Abstract
FokI is a Type IIS restriction endonuclease that recognizes the 5′-GGATG-3′ sequence and cleaves non-specifically at 9 and 13 base pairs away on the top and bottom strands respectively to produce a 5′ overhang. FokI is a bipartite endonuclease with separate recognition and cleavage domains. Because of its bipartite nature, FokI has received considerable interest in generating chimeric nucleases for use in biotechnology, and recently as possible therapeutic agents in gene therapy by initiating homologous gene recombination and repair. Here we show, using single-particle electron microscopic studies, that the FokI active complex prefers a single conformation in which the subunits are arranged in a doughnut shape complex with protein-protein and possibly protein-DNA interactions stabilizing the cleavage complex. Our EM model provides new insights into the activation mechanism of FokI and how non-specific cleavage is avoided.
Keywords: bipartite restriction endonuclease, DNA recognition and cleavage, synaptic complex, single particle analysis, electron microscopy
The Type IIS restriction endonuclease FokI cleaves DNA non-specifically at a fixed distance of 9 and 13 nucleotides downstream of a non-palindromic recognition sequence 5′-GGATG-3′. The enzyme is monomeric (~66 kDa) in solution and has a modular structure consisting of an N-terminal DNA recognition domain and a C-terminal cleavage domain1. The bipartite nature of FokI has made it an excellent candidate for the design of hybrid endonucleases with novel sequence specificities, by attaching the cleavage domain of FokI to the DNA binding domains of transcription factors such as zinc fingers and homeodomains2–4. In particular, the chimeric zinc finger nucleases (ZFNs) have shown potential as therapeutic agents in gene targeting by initiating homologous gene recombination and repair of damaged DNA4–9.
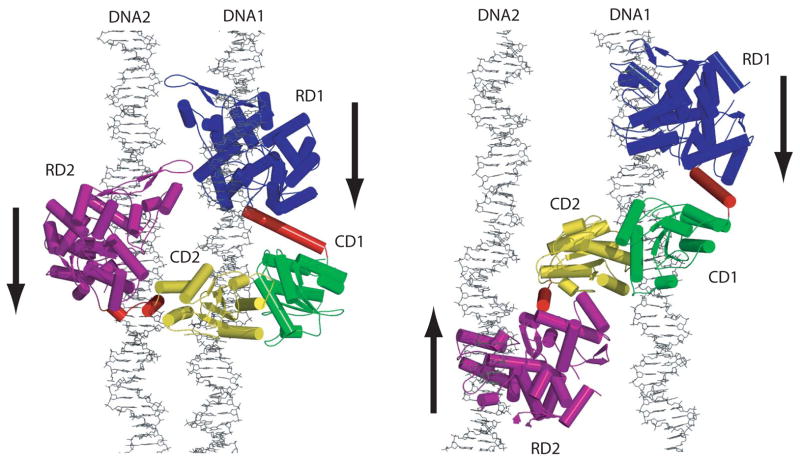
Despite the considerable interest in generating novel nucleases and therapeutics using FokI, a picture of the mechanism of cleavage of the native enzyme has not been fully achieved. An important step toward this goal was the determination of the crystal structure of enzyme bound to its cognate DNA10. The structure determined with a 20-mer oligonucleotide in the absence of divalent metals, showed that the cleavage domain was packed alongside the recognition domain and hence was positioned away from the site of cleavage. Although the structure helped to explain how FokI avoids the “accidental” cleavage of non-cognate DNA sites in the genome, it raised the question of how a monomeric FokI molecule containing a single active site manages to cleave both DNA strands. A partial answer to this question came from the crystal structure of the FokI apoenzyme that revealed a dimer11, as well as kinetic studies that showed a cooperative second-order cleavage mechanism12. Together, these studies suggested that two FokI molecules dimerize on the DNA via their cleavage domains, in a manner similar to the BamHI endonuclease, with which the FokI cleavage domain shares considerable structural homology10. Subsequent biochemical and biophysical studies suggested that a second DNA site was required for successful cleavage 13; 14. The second DNA site acted as an allosteric activator and served to bind the recognition domain of the second FokI molecule. Based on these data, we have proposed two models for the synaptic 2FokI:2DNA complex (Figure 1)13. In the first model, the two DNA sites (DNA1 and DNA2) are in parallel and in the second model, the two DNA sites are arranged anti-parallel. In both cases, the first FokI molecule is bound to DNA1 and its cleavage domain (CD1) is positioned by a simple rotation to cleave the anti-sense strand 13 base pairs away from the recognition site. To allow proper positioning, the linker connecting the recognition and cleavage domains may become α helical, possibly acting as a measuring rod. The second FokI molecule is bound to DNA2 and its cleavage domain (CD2) is positioned to cleave the sense strand of DNA1, 9 base pairs away from its recognition site (Figure 1). The cleavage domains are modeled as a dimer (CD1/CD2) based on the BamHI dimer15. In terms of overall shape, the two models differ in the position of the recognition domain of the second FokI molecule, RD2. In model 1, RD2 is juxtaposed close to RD1, while in model 2, RD1 and RD2 are on opposite sides of the cleavage domains (Figure 1).
Figure 1.
FokI cleavage complexes modeled with both parallel (left) and anti-parallel (right) recognition sites. The models were generated based on the crystal structures of the FokI-20-mer10 and the BamHI-DNA complexes15. The DNAs were extended. Both recognition domains (RD1 and RD2) are bound to their respective recognition sites and the cleavage domains (CD1 and CD2) form a BamHI-like dimer on DNA1. The arrows indicate the direction of the recognition sequence in each DNA. The parallel model provides for protein-protein and possibly protein-DNA interactions, not seen in the anti-parallel model, that could further stabilize the cleavage complex. (Reproduced by permission from Vanamee et al.13)
To see which of these two models is correct, we used electron microscopy (EM) and single particle analysis. Although, the FokI synaptic complex (~130 kDa without DNA) is considered small for such analysis, the very different shapes of the proposed models made it a candidate for negative stain experiments. We were also encouraged by studies demonstrating that macromolecules with molecular weights <100 kDa can be visualized by EM, and that individual domains as small as 10 kDa can be distinguished by negative stain experiments16.
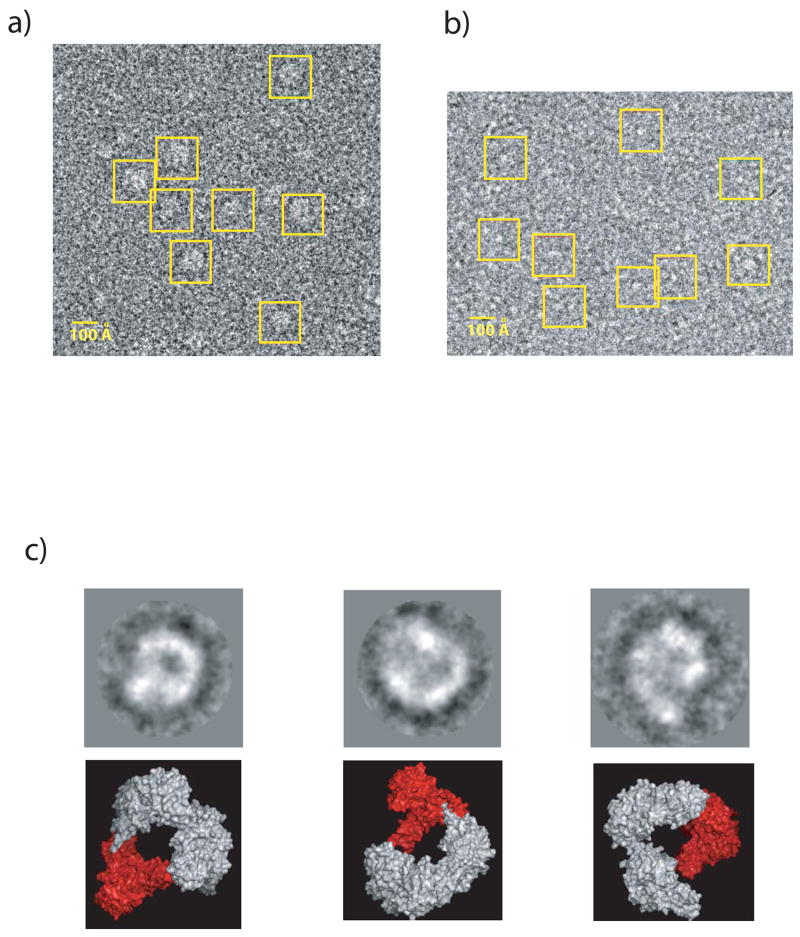
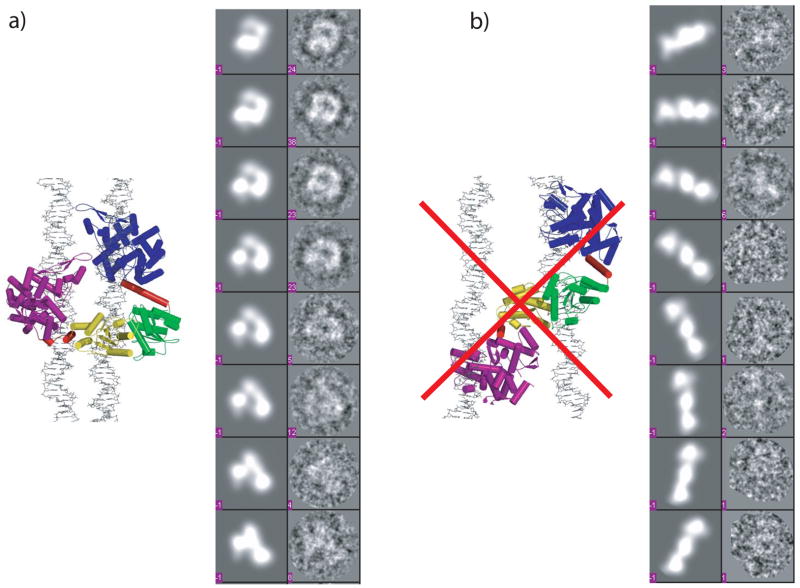
For EM studies, we incubated FokI with an excess of 30-mer oligonucleotide containing a FokI binding site. The incubation was carried out in the presence Ca2+, which we have shown previously to promote the formation of the FokI synaptic complex and to inhibit DNA cleavage13. The complex (at a final concentration of ~3 μM) was then applied to carbon-coated and glow-discharged Cu grids, stained by 2% uranyl acetate, and viewed on a Tecnai F20 electron microscope at 200 keV using a 2 μm defocus. The raw images of the negatively stained complex showed particles uniform in shape and size, suitable for low-resolution analysis (Figure 2a). Dozens of images were then taken with a 4k × 4k charge-coupled device (CCD) camera (at 50,000 × magnification), and the particle selected and analyzed with the program EMAN17. A total of 2,316 particles were initially selected from 12 images using a box size of 104 pixels large enough to accommodate both possible models of the FokI synaptic complex. An initial reference-free classification provided images with recognizable features of model 1 (Figure 2c). Next, we used the two models (Figure 1) to generate 2D projections at a 9° interval that were then matched against the 2D projections of the imaged particles. The resolution of the atomic models was lowered to 5 Å and a low-pass filter of 40 was also applied to better match the low-resolution EM data. The DNA is not observed directly in our experiments as it is often invisible in EM images prepared by uranyl acetate staining18–21. For this reason, the coordinates of the DNA were left out in generating the theoretical projections. The projection matching results showed that only particles corresponding to model 1 are present (Figure 3a), whereas the projection matches generated from model 2 are indistinguishable from noise (Figure 3b). However, not all of the projections generated from model 1 are evenly populated with particles, indicating the presence of a few preferred orientations. In all, we observe six clusters of views that are populated with a significant number of particles. Preferred orientations are expected for particles prepared by negative staining16. As the stain solution dries to a thin layer, any suspended molecular complexes are left stranded on the carbon surface and are found in their more probable orientations. Glow discharging the grids also contributes to preferred orientations. Finally, the axes of the DNA would also contribute towards an in-plane alignment of the FokI complexes onto the carbon of the grids.
Figure 2.
Close-up view of a raw CCD image showing uniform particles of a) the FokI/DNA complex and b) the FokI monomer. a) FokI was mixed with an excess of double stranded oligonucleotide (1:1.2) to ensure that all protein was in complex with DNA. The mixture was diluted to a final concentration of ~3 μM for the electron microscopic studies. 4 μl of such prepared complex was added to carbon-coated 400 mesh square Cu grids. The samples were negatively stained by 2% uranyl acetate. Excess solution was blotted away, and the grids were air-dried prior to loading them into the microscope. Data were collected on a Tecnai F20 electron microscope equipped with a field emission gun (FEG) and operated at 200 keV in low-dose mode. Specimens were imaged at room temperature in a Gatan specimen holder. Dozens of images were taken with a 4k × 4k charge-coupled device (CCD) camera (F415 from Tietz Video and Imaging Processing GmbH, Gauting, Germany) at 50,000 × magnification. b) the FokI monomer was prepared and imaged at identical conditions but in the absence of DNA. c) Reference-free class averages of the FokI/DNA complex on top and the corresponding high-resolution surface models below. In the models, the second recognition domain (RD2) is colored in red. The three class averages from left to right were created from 108, 86, and 75 particles respectively.
Figure 3.
2D projection matches generated a) from model 1 and b) from model 2. The left panels show representative projections generated from the models at 9° intervals and the right panels show the matched classified particles. The numbers in the right panels indicate the number of particles in each projection. The atomic models used to generate the 2D projections are also shown on the sides respectively. Model 2 is crossed out indicating that it is not supported by the EM data. The coordinates of the DNAs were omitted as DNA positively stains with uranyl acetate and cannot be visualized in these experiments. The program EMAN (v.1.7)17 was used for particle selection and data analysis. A total of 2,316 initial particles were selected with a window size of 104 pixels.
We conclude from the single particle analysis that FokI prefers a single active conformation, wherein two FokI monomers adopt a compact “doughnut” shape. Although, we cannot see the DNA in the EM experiment, our earlier analytical ultracentrifugation results as well as a pull-down assay have shown the presence of two DNA molecules in the active complex13. Cleavage studies with a DNA binding deficient N13Y FokI mutant 12; 22 have also shown that DNA bound FokI is a more efficient allosteric activator of DNA cleavage than the unbound form. Thus, under the experimental conditions used to prepare the EM grids, most of the FokI molecules are not only expected to be bound to DNA, but from all previous biophysical and biochemical data they are expected be in a 2FokI:2DNA synaptic complex. Indeed, when we examine negative stain images of FokI in the absence of DNA, the particles are much smaller in size, corresponding to FokI monomers with no sign of dimer formation (Figure 2b).
Importantly, our EM model of the FokI active complex permits intermolecular protein-protein interactions between the two recognition domains (RD1 and RD2). At the same time, there are possible intermolecular protein-DNA interactions that could further stabilize the synaptic complex. Interestingly, there is a loop region (255–259) positioned at the intermolecular interface that is disordered in the FokI apoenzyme structure as well as in the structure of FokI monomer bound to DNA10; 11, and it is possible that this region only becomes ordered upon the formation of the dimeric active complex stabilized by intermolecular interactions.
The EM derived FokI active complex has implications for how the enzyme cleaves its DNA target and avoids accidental cleavage at non-specific sites. First, a FokI monomer binds and scans the DNA in a closed conformation, wherein the cleavage domain is packed alongside the recognition domain. Upon reaching a cognate site, the cleavage domain is released and it dimerizes with the cleavage domain of a second FokI monomer bound to another specific DNA site. From our EM model, this dimerization may be assisted by the intermolecular interactions between the two closely juxtaposed recognition domains that stabilize the FokI active complex. These results have implications for the design of zinc finger nucleases (ZFNs), in which the cleavage domain of FokI is tethered to DNA binding zinc finger domains2; 4. Although, ZFNs have generated much interest as possible therapeutics5–9, a persistent problem has been toxicity, which presumably is partially due to the lack of a “FokI-like” built-in safety mechanism to avoid cleavage at non-specific sites. Based on our model, it would be very challenging to design an intramolecular surface between the zinc fingers and the FokI cleavage domain (for a sequestered state), and then to build-in intermolecular interactions between the zinc-fingers for the release of the cleavage domain. Another strategy would be to modify the FokI recognition domain to target other (or extended) DNA sites, but this is also less than straightforward, as restriction enzymes have proven resilient to alteration in DNA binding specificities10; 23–30.
FokI is not the only restriction endonuclease that requires two DNA sites for successful cleavage. A number of other restriction enzymes have now been shown to bind to two DNA sites simultaneously 14; 31–33. Unlike monomeric FokI that dimerizes transiently to cleave its target site, the Type IIE enzymes such as EcoRII and NaeI are dimeric. Despite these differences, FokI, EcoRII, and NaeI function similarly cleaving only one site during a single turnover requiring the second DNA site for allosteric activation 34; 35. The Type IIF enzymes such as SfiI 31, NgoMIV 36 and Cfr10I 32 are tetrameric and cleave both DNA sites concertedly. Altogether, these unique restriction endonucleases show similarities to various enzymes that bring distant DNA sites together. For example, a FokI-like fold has been identified in TnsA, one of the two proteins of the Tn7 transposase that mediates the release of the transposon 37. EcoRII shows sequence homology to the integrase family of recombinases 38, while NaeI has been shown to possess topoisomerase and recombinase activities 35. SfiI and NgoMIV like recombinases catalyze a four-strand DNA breakage 39. The structures of NgoMIV36 and SfiI 13 in complex with their cognate DNA sites have revealed in both complexes two DNA molecules that are arranged at a 60° (or 120°) angle with respect to each other. Interestingly, junctions in supercoiled DNA are preferentially arranged at a 60° (or 120°) angle 40, and thus it seems that at least some restriction enzymes that require two DNA sites for activity, might have evolved to take advantage of these naturally occurring junctions in supercoiled DNA, as opposed to binding to one DNA site and then waiting for the capture of a second distant DNA site for cleavage to occur. FokI, which is monomeric in solution, could use such junctions as templates to assemble the 2FokI:2DNA cleavage complex. Thus, it will be interesting to test by EM (or other methods) whether FokI prefers to bind such junctions on supercoiled DNA.
Acknowledgments
This work was supported by National Institute of Health Grants, GM44006 to A. K. A., and the New York Structural Biology Center is supported by NIH grant GM66354. The authors thank Steven Ludtke for helpful discussions on data processing using EMAN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci U S A. 1992;89:4275–9. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–90. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci U S A. 1994;91:883–7. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335:447–57. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 5.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 6.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 7.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar S, Allen GC, Thompson WF. Gene targeting in plants: fingers on the move. Trends Plant Sci. 2006;11:159–61. doi: 10.1016/j.tplants.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:2232–7. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 11.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95:10564–9. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95:10570–5. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J Mol Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 14.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J Biol Chem. 2002;277:4024–33. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 15.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–63. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 16.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 18.Miyata T, Oyama T, Mayanagi K, Ishino S, Ishino Y, Morikawa K. The clamp-loading complex for processive DNA replication. Nat Struct Mol Biol. 2004;11:632–6. doi: 10.1038/nsmb788. [DOI] [PubMed] [Google Scholar]
- 19.Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci U S A. 2005;102:13795–800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llorca O, Rivera-Calzada A, Grantham J, Willison KR. Electron microscopy and 3D reconstructions reveal that human ATM kinase uses an arm-like domain to clamp around double-stranded DNA. Oncogene. 2003;22:3867–74. doi: 10.1038/sj.onc.1206649. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Hingorani MM, Patel SS, Egelman EH. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat Struct Biol. 1996;3:740–3. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 22.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–20. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alves J, Ruter T, Geiger R, Fliess A, Maass G, Pingoud A. Changing the hydrogen-bonding potential in the DNA binding site of EcoRI by site-directed mutagenesis drastically reduces the enzymatic activity, not, however, the preference of this restriction endonuclease for cleavage within the site-GAATTC. Biochemistry. 1989;28:2678–84. doi: 10.1021/bi00432a047. [DOI] [PubMed] [Google Scholar]
- 24.Flores H, Osuna J, Heitman J, Soberon X. Saturation mutagenesis of His114 of EcoRI reveals relaxed-specificity mutants. Gene. 1995;157:295–301. doi: 10.1016/0378-1119(94)00863-n. [DOI] [PubMed] [Google Scholar]
- 25.Horton NC, Perona JJ. Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT) J Mol Biol. 1998;277:779–87. doi: 10.1006/jmbi.1998.1655. [DOI] [PubMed] [Google Scholar]
- 26.Lanio T, Jeltsch A, Pingoud A. On the possibilities and limitations of rational protein design to expand the specificity of restriction enzymes: a case study employing EcoRV as the target. Protein Eng. 2000;13:275–81. doi: 10.1093/protein/13.4.275. [DOI] [PubMed] [Google Scholar]
- 27.Osuna J, Flores H, Soberon X. Combinatorial mutagenesis of three major groove-contacting residues of EcoRI: single and double amino acid replacements retaining methyltransferase-sensitive activities. Gene. 1991;106:7–12. doi: 10.1016/0378-1119(91)90559-t. [DOI] [PubMed] [Google Scholar]
- 28.Schottler S, Wenz C, Lanio T, Jeltsch A, Pingoud A. Protein engineering of the restriction endonuclease EcoRV–structure-guided design of enzyme variants that recognize the base pairs flanking the recognition site. Eur J Biochem. 1998;258:184–91. doi: 10.1046/j.1432-1327.1998.2580184.x. [DOI] [PubMed] [Google Scholar]
- 29.Wenz C, Hahn M, Pingoud A. Engineering of variants of the restriction endonuclease EcoRV that depend in their cleavage activity on the flexibility of sequences flanking the recognition site. Biochemistry. 1998;37:2234–42. doi: 10.1021/bi9719197. [DOI] [PubMed] [Google Scholar]
- 30.Whitaker RD, Dorner LF, Schildkraut I. A mutant of BamHI restriction endonuclease which requires N6-methyladenine for cleavage. J Mol Biol. 1999;285:1525–36. doi: 10.1006/jmbi.1998.2409. [DOI] [PubMed] [Google Scholar]
- 31.Bilcock DT, Halford SE. DNA restriction dependent on two recognition sites: activities of the SfiI restriction-modification system in Escherichia coli. Mol Microbiol. 1999;31:1243–54. doi: 10.1046/j.1365-2958.1999.01266.x. [DOI] [PubMed] [Google Scholar]
- 32.Embleton ML, Siksnys V, Halford SE. DNA cleavage reactions by type II restriction enzymes that require two copies of their recognition sites. J Mol Biol. 2001;311:503–14. doi: 10.1006/jmbi.2001.4892. [DOI] [PubMed] [Google Scholar]
- 33.Gormley NA, Hillberg AL, Halford SE. The type IIs restriction endonuclease BspMI is a tetramer that acts concertedly at two copies of an asymmetric DNA sequence. J Biol Chem. 2002;277:4034–41. doi: 10.1074/jbc.M108442200. [DOI] [PubMed] [Google Scholar]
- 34.Reuter M, Kupper D, Meisel A, Schroeder C, Kruger DH. Cooperative binding properties of restriction endonuclease EcoRII with DNA recognition sites. J Biol Chem. 1998;273:8294–300. doi: 10.1074/jbc.273.14.8294. [DOI] [PubMed] [Google Scholar]
- 35.Jo K, Topal MD. DNA topoisomerase and recombinase activities in Nae I restriction endonuclease. Science. 1995;267:1817–20. doi: 10.1126/science.7892605. [DOI] [PubMed] [Google Scholar]
- 36.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat Struct Biol. 2000;7:792–9. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 37.Hickman AB, Li Y, Mathew SV, May EW, Craig NL, Dyda F. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol Cell. 2000;5:1025–34. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 38.Topal MD, Conrad M. Changing endonuclease EcoRII Tyr308 to Phe abolishes cleavage but not recognition: possible homology with the Int-family of recombinases. Nucleic Acids Res. 1993;21:2599–603. doi: 10.1093/nar/21.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wentzell LM, Halford SE. DNA looping by the Sfi I restriction endonuclease. J Mol Biol. 1998;281:433–44. doi: 10.1006/jmbi.1998.1967. [DOI] [PubMed] [Google Scholar]
- 40.Embleton ML, Vologodskii AV, Halford SE. Dynamics of DNA loop capture by the SfiI restriction endonuclease on supercoiled and relaxed DNA. J Mol Biol. 2004;339:53–66. doi: 10.1016/j.jmb.2004.03.046. [DOI] [PubMed] [Google Scholar]