POBN adducts suggests a commonality of outcomes arising from the reduction carried out by the different enzymes. The process illustrated in Fig. 7 lends weight to this conclusion as well. These results provide a bridge between the outcome of chemical and electrochemical reduction of onium ions leading to free radical production (Saeva and Morgan, 1984; Chambers, 1978) and what we see with our in vitro enzymatic system.
Table 1.
Hyperfine splitting constants resulting from the spintrapping of free radicals derived from flavoenzyme-reduced chloroethyl mustards with 4-POBN
| Nitrogena | Hydrogena | ||
|---|---|---|---|
| Enzyme | Mustard | aN(Gb) | aH(G) |
| Porcine cytochrome P450 reductase | HD | 15.46 | 2.54 |
| CEMS | 15.41 | 2.60 | |
| CEES | 15.42 | 2.29 | |
| HN2 | 15.45 | 2.67 | |
| Human cytochrome P450 reductase | HD | 15.40 | 2.68 |
| CEMS | 15.40 | 2.59 | |
| CEES | 15.43 | 2.27 | |
| HN2 | 15.40 | 2.57 | |
| Neuronal nitric oxide synthase reductase domain | HD | 15.40 | 2.69 |
| CEMS | 15.43 | 2.66 | |
| CEES | 15.41 | 2.26 | |
| HN2 | 15.38 | 2.81 | |
| Thioredoxin reductase | HD | 15.42 | 2.70 |
| CEMS | 15.41 | 2.61 | |
| CEES | 15.50 | 2.13 | |
| HN2 | 15.54 | 2.63 | |
Hyperfine splitting constants from computer simulations of the experimental data using Win-Sim software.
Gauss
Identity of the Free Radical Structure
Determination of the free radical structure was done by generating the adduct with the spin trap MNP using CEES (Fig. 1B) rather than sulfur mustard as the source of the radical in the in vitro enzyme system. CEES provided an EPR spectrum qualitatively equivalent to that of sulfur mustard when we used our in vitro system with HCYPOR as the reductase and 4-POBN as the spin trap (Fig. 3A) (Brimfield and Mancebo, 2007). Therefore, we had no reason to expect any difference arising from the use of CEES in place of sulfur mustard with MNP.
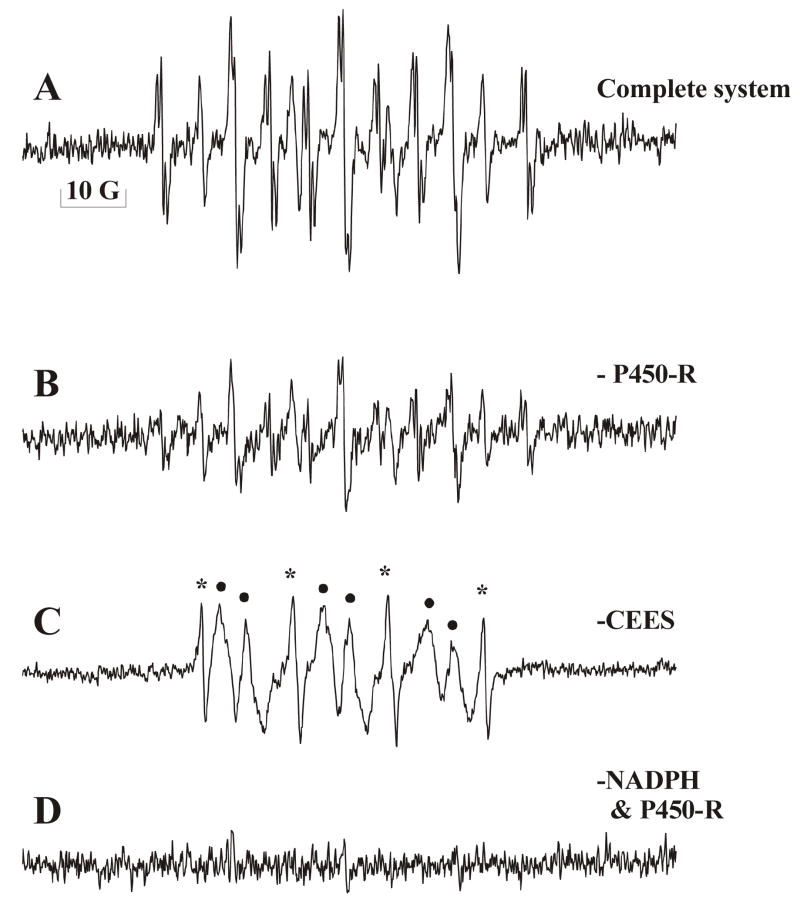
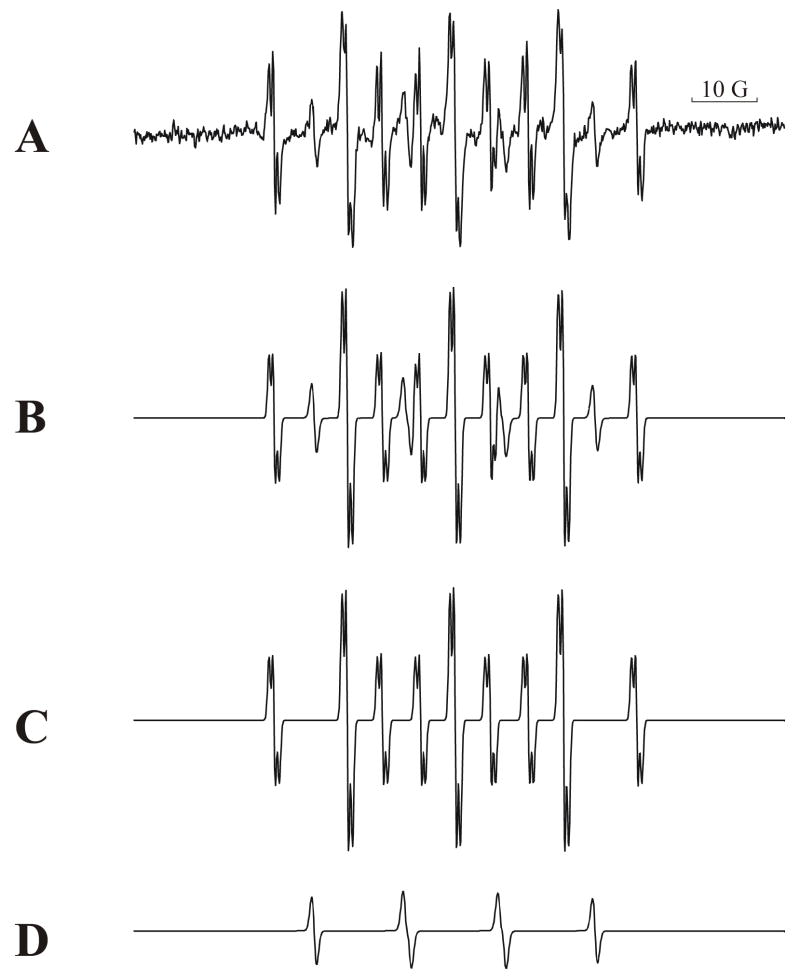
The EPR spectrum of the complete system is shown in Fig. 4A. Figs. 4B to Fig. 4D are the control experiments. Fig. 4C shows the signal generated without CEES, and Fig. 4D shows the effect of leaving out NADPH and HCYPOR. When HCYPOR was left out of the mixture (Fig. 4B) the free radical signal was significantly lower.
Figure 4. The determination of the structure of the mustard radical.
The experimental EPR spectrum of the complete system for the determination of the radical structure containing NADPH, CEES, HCYPOR and MNP is shown in Figure 4A. Figure 4B is the same as 4A but without the HCYPOR. Figure 4C contained no CEES. Figure 4D contained no NADPH or HCYPOR. The 10 Gauss bar is included to define the scale. (See Results)
Without CEES (Fig. 4C) the spectrum showed two components of background signal. One (indicated by *) was identified by computer simulation (Fig. 5D) as the MNP/H• radical commonly appearing in control and spin trapping experiments employing MNP (Kalyanaraman et al., 1979; Mottley et al., 1981), while the other species with the six-line spectrum (indicated by•) was not detected in the complete system. Simulation of the six-line spectrum (Fig. 5C) provided hyperfine splitting constants aH = 3.65 G and aN = 15.8 G with a correlation factor of 0.97. It is likely that this signal was derived from a carbon-centered radical with one hydrogen (•CH(R)2) – possibly a component present in the enzyme preparation. (Note: this control experiment used a doubling of the time constant and a doubling of the conversion time to reduce the noise. The scale was adjusted to be equivalent to the other spectra.)
Figure 5. Simulation of the experimental spectrum shown in Figure 4C using Win Sim software.
In the absence of CEES, the spectrum in Figure 6A was found. The hyperfine splitting constants were simulated, which produced the closly correlating spectrum shown in Figure 5B. One component containing four sharp lines (Figure 5D and 6D) resulted from the hydrogen free radical commonly appearing in spin trapping experiments using MNP. The other component containing six lines only appeared in the absence of CEES and is thought to arise from the formation of a carbon centered radical present in the enzyme preparation. The 10 Gauss bar is included to define the scale. (See Results)
The experimental EPR spectra in Fig. 4 were simulated using Win-Sim software (Fig. 6). Fig. 6A is the experimental spectrum produced by the complete system. Fig. 6B is the computer simulation of Fig. 6A. A correlation factor of 0.96 indicated that the simulation was excellent. Figs. 6C and 6D show the two components of the 6B simulation separately. Fig. 6C is the MNP trapped free radical adduct generated using CEES. The hyperfine structure includes a single nitrogen splitting of aN=16.63 G, splitting of two equivalent hydrogens aHβ=11.24 G representing interaction with the hydrogens on the α carbon and another splitting of two equivalent hydrogens aHγ=0.56 representing the effect of the hydrogens on the β carbon of the chloroethyl group. The CEES radical adduct contributes 84.4% of the entire area under the curve. The proposed reaction and the structure of the free radical generated are shown in Fig. 7. The hyperfine splittings from the simulation are what would be expected from the free radical trapped by MNP in Fig. 7, which features a nitrogen splitting, and splitting by two sets of two equivalent protons contributed by the CEES radical (Makino, 1979). Fig. 6D is the simulation of the spectrum for the hydrogen free radical (MNP/H•) also seen in 5D and commonly appearing in control and spin trapping experiments employing MNP (Kalyanaraman et al., 1979; Mottley et al., 1981), where aN=14.62 G and aH=13.95 G. The hydrogen radical contributes 15.6% of the whole spectrum.
Figure 6. Simulation of the experimental spectra shown in Figure 4 using Win Sim software.
Figure 6A is the original experimental spectrum of the spin trapped MNP-CEES free radical adduct. Figure 6B is the computer simulation of 6A. Figures 6C and 6D show the individual components of the simulation. Figure 6C is the MNP trapped free radical generated by the enzymatic reduction of CEES. Figure 6D is the trapped hydrogen free radical commonly appearing in control and spin trapping experiments employing MNP. Figure 6D contributes 15.6% of the area under the total spectrum shown in Figure 6B. The 10 Gauss bar is included to define the scale.
Discussion
Until now injury from exposure to mustard and related vesicants has been assumed to result from direct alkylation of biological macromolecules by cyclic onium ions. The results of this investigation offer an additional mechanism by which to explain the outcome of mustard exposure of skin, eye and airway epithelium in the form of free radical-related alkylation. It is not necessary to invoke one or the other of these toxicologic processes exclusively. Both direct alkylation and radical driven alkylation could be at work simultaneously. Following the formation of the cyclic sulfonium ions, we cannot exclude the possibility of an enzymatic reduction/free radical mechanism acting in parallel with the traditionally assumed direct alkylation. One can even view flavoenzyme reduction and direct electrophilic alkylation as processes competing for the available cyclic sulfonium ion supply. This is a very interesting perspective. In the event of reductase inhibition, free radical generation would cease, and the available cyclic onium species would form macromolecular adducts via electrophilic alkylation. In that case signs of toxicity would continue to accumulate from non-free radical-related alkylation originating with the cyclic species. However, if toxicity were terminated by reductase inhibition it would point to free radical generation as the sole source of mustard-related tissue damage and provide an additional direction to guide our search for therapeutic strategies.
We can carry this line of reasoning a few steps further. Our results showing enzymatic reduction of cyclic onium ions support work reported several years ago in a series of papers by Sawyer (1996, 1998, 1999) outlining his investigation of nitric oxide synthase (NOS) involvement in sulfur mustard injury. Using primary cultures of chick embryo forebrain neurons exposed to sulfur mustard as a test system, he found that inhibitors of NOS such as L-nitroarginine methyl ester and L-thiocitrulline reduced the damage to cultured neurons from exposure to sulfur mustard in a dose dependent manner. He was working from the point of view that mustard toxicity might result from overproduction of nitric oxide (NO) and that inhibition of NO production would lead to reduced toxicity. Based on the evidence presented here, however, his results seem more consistent with diminished free radical generation due to curtailed onium ion reduction from inhibited NOS reductase domain than to a drop in NO concentration.
Earlier we mentioned the across-the-board inhibition of cytochrome P450 isoforms by sulfur mustard and a reduction in the rate of drug metabolism caused by compounds that uncouple microsomal electron transport. The evolution of free radicals from every chloroethyl mustard tested (Table 2) and the mechanistic implications of that result suggest there might be pharmacokinetic consequences such as reduced clearance rates for therapeutic compounds and disruption of cytochrome P450-dependent levels of endogenous compounds for victims of mustard exposure. This effect has been well documented in rodents (Conney et al., 1967; Fine and Molloy, 1964) and has been noted in a clinical setting, for example, in the case of altered pharmacokinetics for benzodiazepines in the presence of hypnotics or opiates (Olkkola and Ahonen, 2008). Since they are indicated for the control of seizures resulting from exposure to organophosphorus nerve agents, the benzodiazepines would certainly be encountered in the treatment of chemical warfare casualties, and dose adjustments may be necessary in the treatment of nerve agent poisoning in the presence of mustard exposure.
Acknowledgments
This work was supported by the Defense Threat Reduction Agency – Joint Science and Technology Office, Medical S&T Division and, in part, by award number DAMD 17-00-2-008 from the U.S. Army Medical Research and Materiel Command. This research was also supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. Thanks are due to Dr. Carmen Arroyo and Mr. Brendon Gallagher of USAMRICD for helping to get this work started. We are grateful to Professor Bettie Sue Masters of the Department of Biochemistry, University of Texas Health Sciences Center, San Antonio, for the kind gift of recombinant rat brain nNOS reductase domain and recombinant porcine liver NADPH-cytochrome P450 reductase and to Dr. Thomas M. Shea of that laboratory for generous advice. And finally, we would like to express our thanks to Dr. Mark A. Zottola and Dr. Sunil D. Soni of USAMRICD for discussions that were inevitably helpful and enlightening.
Footnotes
The opinions or assertions herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Army or of the Department of Defense
References
- Bartlett PD, Ross SD, Swain CG. Kinetics and mechanism of the reactions of tertiary β– chloroethylamines in solution. III β-chloroethyldiethylamine and tris-β-chloroethylamine. J Am Chem Soc. 1949;71:1415–1419. [Google Scholar]
- Becke AD. Density functional thermochemistry. III The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- Black RM, Brewster K, Clark RJ, Hambrook JL, Harrison JM, Howells DJ. Biological fate of sulphur mustard, 1, 1′-thiobis(chloroethane): isolation and identification of urinary metabolites following intraperitoneal administration to rat. Xenobiotica. 1992;22:405–418. doi: 10.3109/00498259209046652. [DOI] [PubMed] [Google Scholar]
- Brimfield AA, Hodgson E. Observations on the interaction of sulfur mustard with cytochrome P450. Abstract # 781, The Toxicologist. 2005;84:159–160. [Google Scholar]
- Brimfield AA, Mancebo AM. Monofunctional mustards form free radicals when enzymatically reduced making them useful as model compounds for mechanistic study and drug screening for sulfur mustard. Abstract # 356, The Toxicologist. 2007;96:74. [Google Scholar]
- Chambers JQ. Organic sulfur compounds. In: Bard AJ, Lund H, editors. Encyclopedia of Electrochemistry of the Elements, Organic Section. Dekker; New York: 1978. pp. 329–502. [Google Scholar]
- Conney AH, Welch RA, Kuntzman R, Burns JJ. Effects of pesticides on drug and steroid metabolism. Clin Pharmacol Thera. 1967;8:2–10. doi: 10.1002/cpt196781part12. [DOI] [PubMed] [Google Scholar]
- Elias S, Arner J, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin reductase. Meth Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- Fine BC, Molloy JO. Effects of insecticide synergists on duration of sleep induced in mice by barbiturates. Nature (London) 1964;204:789–791. doi: 10.1038/204789b0. [DOI] [PubMed] [Google Scholar]
- Gray JP, Heck DE, Mishin V, Smith JS, Hong JH, Thiruchelvam M, Cory-Slechta DA, Laskin DL, Laskin JD. Paraquat increases cyanide-insensitive respiration in murine lung epithelial cells by activating an NAD(P)H: paraquat oxidoreductase. J Biol Chem. 2007;282:7939–7949. doi: 10.1074/jbc.M611817200. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 2. Oxford University Press; New York: 1989. p. 300. [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 3. Oxford University Press; New York: 1999. pp. 511–514. [Google Scholar]
- Kalyanaraman B, Perez-Reyes E, Mason RP. The reduction of nitroso-spin traps in chemical and biological systems. Tetrahedron Lett. 1979;50:4809–4812. [Google Scholar]
- Kang S-I, Spears CP. Linear free energy relationships and cytotoxicities of para-substituted 2-haloethyl selenides and bis (2-chloroethyl) selenides. J Med Chem. 1987;30:597–602. doi: 10.1021/jm00387a003. [DOI] [PubMed] [Google Scholar]
- Lee IP, Lucier GW. The potentiation of barbiturate-induced narcosis by procarbazine. J Pharmacol Exp Thera. 1976;196:586–593. [PubMed] [Google Scholar]
- Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a function of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Lowe PA. Synthesis of sulfonium salts. In: Stirling CJM, editor. Chemistry of the Sulphonium Group. Wiley; New York: 1981. pp. 267–312. [Google Scholar]
- Makino K. Studies on the spin trapped radicals in γ-irradiated aqueous solutions of 2-methyl-2-nitrosopropane by high performance liquid chromatography and ESR spectroscopy. J Phys Chem. 1979;84:2520–2523. [Google Scholar]
- Marshall EK, Lynch V, Smith HW. On dichlorodiethylsulfide (mustard gas) II. Variations in susceptibility of the skin to dichlorodiethylsulfide. J Pharmacol Exp Ther. 1919;12:291–301. [Google Scholar]
- Mottley C, Kalyanaraman B, Mason RP. Spin trapping artifacts due to the reduction of nitroso spin traps. FEBS Lett. 1981;130:12–14. [Google Scholar]
- Muakkassah SF, Bidlack WR, Yang WCT. Mechanism of the inhibitory action of isoniazid on microsomal drug metabolism. Biochem Pharmacol. 1981;30:1651–1658. doi: 10.1016/0006-2952(81)90393-2. [DOI] [PubMed] [Google Scholar]
- Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000;346:1–8. [PMC free article] [PubMed] [Google Scholar]
- O’Donnell VB, Smith GCM, Jones OTG. Involvement of phenyl radicals in iodonium compound inhibition of flavoenzymes. Mol Pharmacol. 1994;46:778–785. [PubMed] [Google Scholar]
- Olkkola KT, Ahonen J. Midazolam and other Benzodiazepines. Handb Exp Pharmacol. 2008;182:335–60. doi: 10.1007/978-3-540-74806-9_16. [DOI] [PubMed] [Google Scholar]
- Papirmeister B, Feister AJ, Robinson SI, Ford RD. Medical Defense Against Mustard Gas: Toxic Mechanisms and Pharmacological Implications. CRC Press; Boca Raton: 1991. [Google Scholar]
- Reed DJ. Effects in vivo of lymphoma ascites tumors and procarbazine, alone and in combination, upon hepatic drug-metabolizing enzymes of mice. Biochem Pharmacol. 1976;25:153–156. doi: 10.1016/0006-2952(76)90283-5. [DOI] [PubMed] [Google Scholar]
- Ross WCJ. Biological Alkylating Agents, Fundimental Chemistry and the Design of Compounds for Selective Toxicity. Butterworths; London: 1962. Ammonium and sulfonium compounds, 2-chloroethyl sulphides, 2-chloroethyl amines; pp. 9–12. [Google Scholar]
- Sabourin CLK, Petrali JP, Casillas RP. Alterations in inflammatory cytokine gene expression in sulfur mustard-exposed mouse skin. J Biochem Molec Toxicol. 2000;14:291–302. doi: 10.1002/1099-0461(2000)14:6<291::AID-JBT1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Saeva FD, Morgan BP. Mechanism of one-electron electrochemical reductive cleavage reactions of sulfonium salts. J Am Chem Soc. 1984;106:4121–4125. [Google Scholar]
- Sawyer TW, Lundy PM, Weiss MT. Protective effect of an inhibitor of nitric oxide synthase on sulfur mustard toxicity in vitro. Toxicol App Pharmacol. 1996;141:138–144. doi: 10.1006/taap.1996.0270. [DOI] [PubMed] [Google Scholar]
- Sawyer TW. Characterization of the protective effects of L-nitroarginine methyl ester (L-NAME) against the toxicity of sulfur mustard in vitro. Toxicol. 1998;131:21–32. doi: 10.1016/s0300-483x(98)00120-6. [DOI] [PubMed] [Google Scholar]
- Sawyer TW. Synergistic protective effects of selected arginine analogues against sulfur mustard toxicity in neurone culture. Toxicol App Pharmacol. 1999;155:169–176. doi: 10.1006/taap.1998.8588. [DOI] [PubMed] [Google Scholar]
- Smith WJ, Neally EW, Clark OE, Cowen FM. Human epidermal keratinocytes exposed in vitro to the vesicating agent sulfur mustard express markers of apoptosis and inflammation. In Vitro. 2001;37:47A. [Google Scholar]
- Somani SM. Toxicokinetics and toxicodynamics of mustard. In: Somani SM, editor. Chemical Warfare Agents. Academic Press Inc.; New York: 1992. pp. 28–35. [Google Scholar]
- Stasko A, Rapta P, Brezova V, Nuyken O, Vogel R. Photochemically and electrochemically initiated radical decomposition of sulfonium salts (A spin trap study) Tetrahedron. 1993;49:10917–10924. [Google Scholar]
- Steuhr DJ, Fasehun OA, Kwon NS, Gross SS, Gonzalez JA, Nathan CF. Inhibition of macrophage and endothelial cell nitric oxide synthase by diphenyleneiodonium and its analogs. FASEB J. 1991;5:98–103. doi: 10.1096/fasebj.5.1.1703974. [DOI] [PubMed] [Google Scholar]
- St Quintin TD, Leslie DR, Collins JG. Hydrolysis of sesquimustards. Austral J Chem. 2003;56:309–313. [Google Scholar]
- Strobel HW, Dignam JD. Purification and properties of NADPH-cytochrome P-450 reductase. Meth Enzymol. 1978;LII:89–96. doi: 10.1016/s0076-6879(78)52009-0. [DOI] [PubMed] [Google Scholar]
- Testa B. Reductions catalyzed by cytochrome P450 and other oxidoreductases. In: Testa B, Caldwell J, editors. The Metabolism of Drugs and Other Xenobiotics, Biochemistry of Redox Reactions. Academic Press; New York: 1995. pp. 434–437. [Google Scholar]
- Tew DG. Inhibition of cytochrome P450 reductase by the diphenyliodonium cation kinetic analysis and covalent modifications. Biochem. 1993;32:10209–10215. doi: 10.1021/bi00089a042. [DOI] [PubMed] [Google Scholar]
- Vasquez-Vivar J, Martasek P, Hogg N, Karoui H, Masters BS, Pritchard KA, Jr, Kalyanaraman B. Electron spin resonance spin-trapping detection of superoxide generated by neuronal nitric oxide synthase. Meth Enzymol. 1999;301:169–177. doi: 10.1016/s0076-6879(99)01080-0. [DOI] [PubMed] [Google Scholar]
- Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim J-JP. Three-dimentional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YC, Szanfraniec LL, Beaudry WT, Ward JR. Kinetics and mechanisms of hydrolysis of 2-chloroethyl sulfides. J Org Chem. 1988;53:3293–3297. [Google Scholar]